Summary
Reactive oxygen species (ROS) activate NF-E2-related transcription factor 2 (Nrf2), a key transcriptional regulator driving antioxidant gene expression and protection from oxidant injury. Here we report that in response to elevation of intracellular ROS above a critical threshold, Nrf2 stimulates expression of transcription Kruppel-like factor 9 (Klf9), resulting in further Klf9-dependent increases in ROS and subsequent cell death. We demonstrated that Klf9 independently causes increased ROS levels in various types of cultured cells and in mouse tissues and is required for pathogenesis of bleomycin-induced pulmonary fibrosis in mice. Mechanistically, Klf9 binds to the promoters and alters the expression of several genes involved in the metabolism of ROS, including suppression of thioredoxin reductase 2, an enzyme participating in ROS clearance. Our data reveal an Nrf2-dependent feed-forward regulation of ROS and identify Klf9 as a novel ubiquitous regulator of oxidative stress and lung injury.
Introduction
Oxidative stress in the cell is caused by an imbalance between formation of free radicals and their removal by enzymatic and non-enzymatic antioxidant molecules (Finkel and Holbrook, 2000). This imbalance may originate from internal sources such as mitochondrial dysfunction (Finkel and Holbrook, 2000), or from external sources such as exposure to hydrogen peroxide (H2O2).
Transcription factor Nrf2 is a major sensor of oxidative stress in the cell (Itoh et al., 1999). Under basal conditions, Nrf2 is sequestered by cytoplasmic Kelch-like-ECH-associated protein 1 (Keap1) and targeted to proteasomal degradation (Itoh et al., 1999; Wakabayashi et al., 2003). However, under conditions of oxidative stress, Nrf2-Keap1 interaction is disrupted in a dose-dependent manner (Itoh et al., 1999). This leads to Nrf2 translocation to the nucleus where it activates transcription of antioxidant and detoxifying genes by binding to the antioxidant response elements (ARE) in their regulatory regions (Itoh et al., 1999).
Kruppel-like factor 9 (Klf9) is a ubiquitously expressed member of the Sp1 C2H2-type zinc-finger family of transcription factors (Kikuchi et al., 1996). It has been shown to regulate animal development (Morita et al., 2003; Zeng et al., 2008) and differentiation of various cell types, including B-cells, keratinocytes adipocytes and neurons (Bonett et al., 2009; Good and Tangye, 2007; Sporl et al., 2012). Klf9 levels can be increased by several stress-inducing agents, such as the proteasomal inhibitor bortezomib and the histone deacetylase inhibitor panobinostat, and Klf9 in turn mediates their cytotoxicity (Mannava et al., 2012).
Here, we present data identifying Klf9 as a key inducer of cellular oxidative stress that modulates cell death and oxidant-dependent tissue injury. Paradoxically, Klf9 is upregulated by Nrf2 under conditions of excessive oxidative stress, thus suggesting novel functions and modalities of action of Nrf2 in the cell.
Results
Klf9 expression is induced by oxidative stress
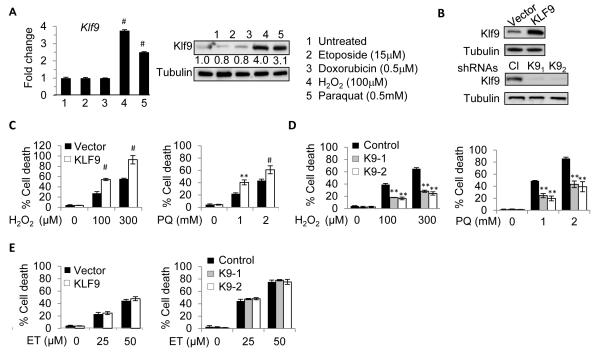
Recently, we have shown that Klf9 expression is induced by proteotoxic stress (Mannava et al., 2012). To identify other types of stress increasing Klf9 expression, we treated NIH3T3 mouse fibroblasts for 8 hours with sub-lethal amounts of chemical agents inducing primarily DNA damage (etoposide and doxorubicin) or oxidative stress (hydrogen peroxide (H2O2) and 1, 1′-Dimethyl-4, 4′-bipyridinium dichloride (paraquat)). Expression analysis via quantitative reverse-transcription PCR (Q-RT-PCR) and immunoblotting revealed that oxidative stress induces Klf9 mRNA and protein levels (Figure 1A).
Figure 1. Klf9 is induced by oxidative stress and promotes oxidative stress-induced cell death.
(A) NIH3T3 cells were treated with indicated agents for 8 hours followed by immunoblot (left panels) and Q-RT-PCR (right panels) analysis. Numbers under Klf9 panel indicate fold induction of Klf9 signal (normalized by tubulin signal) compared to corresponding signals in untreated cells. (B) NIH3T3 cells were transduced with the indicated constructs followed by immunoblot analysis with indicated antibodies. Cl - control shRNA, K91 and K92 are Klf9 shRNAs. (C-E) Cells transduced as in (B) were treated as indicated for 16 hours followed by viability assay (trypan blue exclusion), PQ=paraquat, ET=etoposide. The data are presented as the mean values of triplicates ± S.E.M. p-values were determined by Student’s t-test. (*p<0.05), (**p<0.001), (#p<0.0001). Each experiment was performed at least two times with consistent results. See also Figure S1.
To determine whether upregulation of Klf9 plays a functional role in oxidative stress-induced cell death, we modulated Klf9 levels in NIH3T3 cells via ectopic expression of its cDNA or Klf9-specific shRNAs (Figure 1B), followed by treatment with H2O2 or paraquat. Depletion or overexpression of Klf9 did not affect proliferation of NIH3T3 cells (Figure S1A, B). At the same time, Klf9 overexpression sensitized, whereas depletion of Klf9 provided resistance to oxidative stress-induced cell death (Figure 1C, D). These findings were confirmed in a clonogenic cell survival assay where cells were exposed to a higher dose of H2O2 (400 μM) (Figure S1C). Of note, the specificity of Klf9 shRNAs was verified with shRNA-resistant human KLF9 cDNA (Figure S1D). On the contrary, depletion or overexpression of Klf9 did not alter NIH3T3 cell susceptibility to the DNA damaging agent etoposide (Figure S1E). Immortalized Klf9 knock-out mouse embryonic fibroblasts (MEFs) also demonstrated higher resistance to H2O2 and paraquat but not etoposide compared to their wild-type counterparts (Figure S1F, G). Thus, Klf9 is upregulated by oxidative stress and promotes oxidative stress-induced cell death.
Klf9 can cause accumulation of ROS
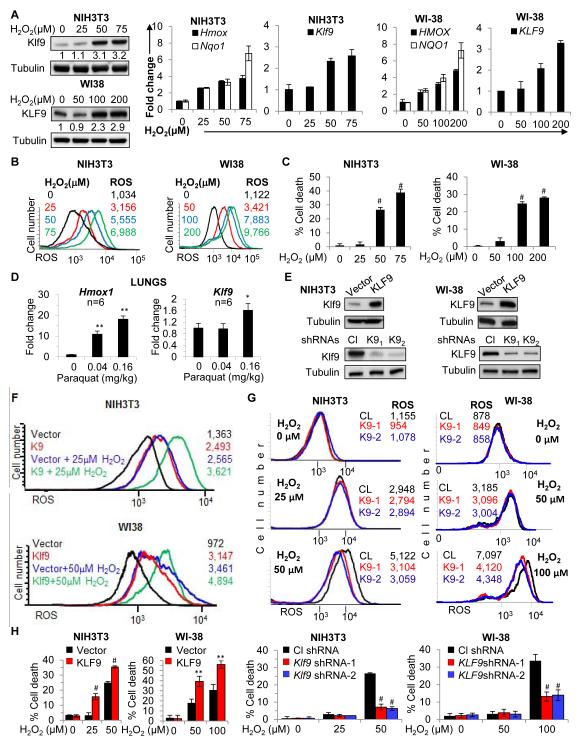
To understand the mechanisms of Klf9-mediated response to oxidative stress, we first studied the dose-dependent induction of Klf9 by H2O2. Treatment of mouse and human fibroblasts with H2O2 for 2 hours induced expression of Klf9 at concentrations greater than 50μM and 100μM, respectively (Figure 2A); these concentrations corresponded to a ~5 and ~7 fold increase in intracellular ROS, respectively (Figure 2B). At the same time, oxidative stress response genes Nqo1 and Hmox1 (Alam et al., 1999; Nioi et al., 2003), were upregulated in mouse and human fibroblasts at lower concentrations of H2O2 (25μM and 50μM, respectively, Figure 2A, right panel). We further investigated the dynamics of Klf9 upregulation by H2O2 in NIH3T3 cells and found the same pattern of expression of Klf9, Nqo1 and Hmox1 genes at later time points in H2O2-treated cells (Figure S2B). Importantly, treatment of mouse and human fibroblasts with H2O2 at concentrations sufficient for upregulation of Klf9 (50 μM and 100μM, respectively), led to cell death 16 hours post-treatment, whereas lower concentrations of H2O2 (still sufficient for induction of Hmox1 and Nqo1 [Figure 2A]) were well tolerated by the cells (Figure 2C).
Figure 2. Klf9 upregulates intracellular ROS levels.
(A) NIH3T3 and WI38 cells were treated with indicated amounts of H2O2 for 2 hours followed by immunoblotting with indicated antibodies (left panel) and Q-RT-PCR analysis with indicated probes (right graphs). Numbers under Klf9 panel indicate fold induction of Klf9 signal (normalized by tubulin signal) compared to corresponding signals in untreated cells. (B) Cells treated as in (A) were stained with H2DCFDA followed by FACS analysis to determine intracellular ROS (ROS-FACS). (C) Cells treated as in (A) for 16 hours were subjected to trypan blue exclusion cell viability assay. (D) C57BL/6J mice were euthanized 6 hours after treatment with the indicated amounts of paraquat (n=6). Lungs were dissected, RNA was isolated and Q-RT-PCR analysis was performed with indicated probes. (E) Cells were transduced with the indicated constructs followed by immunoblot analysis with indicated antibodies. Cl -control shRNA, K91 and K92 are Klf9 shRNAs. Cells transduced as in (E) were treated with indicated amounts of H2O2 followed by ROS-FACS analysis 2 hours post-treatment (F, G) and trypan blue exclusion assay 16 hours post-treatment (H). The data are presented as the mean values of triplicates ± S.E.M. p-values were determined by Student’s t-test. (*p<0.05), (**p<0.001), (#p<0.0001). Each experiment was performed at least two times with consistent results. See also Figure S2.
To evaluate whether regulation of Klf9 occurs post-transcriptionally, NIH3T3 cells were incubated with transcription inhibitor actinomycin D (5μg/ml) for 30 minutes followed by treatment with 50μM of H2O2. Cells were collected 2 hours after treatment, followed by assessment of Klf9 mRNA and protein levels. As shown in Figure S2C, actinomycin D completely suppressed H2O2-dependent increase of Klf9 mRNA and protein levels, suggesting that induction of Klf9 by H2O2 does not occur post-transcriptionally.
To validate oxidative stress-dependent Klf9 expression in vivo, we induced oxidative stress in wild-type mice by intranasal application of paraquat as previously described (Tomita et al., 2007). Mice were euthanized 6 hours after application, followed by surgical dissection of lungs, isolation of total RNA and assessment of Klf9 and Hmox1 levels by Q-RT-PCR. Notably, the dose of paraquat required to induce Klf9 was greater than the one sufficient for upregulation of Hmox1 (Figure 2D).
Next, we were interested in the mechanisms of Klf9-dependent modulation of H2O2 cytotoxicity. One possibility could be that Klf9 increases accumulation of intracellular ROS. To test this hypothesis, we measured ROS in mouse and human fibroblasts either overexpressing or depleted of Klf9 (Figure 2E-G). Klf9 overexpression resulted in a significant 200%-300% increase in the amount of intracellular ROS (Figure 2F). To determine whether this would affect cells proliferation, we incubated NIH3T3 cells with a non-lethal dose (2.5 μM) of arsenic trioxide (ATO), a well-characterized oxidative stress inducer, for 4 days, with daily replacement of ATO-containing media (i.e. the same time interval that was used for measuring ROS after infection with Klf9-expressing vector). The above treatment did not affect cell proliferation (Figure S2D), and ROS levels measured on day 4 were comparable with ROS levels in Klf9-overexpressing cells (Figures S2E, 2F upper panel). These results are consistent with previous reports on several types of tumor cells (Adams et al., 2013).
Depletion of Klf9 did not change basal ROS levels in human and mouse cells (Figure 2G upper panel). Moreover, treatment of mouse or human fibroblasts with low concentrations of H2O2 (below the threshold amounts required for Klf9 induction) increased ROS similarly in control and Klf9-depleted cells (Figure 2G middle panel). On the contrary, at higher concentrations of H2O2, Klf9-depleted cells possessed less ROS compared to control cells (Figure 2G lower). Accordingly, Klf9 overexpression decreased, whereas depletion of Klf9 increased the threshold H2O2 concentrations necessary for induction of cell death (Figure 2H). In summary, if cells were treated with H2O2 amounts that did not upregulate Klf9, its depletion did not affect H2O2-induced ROS; however, if cells were treated with H2O2 amounts that upregulated Klf9, its depletion suppressed H2O2-induced ROS. Thus, Klf9 is induced by ROS and, in a feed-forward mechanism, further augments ROS accumulation and promotes cell death.
Recently, we reported that in multiple myeloma cells KLF9 regulates bortezomib-induced, but not endogenous levels, of the pro-apoptotic protein NOXA (Mannava et al., 2012). Manipulation of Klf9 expression in mouse fibroblasts did not affect endogenous levels of Noxa, nor was Noxa induced after 2 hours of incubation with H2O2 (Figure S2F). Moreover, suppression of Noxa via specific shRNAs did not alter Klf9-induced induction of ROS (Figures S2G), suggesting that Klf9 modulates ROS via Noxa-independent pathways.
Klf9 is upregulated by Nrf2
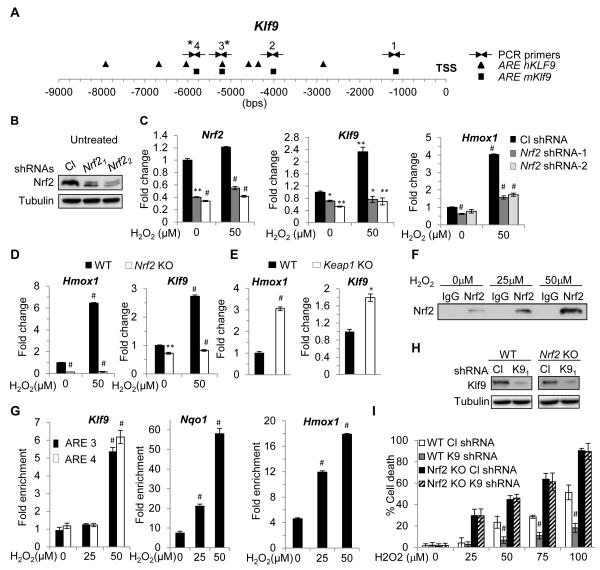
In order to identify potential transcriptional regulators of Klf9, we analyzed the promoters of mouse and human Klf9 genes for binding sequences of transcription factors regulating the oxidative stress response. We focused on the region 10 Kb upstream and 1 Kb downstream of Klf9 transcription start site. Unexpectedly, we found that these regions contained several conserved antioxidant response elements (ARE, 5′-RTGAYnnnGCR-3′, (Wasserman and Fahl, 1997)) which are binding sites for Nrf2, a major regulator of anti-oxidant defense in the cell (Itoh et al., 1997; Itoh et al., 1999) (Figure 3A). To test whether Klf9 expression would be modulated by Nrf2, we depleted Nrf2 levels in NIH3T3 cells via shRNAs (Figure 3B). Partial depletion of Nrf2 suppressed Klf9 levels in untreated cells and blunted its induction by H2O2 (Figure 3C). A similar pattern of regulation was observed for the Nrf2-responsive gene Hmox1 (Alam et al., 1999), although in untreated cells its levels were affected more than Klf9 levels by Nrf2 knock-down (Figure 3C). Furthermore, using Nrf2 knock-out MEFs, Keap1 knockout MEFs, Keap1-knock-out-Nrf2-knockdown MEFs and Keap1 knockdown NIH3T3 cells, we confirmed the Nrf2- and Keap1-dependent pattern of Klf9 expression (Figure 3D, E; Figure S3A-D). Noteworthy, Keap-1 depletion in two different systems resulted in lower levels of Klf9 induction compared to treatment with toxic doses of H2O2 (Figures 3E, S3A, B, D). These results are in agreement with previous reports showing that Nrf2 and its downstream targets could be regulated via both Keap1-dependent and Keap1-independent mechanisms (Chowdhry et al., 2013). Consistently, ROS levels in Keap1-depleted cells were lower than in control cells (Figure S3C). In Keap1-inactivated cells, the basal levels of Nrf2 are upregulated and this leads to the accumulation of antioxidant enzymes with subsequent decrease of intracellular ROS. At the same time Klf9 amounts are also upregulated, however, below the toxic levels. In this scenario, oxidative stress-inducing agents may be more efficient in upregulating Klf9 to the toxic levels in Keap1-inactive cells than in wild-type cells and cause higher toxicity. To test this hypothesis, we treated cells from several human lung cancer lines with wildtype or mutant KEAP1 protein with different doses of ATO. As shown in Figure S3E, cells with inactive KEAP1 were more resistant to ATO than their counterparts, suggesting that upregulation of Klf9 to the “toxic” levels is not enough to oversaturate original excess of antioxidant molecules in Keap1-inactivated cells.
Figure 3. Nrf2 transcriptionally activates Klf9 in response to threshold levels of ROS.
(A) Schematic representation of human and mouse Klf9 promoters. Squares and triangles represent human and mouse Nrf2 binding sites (ARE), respectively. TSS-transcription start site. Arrows indicate PCR primers. (B) NIH 3T3 cells were transduced with control (Cl) or two Nrf2-specific shRNAs (Nrf21 or Nrf22) followed by immunoblotting analysis with indicated antibodies. (C) NIH3T3 cells transduced as in (B) were treated with indicated amounts of H2O2 for 2 hours followed by Q-RT-PCR analysis with indicated probes. (D) Wildtype (WT) or Nrf2 knock-out (Nrf2 KO) MEFs were treated with indicated amounts of H2O2 followed by Q-RT-PCR analysis with indicated probes. (E) WT or Keap1 knock-out (Keap1 KO) MEFs were analyzed using Q-RT-PCR for the expression of indicated genes. (F) NIH 3T3 cells were treated with indicated amounts of H2O2 for 2 hours. Cells were fixed and sheered cross-linked chromatin was prepared as detailed in the Extended Experimental Procedures. The chromatin was precipitated using control (IgG) or Nrf2-specific antibodies (Nrf2). A portion of the immunoprecipitate was probed in immunoblotting with Nrf2-specific antibodies. (G) DNA isolated from the precipitated materials described in (F) was analyzed in Q-PCR with indicated primers (see Extended Experimental Materials). The Hmox1-Nqo1- and Klf9-specific signals from Nrf2-precipitated DNA were normalized by those from IgG precipitated DNA. (H) Wildtype (WT) and Nrf2 knock-out (Nrf2 KO) MEFs were infected with control shRNA (Cl shRNA) or Klf9 shRNA1 (K9 shRNA). Two days post infection Klf9 expression was assessed in the cells by immunoblotting with indicated antibodies. (I) In parallel, cells were treated with indicated amounts of H2O2 and tested by trypan blue exclusion cell viability assay 16 hours after treatment. The data are presented as the mean values triplicates ± S.E.M. p-values were determined by Student’s t-test. (*p<0.05), (**p<0.001), (#p<0.0001). Each experiment was performed at least two times with consistent results. See also Figure S3.
Next, we treated NIH3T3 cells for 12 hours with increasing amounts of sulforaphane, a classic Nrf2 activator and oxidative stress agent (Higgins et al., 2009) (Figure S3F). Like H2O2, non-toxic amounts of sulforaphane (10μM) did not upregulate Klf9 levels however treatment with higher doses (12μM-14μM) led to Klf9 upregulation (Figure S3F middle panel), which was accompanied with cell death 24hrs post treatment (Figure S3F bottom panel).
To understand the dose-dependent regulation of Klf9 expression by Nrf2, we performed chromatin immunoprecipitation from NIH3T3 cells treated for 2 hours with increasing amounts of H2O2 using isotype- (IgG) or Nrf2-specific antibodies (Figure 3F). Increased binding of Nrf2 to the Klf9 promoter (~5,600 bps upstream of its transcription start site) occurred only in cells treated with dose of H2O2 sufficient to induce transcription of Klf9 (50μM) (Figure 3G). However, increased binding of Nrf2 to Nqo1 and Hmox1 regulatory sequences was readily detectable in cells treated with lower amounts of H2O2 (25μM) (Figure 3G), which is in agreement with Klf9, Nqo1 and Hmox1 expression data (Figure 2A).
A possible explanation to these results is that the Klf9 regulatory region containing ARE3 and ARE4 (Figure 3A) interacts with Nrf2 less efficiently than the ARE-containing regulatory regions of Nqo1 or Hmox1 genes. To test this hypothesis, we cloned into a luciferase-reporter vector (pGL3-promoter) 34 bps DNA regions containing AREs of Hmox1 (Alam et al., 1995), Nqo1 (Nioi et al., 2003), Klf9 ARE3 and ARE4 as well as ARE3 and ARE4 containing mutated Nrf2-binding sites (mARE3 and mARE4) (Figure S3G). Control empty-vector and vectors containing the above AREs were transfected into NIH3T3 cells followed by treatment with 100μM of H2O2 or 14 μM of sulforaphane. As shown in Figure S3H, treatment with the above agents induced luciferase expression in cells containing constructs with Hmox1 or Nqo1 AREs several folds more efficiently than cells with constructs carrying ARE3 or ARE4. Importantly, oxidative stress-induced upregulaiton of ARE3- or ARE4-driven luciferase expression was not detected in mARE3 or mARE4 constructs. These results are in good agreement with the data described above (Figures S2B; 3F, G).
The transcription factor Bach1 has been shown to suppress the induction of several Nrf2 target genes via interaction with the ARE sites in their promoters (Sun et al., 2002). Thus, we asked whether Klf9 would be a Bach1 target to explain the observed pattern of expression. Treatment of NIH3T3 cells with Bach1-specific siRNA resulted in substantial depletion of Bach1 protein, however Klf9 mRNA and protein levels did not change significantly (Figure S3I). Moreover, analysis of a published database of global ChIP experiments (Warnatz et al., 2011) revealed that the Klf9 promoter was not among the detected interactors of Bach1. Therefore, Klf9 is unlikely to be a Bach1 target.
To identify whether Klf9 plays a role in the regulation of oxidative stress-induced death in Nrf2-deficient cells, we infected wild-type and Nrf2-knock-out MEFs with control or Klf9 shRNA followed by incubation with various concentrations of H2O2. Contrary to wild-type cells, depletion of Klf9 did not protect Nrf2-knock-out MEFs from H2O2-induced cell death (Figure 3I), which is in agreement with our findings of Nrf2-dependent upregulation of Klf9 and Klf9-induced accumulation of ROS.
To determine whether knock-out of Klf9 influences basal or inducible Nrf2 activity, we measured Nqo1 mRNA levels in untreated wild-type and Klf9 knock-out MEFs and in the same cells treated with 10μM and 14μM of sulforaphane. No difference in Nqo1 mRNA levels was detected between untreated cells of either genotype, suggesting that Klf9 depletion does not affect basal Nrf2 activity (Figure S3J). Treatment with amounts of sulforaphane insufficient for Klf9 induction (10μM) upregulated Nqo1 mRNA similarly in both cell types thus suggesting that inducible Nrf2 activity is not affected by Klf9 deficiency (Figure S3J). Instead, treatment with 14μM of sulforaphane resulted in lower Nqo1 mRNA levels in Klf9 knock-out than wild-type MEFs (Figure S3J), however this difference was most likely due to suppression of ROS levels in Klf9-deficient cells (Figure 2G). Therefore, Klf9 deficiency does not appear to affect basal or inducible Nrf2 activity.
Klf9 suppresses expression of thioredoxin reductase 2
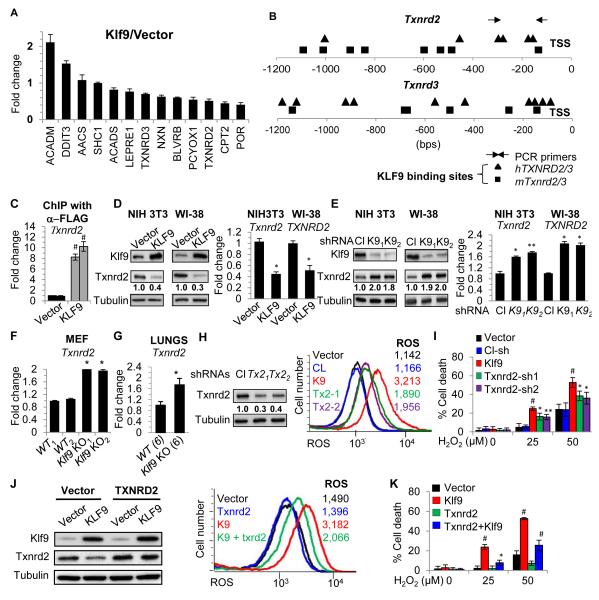
To identify Klf9 transcriptional targets, we performed global chromatin immuno-precipitation-sequencing (ChIP-seq) assay in NIH3T3 cells transduced with control vector or vector expressing FLAG-Klf9 cDNA. Logarithmically growing cells were fixed and cross-linked, and DNA that co-precipitated with FLAG-specific antibodies from both types of cells was sequenced using Illumina Genome Analyzer II. The significant peaks obtained in at least two sequence runs from two pairs of independently transduced “vector” and “FLAG-Klf9” cells were analyzed using the Integrative Genomics Viewer (IGV) program (Robinson et al., 2011), and FLAG-Klf9-specific peaks were aligned to the mouse genome. Among 614 Klf9-specific peaks, 176 corresponded to the regions of Polymerase lI genes within 5kbs from their transcription start sites. DAVID-based gene ontology analysis (Huang da et al., 2009) of the ChIP-seq data revealed an enrichment of genes involved in regulation of intracellular ROS (13 total, Table S1, S2). Q-RT-PCR analysis validated the Klf9-dependent expression of several of the 13 identified genes (Figure 4A). It is noteworthy that none of these genes are considered as bona-fide Nrf2 targets (Campbell et al., 2013; Chorley et al., 2012). Accordingly, unlike Nrf2 target genes Hmox1, Nqo1, and Txnrd1 which were induced by H2O2 treatment, expression of at least two potential Klf9 target genes thioredoxin reductases 2 and 3 (Txnrd2 and Txnrd3) was suppressed by this treatment, but only at concentrations that upregulated Klf9 and induced cell death (50μM and above) (Figure S4AB). We assessed the levels of Txnrd-1, −2, and −3 in NIH3T3 cells treated with increasing amounts of H2O2 at two time points and confirmed the pattern of their expression described in Figure S4A (Figure S4C).
Figure 4. Txnrd2 is a target of Klf9.
(A) RNA isolated from NIH3T3 cells infected with empty vector “Vector” or KLF9 cDNA “KLF9” was probed in Q-RT-PCR with indicated primers. (B) Schematic representation of human and mouse Txnrd2 and Txnrd3 promoters. Squares and triangles represent human and mouse Klf9 binding sites, respectively. TSS-transcription start site. Arrows indicate PCR primers. (C) ChIP with FLAG-specific antibodies was performed on NIH3T3 cells expressing empty vector or FLAG-KLF9 cDNA. The obtained DNA samples were probed by Q-PCR with primers specific to the promoter region of mouse Txnrd2 gene. (D-E) Cells were transduced with the indicated constructs followed by immunoblot analysis (left panels) or Q-RT-PCR (right panels) with indicated antibodies and probes, respectively (Cl - control shRNA, K91 and K92 are Klf9 shRNAs). RNA isolated from wildtype and Klf9 knock-out MEFs (F) or mice (6 per group) (G) was probed in Q-RT-PCR with mouse Txnrd2 probe. (H) NIH3T3 cells were transduced with the indicated constructs followed by immunoblot analysis (left panel) (Cl-control, Tx21 and Tx22 are Txnrd2 shRNAs) and ROS-FACS analysis along with NIH3T3 cells expressing empty vector “Vector” or a vector encoding KLF9 cDNA “KLF9” (right panel). (I) Cells described in (H) were treated with indicated amounts of H2O2 and cell viability was assessed by trypan blue exclusion assay 16 hours post-treatment. NIH 3T3 cells expressing indicated constructs were probed in immunoblotting with indicated antibodies (J, left panel) or FACS-ROS analysis (J, right panel) or tested for trypan blue exclusion (K). The data are presented as the mean values of triplicates ± S.E.M. p-values were determined by Student’s t-test. (*p<0.05), (**p<0.001), (#p<0.0001). Each experiment was performed at least two times with consistent results. See also Figure S4 and Table S1,S2.
Next, we were interested in the possible functional involvement of the identified genes in Klf9-dependent regulation of ROS. We selected thioredoxin reductase 2 (Txnrd2), as its product has been implicated in the regulation of ROS in various cell types (Arner, 2009).
The promoters of mouse and human Txnrd2 and Txnrd3 genes contain several conserved Klf9-binding sites (5′-CA/GCCC-3′) (Figure 4B). We confirmed Klf9 binding to the promoter of mouse Txnrd2 gene by probing the original ChIP material with Txnrd2 promoter-specific primers in Q-PCR (Figure 4C). Overexpression or depletion of Klf9 resulted in a 2-2.5-fold decrease or 1.6-2-fold increase in the expression of Txnrd2, respectively, as evidenced by Q-RT-PCR and immunoblotting (Figure 4D, E). Accordingly, Txnrd2 mRNA levels were ~2 fold higher in MEFs derived from Klf9 knockout mice than in wild-type (Figure 4F). Additionally, levels of Txnrd2 (and Txnrd3) were higher in Nrf2-knock-out MEFs compared to wild-type (Figure S4D). Moreover, a statistically significant difference in the expression of Txnrd2 mRNA was detected between lungs of age-matched wild-type and Klf9 knock-out mice (Figure 4G). Taken together, these data demonstrate that Klf9 binds to the Txnrd2 promoter and suppresses its expression.
To address the role of Txnrd2 in Klf9-dependent regulation of ROS, we first depleted Txnrd2 in NIH3T3 cells via shRNA to approximately the levels detected in Klf9-overexpressing cells (compare Figure 4H and 4D). Txnrd2 depletion resulted in elevation of intracellular ROS to about 37% of the increase achieved by Klf9 overexpression in these cells (Figure 4H, right panel). Accordingly, partial depletion of Txnrd2 rendered NIH3T3 cells more sensitive to H2O2 induced toxicity, although to a lesser degree as compared to cells overexpressing Klf9 (Figure 4I). Next, NIH3T3 cells were transduced with empty vector or a vector expressing Txnrd2 cDNA. Three days after infection the cells were superinfected with empty vector or a vector expressing Klf9 cDNA (Figure 4J). The resulting populations were tested for intracellular ROS and H2O2 resistance. Ectopic expression of Txnrd2 did not significantly change the basal levels of ROS compared to vector-infected cells (Figure 4J, right panel). However, Klf9-dependent elevation of ROS was partially (~48%) suppressed by ectopic expression of Txnrd2 (Figure 4J, right panel). Moreover, overexpression of Txnrd2 decreased Klf9-dependent sensitivity to H2O2 in studied cells (Figure 4K). Similar results were obtained with WI38 cells (Figure S4E, F).
Accordingly, depletion of KLF9 in human melanoma (SK-Mel-103), breast cancer (MCF7) and colon cancer (HCT-116) cells resulted in upregulation of TXNRD2, suppression of H2O2-dependent ROS and increased resistance to H2O2-induced cell death (Figure S4G-I). Thus, Klf9 reduces ROS defenses via suppression of Txnrd2 in non-transformed and transformed cells of different types.
Klf9 deficient mice demonstrate resistance to bleomycin-induced oxidative stress and pulmonary fibrosis
To test whether deficiency of Klf9 leads to suppression of oxidative stress in vivo, we investigated the role of Klf9 in bleomycin-induced pulmonary fibrosis in mice. It has been well established that increased amounts of ROS promote the pathogenesis of pulmonary fibrosis in humans and in laboratory animals (Cheresh et al., 2013). The chemotherapeutic agent bleomycin has been shown to cause oxidative lung injuries in mice, and bleomycin-treated mice are considered to be a classical model of idiopathic pulmonary fibrosis (Mouratis and Aidinis, 2011).
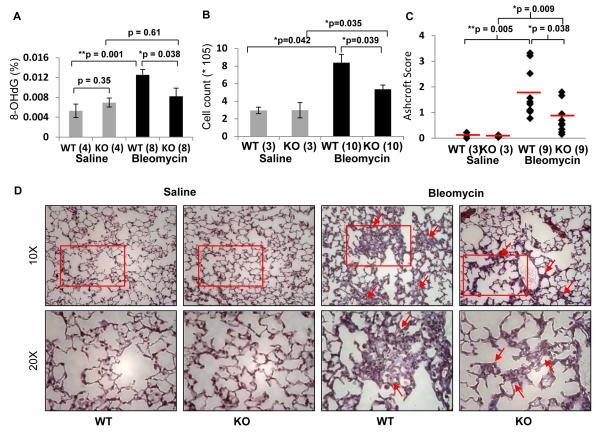
Age-matched wild-type and Klf9 knock-out mice were intratracheally instilled with either saline or a saline solution of bleomycin sulfate (2.5 mg/kg). Mice were euthanized 10 days after instillation and the content of 8-oxo-2′-deoxyguanosine (8-OHdG), a marker of oxidative DNA damage, was determined in the DNA isolated from lungs of treated animals. Bleomycin treatment induced a statistically significant increase in the levels of 8-OHdG in the lungs of wild-type mice (p=0.0003), but did not significantly affect the levels of 8-OHdG in the lungs of Klf9 knock-out mice (Figure 5A). A second cohort of saline- or bleomycin-treated mice was analyzed 19 days after treatment for the accumulation of cells in the bronchoalveolar lavage fluid (BALF) and for the accumulation of collagen (markers of lung fibrosis). We observed a significant increase in the number of total BALF cells in all animals treated with bleomycin (Figure 5B). However, such an increase was significantly lower in Klf9 knock-out mice as compared to wild type counterparts (p <0.05). The degree of fibrosis in lung histological sections stained with Masson’s trichrome was quantified using the Ashcroft scale (Ashcroft et al., 1988) by a pathologist blinded to the study groups (Figure 5C,D). No sign of fibrosis was detected in wild-type or Klf9 knock-out mice treated with the saline solution. Treatment with bleomycin led to pulmonary fibrosis in animals of both genotypes, although the degree of the fibrosis was significantly higher in wild-type mice as compared to knock-out counterparts (p=0.038). Thus, Klf9 deficiency provides resistance to bleomycin-induced oxidative stress and pulmonary fibrosis in mice.
Figure 5. Klf9 deficiency suppresses bleomycin-induced oxidative stress and fibrosis in lungs.
(A) Indicated number of wild-type and Klf9 knock-out mice were euthanized 10 days after treatment with a single intratracheal dose of saline or bleomcyin (2.5 mg/Kg). Lungs were dissected, DNA was isolated, and levels of 8-hydroxy-2′-deoxyguanosine (8-OHdG) were assessed as described in Experimental Procedures. Data are represented as percent of 8-OHdG of total DNA analyzed (200ng). (B) Bronchoalveolar lavage fluid (BALF) was collected from indicated number of animals 19 days after treatment, processed and analyzed for total cell counts using a hemocytometer. (C-D) Mice treated, as in (A), were euthanized 19 days post treatment. Lungs were inflated and fixed with 4% paraformaldehyde, processed, and embedded in paraffin wax. Fibrosis scores based on histopathological assessment of Masson’s trichrome-stained sections (C) and representative histology of Masson’s trichrome-stained slides (D) of each genotype and condition are shown. The data are presented as the mean values ± S.E.M. Comparisons were made using the Student’s t-test or a nonparametric Mann-Whitney U test. (*p<0.05), (**p<0.001).
Discussion
In the present manuscript we report the identification of transcription factor Klf9 as a novel regulator of intracellular ROS. Klf9 is a ubiquitously expressed protein with relatively understudied function. Our data suggests that Klf9 can cause accumulation of intracellular ROS at least in part by transcriptional suppression of the thioredoxin reductase 2 gene (Txnrd2). Txnrd2 is a mitochondrial selenoprotein that plays a critical role in defense against oxidative damage (Arner, 2009).
The family of thioredoxin reductases consists of 3 members that share domain structure but differ in subcellular localization: Txnrd1 in the cytoplasm, Txnrd2 in the mitochondria, thus suggesting largely non-overlapping spectra of substrates for each reductase. Localization of Txnrd3 has not been characterized. Mitochondrial thioredoxin and Txnrd2 are considered the major regulator of ROS in mitochondria (Arner, 2009).
The fact that Txnrd2 deficiency only partially recapitulates Klf9-dependent phenotypes suggests that other Klf9-target genes (possibly some of the ones identified in our study) could be functionally involved in regulation of Klf9-dependent oxidative stress.
(Higgins et al., 2009) demonstrated that pre-treatment of mouse fibroblasts with non-lethal amounts of sulforaphane (3μM) increased their resistance to subsequent treatment with paraquat or hydrogen peroxide in Nrf2-dependent manner. These data are in agreement with our findings on the dynamics of Klf9 upregulation by sulforaphane and hydrogen peroxide. We showed that in NIH3T3 cells, low dose of sulforaphane (10μM, Figure S3F) or hydrogen peroxide (25μM, Figures 2A, S2B) did not induce Klf9 but did upregulate Nqo1. At toxic doses, both sulforaphane (12-14 μM) and hydrogen peroxide (≥ 50μM) induced expression of Klf9 and continued upregulation of Nqo1. At relatively high concentrations, sulforaphane is known to cause cell death which at least in part is attributed to oxidative stress (Singh et al., 2005). It would be interesting to determine if Klf9 plays any role in this process. Therefore, pre-treatment of cells with low dose of sulforaphane described by Higgins et al, most likely increased levels of antioxidant molecules without induction of Klf9.
The existence of a feed-forward regulatory loop triggering cell death in response to excessive damage of cellular macromolecules has been described for several types of insults (Biton and Ashkenazi, 2011; Rocourt et al., 2013). Our data suggest a model of Nrf2-dependent feed-forward regulation of ROS. Under conditions of low oxidative stress, Nrf2-dependent antioxidant defense is activated ultimately resulting in reduced ROS levels to the amounts that are not detrimental for cell survival. However, upon further elevation of ROS, Nrf2 continues to accumulate in the nucleus leading to Nrf2 binding to the Klf9 promoter, transcriptional upregulation of Klf9 and consequently further increases in ROS levels. This Klf9-dependent accumulation of ROS is sufficient to cause cell death.
Our data suggest that Klf9 induction requires high amounts of Nrf2. Indeed, Klf9 expression was induced in Keap1 knock-out fibroblasts without H2O2 treatment (Figure 3E), and suppressed to the original levels by partial depletion of Nrf2 via shRNA (Figure S3D). Moreover, Nrf2 binding to the Klf9 promoter occurred at high levels of Nrf2 induced by 50μM H2O2, whereas lower amounts of Nrf2 (induced by 25 μM H2O2) did not result in efficient binding to Klf9 regulatory regions. (Figure 3F).
In a transient reporter assay, Klf9 ARE-containing sequences did not respond to oxidative stress as efficiently as ARE regions from the enhancers of Hmox1 or Nqo1 genes (Figure S3H), thus reproducing a difference in pattern of ROS-induced expression of Klf9 and Hmox1 or Nqo1 genes. These data suggest that the observed differential interactions of studied genes with Nrf2 and their response to oxidative stress are most likely due to variations in the individual base pair composition of corresponding ARE regions. Indeed, despite the existence of a common core (5′-RTGAYnnnGCR-3′) the integrity of which is crucial for activity of all functional AREs, the nucleotide sequences flanking the core can strongly affect the activity of AREs from different genes (Nioi et al., 2003). Moreover, bases in Nqo1 ARE not important for its activity were found important for activities of AREs from other Nrf2-target genes when inserted in corresponding positions, and vice versa (Nioi et al., 2003) Accordingly, ARE-containing sequences from different Nrf2 targets differed in ability to function in transient expression reporter assays in response to oxidative stress (Mulcahy et al., 1997). Such diversity among ARE sequences reflects their complex regulation that ultimately results in a fine-tuned orchestration of the anti-oxidant response.
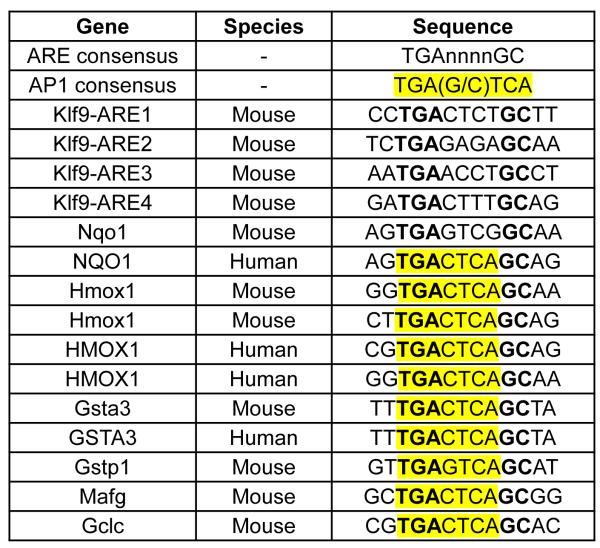
AP-1 transcription factors are one of the most extensively studied transcription regulators interacting with ARE, since AP-1 binding site 5′-TGA(G/C)TCA-3′ is often embedded into ARE core sequence 5′-RTGAYnnnGCR-3′ (Li and Jaiswal, 1992; Nguyen et al., 2003). The role of an embedded AP-1 binding site in the regulation of ARE-driven transcription is controversial as its presence has been reported to suppress (Venugopal and Jaiswal, 1996) or facilitate (Soriano et al., 2009) Nrf2-dependent transcription. Nonetheless, it is noteworthy that AREs in mouse Klf9 promoter do not contain embedded AP-1 sites unlike AREs in promoters of other Nrf2 targets including Nqo1, Hmox1, Txnrd1, etc. (Figure 6), therefore providing another possible explanation for the functional difference between Klf9 AREs and AREs of studied Nrf2 targets.
Figure 6. Sequence comparison of ARE-containing DNA regions of different Nrf2 targets.
Depiction of AREs and flanking sequences from mouse Klf9 regulatory regions and previously reported Nrf2 targets (Wang et al., 2007). Highlighted is the AP-1 site embedded in some of the AREs.
Interestingly, genes identified by us as Klf9 targets via ChIP-Seq are not considered to be bona fide Nrf2 targets (Campbell et al., 2013; Chorley et al., 2012). These observations suggest that depending on ROS levels, Nrf2 induces accumulation or removal of ROS via two separate regulatory circuits that are Klf9-dependent and – independent, respectively.
Pulmonary fibrosis (PF) is a chronic disease of variable etiology, and oxidative stress has long been characterized as one of the major drivers of PF (Cheresh et al., 2013). Yet, only few genes involved in control of ROS have been implicated in the pathogenesis of PF in mouse knock-out models (Cheresh et al., 2013), including Nrf2 as a sole transcription factor among these genes (Kikuchi et al., 2010). Furthermore, Nrf2 was activated and its targets including Nqo1 were upregulated in mice treated with bleomycin (Cho et al., 2004). Therefore, functional involvement of Klf9, and potentially its targets, in pathogenesis of PF uncovers a novel regulatory network of this disease and may offer new targets and/or prognostic markers.
Klf9 has been implicated in control of several aspects of mouse development and its pro-oxidant mode of action may shed light on their mechanisms. For example, Klf9 knock-out female mice demonstrate delayed parturition compared to wild-type animals (Zeng et al., 2008). ROS represent central signaling molecules in physiological processes of female reproduction system including parturition, and their potential downregulation in Klf9 knock-out mice may contribute to the reported phenotype. Additionally, increased Klf9 expression was observed in cerebellum of postnatal mice, whereas Klf9 knock-out mice demonstrate defects in motor coordination (Morita et al., 2003). Accordingly, treatment of developing rats with antioxidants resulted in inhibition of ROS in cerebellum and alterations in motor coordination (Coyoy et al., 2013). Furthermore, it has been shown that the Keap1 knock-out mice die postnatally due to a hyperkeratotic proliferative disorder (Wakabayashi et al., 2003). Therefore, it would be of interest to determine the role of Klf9 in this process by creating double knock-out animals.
Several reports have demonstrated that Klf9 may act as a tumor suppressor (Tetreault et al., 2013), and our data could account for these observations. Indeed, it has been recently reported that Nrf2 can act as an oncogene by protecting cancer cells from excessive oxidative stress (DeNicola et al., 2011). Since Klf9 depletion suppresses oxidative stress as well, Klf9 downregulation may promote cancer progression and potentially resistance to treatment. Thus, it will be of interest to evaluate the prognostic value of KLF9 expression in human cancers in general and in Nrf2 gain-of–function malignancies in particular. Moreover, from a therapeutic standpoint, dual targeting of Nrf2 and Klf9 could be promising both in oxidative injury and in cancer treatment.
In summary, for the first time our data provide molecular mechanisms underlying Nrf2-dependent feed-forward response to excessive oxidative stress and identify its novel ubiquitous executioner, Klf9.
EXPERIMENTAL PROCEDURES
Cell lines and reagents
WI38 and NIH3T3 cells were obtained from ATCC. H358, H460 and H1650, cells were provided by Dr. Elena Kurenova, RPCI. H1299 and A549 were provided by Dr. Andrei Gudkov, RPCI. Cells were cultured in DMEM supplemented with 10% fetal calf serum, 2mM glutamine and penicillin-streptomycin antibiotics. Nrf2 and Keap1 knock-out MEFs were obtained from Dr. Masayuki Yamamoto (University of Tsukuba, Japan) and cultured as described previously (Li et al., 2012). Immortalized wildtype and Klf9 knockout mouse embryonic fibroblasts MEFs were derived from corresponding MEFs via infection with p53 shRNA (a gift from Dr. Andrei Gudkov, RPCI). Hydrogen peroxide (H2O2), sulforaphane, methyl viologen dichloride hydrate (paraquat), bleomycin, etoposide, actinomycin D and cycloheximide were purchased from Sigma-Aldrich, St. Louis, MO, USA.
Plasmids and Infection
Lentiviral and retroviral infection protocols were described previously (Mannava et al., 2008). All infected cells were selected for resistance to respective selectable markers and used for assays. For a list of plasmids used in this study see the Extended Experimental Procedures.
Dual Luciferase Reporter Assay
Annealed oligonucleotides containing ARE sequences were cloned into pGL3-promoter plasmid (Promega, USA) via KpnI and NheI sites and the inserts were verified by sequencing. For a list of oligonucleotides see the Extended Experimental Procedures. The obtained constructs were mixed with pRL-SV40 plasmid expressing the renilla luciferase gene (Promega, USA). NIH3T3 cells were transfected in triplicates with the plasmid mixtures using Superfect reagent (Qiagen, Valencia CA). 48 hours after transfection, firefly luciferase and renilla signals were detected via dual-luciferase assay kit (Promega, USA). Firefly luciferase signals were normalized by corresponding renilla signals.
Immunoblotting
Membranes were developed with alkaline phosphatase-conjugated secondary antibodies; signals were visualized using the Alpha-Innotech FluorChem HD2 imaging system (Alpha Innotech Corporation, San Leandro, CA) and quantified using ImageQuant software (GE Healthcare Life Sciences). Antibodies were purchased from Santa Cruz Biotechnology, Santa Cruz, CA (dilutions 1÷250): Klf9 sc-12996, , Txnrd2 – sc-46279, Nrf2 – sc-722, Bach1 – sc-14700; Noxa – sc-56169, and α-tubulin: sc-8035 (1÷1000).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was carried as per manufacturer’s instructions (EZ-ChIP™ - Chromatin Immunoprecipitation Kit, Millipore, Billerica, MA). Antibodies and a detailed protocol are described in the Extended Experimental Procedures
Gene Ontology
Gene Ontology (GO) analysis was performed on the ChIP-seq data set using the DAVID web server (http://david.abcc.ncifcrf.gov/) (Huang da et al., 2009) with default setting; p values are calculated by Fisher’s exact test with Bonferroni correction.
Quantitative Real Time PCR
Total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was prepared using cDNA reverse transcription kit (Invitrogen). Quantitative reverse transcription PCR was performed on 7900HT Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA) using TaqMan Universal Master Mix II (Applied Biosystems). PCR data were analyzed using sequence detection software 2.4 (Applied Biosystems). For a list of PCR primers and probes, see the Extended Experimental Procedures. All reactions were performed in triplicate and the experiments were repeated at least twice.
Mice
Animal experiments were conducted according to NIH guidelines and were approved by IACUC at RPCI. Klf9 knock-out mice were previously described (Morita et al., 2003).
Paraquat treatment
14-16 week old male C57BL/6J wild type or KLF9 knock-out mice were administered with saline or up to 20μL Paraquat (PQ) solution (0.04, 0.16 mg mg/kg; Sigma Chemical Co., St. Louis, MO) directly to the nasal cavity by placing drops of the solution into the nares (i.n.). At 6 h post i.n. administration, the mice were anesthetized, the lungs were removed and RNA was isolated according to standard protocol.
Bleomycin treatment
14-16 week old female KLF9 knock-out mice and age-matched wild type female mice (C57BL/6J) were anesthetized and subjected to intratracheal instillation of saline or 2.5 mg/Kg Bleomycin sulfate saline solution (Sigma Aldrich).
Bronchoalveolar lavage fluid (BALF) cell count
Nineteen days post intratracheal administration of bleomycin, the mice were euthanized and BALF was collected by cannulating the trachea and lavaging the lung with 1mL of sterile PBS more than five times to recover a final volume of 5mL. The total cell number was determined using a hemocytometer. More than 200 cells were counted for each sample.
Detection of 8-hydroxy-2′-deoxyguanosine (8-OHdG)
Genomic DNA was isolated from frozen lung samples (10-30mg) according to the manufacturer’s instructions (Promega, USA). 8-OHdG was measured fluorometrically in 200ng of isolated DNA according to manufacturer’s instructions using EpiQuik™ 8-OHdG DNA damage quantification kit (Epigentek, Farmingdale, NY).
Histology
Nineteen days post intratracheal administration of bleomycin, the mice were euthanized and the lungs were inflated and fixed with 4% paraformaldehyde. The lungs were surgically resected and embedded in paraffin wax. Five-micron lungs sections were cut and stained with Hematoxylin and Eosin or Masson’s Trichrome. The extent of lung injury and fibrosis was graded by a pathologist, blinded to the treatment groups, on a scale of 0 for normal lung to 8 for severe distortion of structure and large tissue areas as previously reported by Ashcroft and colleagues (Ashcroft et al., 1988). The major criteria examined included interstitial thickening of alveolar or bronchiolar walls, collagen deposition, and inflammatory cell infiltration.
Statistical Analysis
Each experiment was reproduced at least two times with consistent results. Comparisons were made using the Student’s t-test or a nonparametric Mann-Whitney U test. A two-tailed P-value <0.05 was considered statistically significant for all analyses.
Supplementary Material
Highlights.
 High intracellular ROS trigger Nrf2-dependent increase in ROS levels
High intracellular ROS trigger Nrf2-dependent increase in ROS levels
 Nrf2 binds to the promoter and activates expression of Klf9 gene
Nrf2 binds to the promoter and activates expression of Klf9 gene
 Klf9 reduces oxidative stress resistance by suppressing Trxrd2
Klf9 reduces oxidative stress resistance by suppressing Trxrd2
 Klf9 deficiency reduces bleomycin-induced pulmonary fibrosis in mice
Klf9 deficiency reduces bleomycin-induced pulmonary fibrosis in mice
ACKNOWLEDGEMENTS
This work has been supported by NIH grants CA120244 (M.A.N), R01AI079253 (B.H.S.) and American Cancer Society grant RSG-10-121-01 (M.A.N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
REFERENCES
- Adams DJ, Boskovic ZV, Theriault JR, Wang AJ, Stern AM, Wagner BK, Shamji AF, Schreiber SL. Discovery of Small-Molecule Enhancers of Reactive Oxygen Species That are Nontoxic or Cause Genotype-Selective Cell Death. ACS Chemical Biology. 2013;8:923–929. doi: 10.1021/cb300653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam J, Camhi S, Choi AM. Identification of a second region upstream of the mouse heme oxygenase-1 gene that functions as a basal level and inducer-dependent transcription enhancer. J Biol Chem. 1995;270:11977–11984. doi: 10.1074/jbc.270.20.11977. [DOI] [PubMed] [Google Scholar]
- Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap‘n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- Arner ES. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim Biophys Acta. 2009;6:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol. 1988;41:467–470. doi: 10.1136/jcp.41.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton S, Ashkenazi A. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell. 2011;145:92–103. doi: 10.1016/j.cell.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Bonett RM, Hu F, Bagamasbad P, Denver RJ. Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: implications for neural development and plasticity. Endocrinology. 2009;150:1757–1765. doi: 10.1210/en.2008-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MR, Karaca M, Adamski KN, Chorley BN, Wang X, Bell DA. Novel hematopoietic target genes in the NRF2-mediated transcriptional pathway. Oxid Med Cell Longev. 2013;120305:25. doi: 10.1155/2013/120305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheresh P, Kim SJ, Tulasiram S, Kamp DW. Oxidative stress and pulmonary fibrosis. Biochim Biophys Acta. 2013;7:5. doi: 10.1016/j.bbadis.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Reddy SP, Yamamoto M, Kleeberger SR. The transcription factor NRF2 protects against pulmonary fibrosis. Faseb J. 2004;18:1258–1260. doi: 10.1096/fj.03-1127fje. [DOI] [PubMed] [Google Scholar]
- Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhry S, Zhang Y, McMahon M, Sutherland C, Cuadrado A, Hayes JD. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyoy A, Olguin-Albuerne M, Martinez-Briseno P, Moran J. Role of reactive oxygen species and NADPH-oxidase in the development of rat cerebellum. Neurochemistry international. 2013;62:998–1011. doi: 10.1016/j.neuint.2013.03.009. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000:408. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Good KL, Tangye SG. Decreased expression of Kruppel-like factors in memory B cells induces the rapid response typical of secondary antibody responses. Proc Natl Acad Sci U S A. 2007;104:13420–13425. doi: 10.1073/pnas.0703872104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins LG, Kelleher MO, Eggleston IM, Itoh K, Yamamoto M, Hayes JD. Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol Appl Pharmacol. 2009;237:267–280. doi: 10.1016/j.taap.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi N, Ishii Y, Morishima Y, Yageta Y, Haraguchi N, Itoh K, Yamamoto M, Hizawa N. Nrf2 protects against pulmonary fibrosis by regulating the lung oxidant level and Th1/Th2 balance. Respir Res. 2010;11:1465–9921. doi: 10.1186/1465-9921-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y, Sogawa K, Watanabe N, Kobayashi A, Fujii-Kuriyama Y. Purification and characterization of the DNA-binding domain of BTEB, a GC box-binding transcription factor, expressed in Escherichia coli. J Biochem. 1996;119:309–313. doi: 10.1093/oxfordjournals.jbchem.a021240. [DOI] [PubMed] [Google Scholar]
- Li Y, Jaiswal AK. Regulation of human NAD(P)H:quinone oxidoreductase gene. Role of A. 1992;1 binding site contained within human antioxidant response element. J Biol Chem 267:15097–15104. [PubMed] [Google Scholar]
- Li Y, Paonessa JD, Zhang Y. Mechanism of chemical activation of Nrf2. PLoS One. 2012;7:25. doi: 10.1371/journal.pone.0035122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannava S, Zhuang D, Nair JR, Bansal R, Wawrzyniak JA, Zucker SN, Fink EE, Moparthy KC, Hu Q, Liu S, et al. KLF9 is a novel transcriptional regulator of bortezomib- and LBH589-induced apoptosis in multiple myeloma cells. Blood. 2012;119:1450–1458. doi: 10.1182/blood-2011-04-346676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Kobayashi A, Yamashita T, Shimanuki T, Nakajima O, Takahashi S, Ikegami S, Inokuchi K, Yamashita K, Yamamoto M, et al. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol Cell Biol. 2003;23:2489–2500. doi: 10.1128/MCB.23.7.2489-2500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouratis MA, Aidinis V. Modeling pulmonary fibrosis with bleomycin. Curr Opin Pulm Med. 2011;17:355–361. doi: 10.1097/MCP.0b013e328349ac2b. [DOI] [PubMed] [Google Scholar]
- Mulcahy RT, Wartman MA, Bailey HH, Gipp JJ. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J Biol Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annual Review of Pharmacology and Toxicology. 2003;43:233–260. doi: 10.1146/annurev.pharmtox.43.100901.140229. [DOI] [PubMed] [Google Scholar]
- Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. The Biochemical journal. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011 Jan;29(1):24–6. doi: 10.1038/nbt.1754. 2011. doi: 10.1038/nbt.1754.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocourt CR, Wu M, Chen BP, Cheng WH. The catalytic subunit of DNA-dependent protein kinase is downstream of ATM and feeds forward oxidative stress in the selenium-induced senescence response. J Nutr Biochem. 2013;24:781–787. doi: 10.1016/j.jnutbio.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Singh SV, Srivastava SK, Choi S, Lew KL, Antosiewicz J, Xiao D, Zeng Y, Watkins SC, Johnson CS, Trump DL, et al. Sulforaphane-induced Cell Death in Human Prostate Cancer Cells Is Initiated by Reactive Oxygen Species. Journal of Biological Chemistry. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- Soriano FX, Baxter P, Murray LM, Sporn MB, Gillingwater TH, Hardingham GE. Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol Cells. 2009;27:279–282. doi: 10.1007/s10059-009-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporl F, Korge S, Jurchott K, Wunderskirchner M, Schellenberg K, Heins S, Specht A, Stoll C, Klemz R, Maier B, et al. Kruppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc Natl Acad Sci U S A. 2012;109:10903–10908. doi: 10.1073/pnas.1118641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, et al. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. Embo J. 2002;21:5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetreault MP, Yang Y, Katz JP. Kruppel-like factors in cancer. Nature reviews Cancer. 2013;13:701–713. doi: 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]
- Tomita M, Okuyama T, Katsuyama H, Miura Y, Nishimura Y, Hidaka K, Otsuki T, Ishikawa T. Mouse model of paraquat-poisoned lungs and its gene expression profile. Toxicology. 2007;231:200–209. doi: 10.1016/j.tox.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Human molecular genetics. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnatz HJ, Schmidt D, Manke T, Piccini I, Sultan M, Borodina T, Balzereit D, Wruck W, Soldatov A, Vingron M, et al. The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J Biol Chem. 2011;286:23521–23532. doi: 10.1074/jbc.M111.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proceedings of the National Academy of Sciences. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z, Velarde MC, Simmen FA, Simmen RC. Delayed parturition and altered myometrial progesterone receptor isoform A expression in mice null for Kruppel-like factor 9. Biol Reprod. 2008;78:1029–1037. doi: 10.1095/biolreprod.107.065821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.