Summary
MEK inhibitors are clinically active in BRAFV600E melanomas, but only marginally so in KRAS-mutant tumors. Here we found that MEK inhibitors suppress ERK signaling more potently in BRAFV600E- than in KRAS-mutant tumors. To understand this, we performed an RNAi screen in a KRAS-mutant model and found that CRAF knockdown enhanced MEK inhibition. MEK activated by CRAF was less susceptible to MEK inhibitors than when activated by BRAFV600E. MEK inhibitors induced RAF-MEK complexes in KRAS mutant models, and disrupting such complexes enhanced inhibition of CRAF-dependent ERK signaling. Newer MEK inhibitors not only target MEK catalytic activity, but also impair its reactivation by CRAF: either by disrupting RAF-MEK complexes or by interacting with Ser 222 to prevent RAF-mediated phosphorylation in the complex.
Introduction
Oncogenic KRAS mutations are common in cancer. Active RAS mediates its effects on tumor formation through a number of effector proteins, including RAF, PI3K and RAL (Blasco et al., 2011; Gonzalez-Garcia et al., 2005; Gupta et al., 2007; Kolch et al., 1991). Active RAS causes dimerization and activation of RAF kinases. This initiates a signaling cascade in which RAF phosphorylates and activates MEK, which, in turn phosphorylates and activates ERK (reviewed in Schubbert et al., 2007; Wellbrock et al., 2004). Under physiologic conditions, the amplitude and duration of ERK signaling are regulated by ERK-dependent feedback inhibition of multiple components of the pathway, including receptors, exchange factors, CRAF and ERK itself (Dong et al., 1996; Dougherty et al., 2005; Douville and Downward, 1997).
The importance of ERK signaling in cancers with mutant RAS has been demonstrated in experimental systems in which genetic and pharmacologic manipulation shows that this cascade is required for tumor initiation and maintenance (reviewed in Pylayeva-Gupta et al., 2011). The widespread importance of ERK signaling in cancer is also demonstrated by the frequent occurrence of mutations in other members of this pathway, especially BRAF mutations that occur frequently in melanomas, thyroid and other cancers (Davies et al., 2002).
RAF and MEK inhibitors have been developed as potential therapeutics in an effort to inhibit the growth of tumors dependent on ERK signaling (Bollag et al., 2012; McCubrey et al., 2010; Sebolt-Leopold et al., 1999). RAF inhibitors inhibit ERK signaling in tumors harboring BRAFV600E/K mutations (Heidorn et al., 2010; Joseph et al., 2010; Poulikakos et al., 2010), and have remarkable therapeutic activity in melanomas harboring these mutations (Bollag et al., 2010; Chapman et al., 2011). In other tumors, however, including those with mutant RAS, RAF inhibitors transactivate RAF dimers and stimulate ERK signaling. In contrast, allosteric MEK inhibitors suppress ERK signaling in all normal and tumor cells (Pratilas et al., 2008; Solit et al., 2006). Yet, whereas MEK inhibitors have significant antitumor activity in BRAFV600E tumors (Flaherty et al., 2012), their effectiveness is marginal in tumors with KRAS mutations. We have now investigated the basis for this genotype-specific differential sensitivity.
Results
KRAS mutant tumors are less sensitive to MEK inhibitors than BRAF mutant tumors
We examined the Genomics of Drug Sensitivity in Cancer (GDSC) dataset (Yang et al., 2013) to correlate the sensitivity of tumor cells to MEK inhibitors with cancer genotype. The mean IC50s for three such compounds, i.e. selumetinib, RDEA119 and PD0325901, were compared in tumors harboring BRAF or RAS mutations and those with wild type alleles for these genes. Tumors of various lineages were included in this analysis. Sensitivity to MEK inhibition was correlated with the presence of oncogenic mutations and with the particular oncoprotein responsible for activating the pathway (Figure S1A and below). The mean IC50 for each compound was higher in KRAS mutant tumors than in BRAF-mutant tumors, whereas NRAS mutant tumors had an intermediate sensitivity.
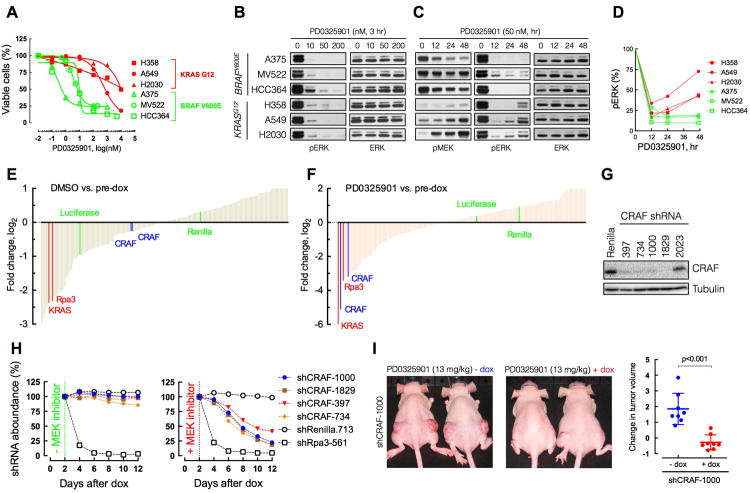
In order to investigate the reason for the reduced sensitivity of KRAS mutant tumors to MEK inhibitors, we first confirmed the mutation-dependent sensitivity to PD0325901 in a group of melanoma (M) and lung (L) cancer cell lines harboring BRAFV600E (A375M, MV522L and HCC364L) or KRASG12C/S (H358L, A549L and H2030L) mutations. As predicted, the latter were significantly less sensitive than the former (Figure 1A). Three hours after treatment, 40-50 nM PD0325901 was found to inhibit ERK phosphorylation more than 95% both in KRAS and BRAF mutant tumors (Figure 1B and S1B). We used this dose to ask whether the difference in sensitivity between the genotypes was associated with a difference in the durability of inhibition of ERK signaling over time. In KRAS mutant lung cancer cell lines prolonged PD0325901 exposure was unable to produce sustained ERK inhibition as indicated by a rebound in ERK phosphorylation after 24-48 hours (Figure 2C). The magnitude of this rebound ranged from 25% to 75% of the pretreatment ERK phosphorylation (Figure 2D) and also occurred in KRAS mutant pancreatic cancer cells (Figure S1B). In contrast, much less pERK rebound was observed in BRAFV600E cell lines (Figure 2C and D). These data imply that the reduced efficacy of PD0325901 in KRAS mutant cancers may result from an inability to sustain pathway inhibition.
Figure 1. KRAS mutant tumors are less sensitive to allosteric MEK inhibitors than BRAFV600E tumors.
(A) BRAFV600E and KRAS-mutant tumor cell lines were treated with increasing doses of PD0325901 for three days to determine the effect on proliferation. A representative example of three independent experiments (each performed in triplicate) is shown as means +/- SEM.
(B-D) The indicated tumor cell lines were treated with increasing amounts of PD0325901 for three hours (B) or with 50 nM of PD0325901 for the indicated times (C). Lysates were assayed by immunoblotting to determine the level of MEK and ERK phosphorylation. The bands were quantified by densitometry and the pERK level after treatment was normalized to the pretreatment pERK level (D). A representative example of three independent experiments for each cell line is shown.
(E, F) Fold change in shRNA abundance in DMSO-treated (E) or PD0325901 (F)-treated cells. Note the selective depletion in shRNAs targeting CRAF with PD0325901 treatment. shRNAs targeting Rpa3 or KRAS were used as positive controls, whereas those targeting luciferase or renilla were used as negative controls. Bars represent the mean fold change from an experiment performed in triplicate.
(G) KRPC cells were transduced with the indicated CRAF shRNAs and lysates were subjected to immunoblotting to determine the knockdown in CRAF expression.
(H) KRPC cells infected with four different CRAF shRNAs were treated with dox to induce CRAF knockdown and PD0325901 (+MEK inhibitor) or DMSO (-MEK inhibitor) for the indicated times. The data were normalized to the shRNA abundance in cells after 48 hr of treatment with dox.
(I) KRPC cells were stably infected with the indicated dox-inducible CRAF shRNA followed by implantation in athymic mice. The mice were treated with MEK inhibitor PD0325901 in the presence or absence of dox. The effect of CRAF knockdown (+dox) in the ability of the MEK inhibitor to inhibit KRAS tumor growth is represented as mean +/-SEM (n=6)
See also Figure S1.
Figure 2. CRAF expression attenuates the effects of MEK inhibition in KRAS-mutant tumors.
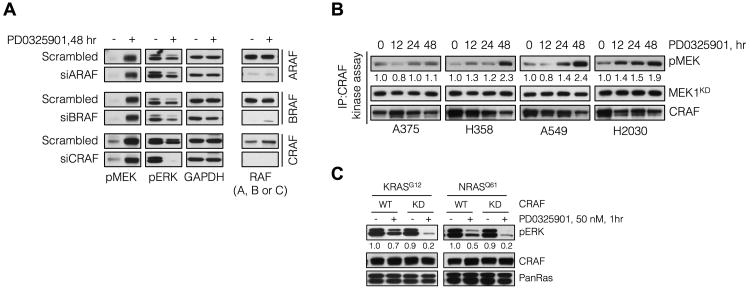
(A) KRAS-mutant lung cancer cells (A549) were transfected with siRNA pools targeting each of the RAF isoforms and then treated with 50 nM of PD0325901 for 48 hr. Whole cell lysates were evaluated by immunoblotting to determine the effect on ERK signaling. A representative example of three independent experiments for each siRNA is shown.
(B) The indicated cell lines were treated with 50 nM of PD0325901 as shown. CRAF was immunoprecipitated from whole cell lysates and subjected to a kinase assay using an inactive MEK1 (K97R, KD) as the substrate. CRAF activity was determined by immunubloting with a phospho-MEK antibody. A representative example of two or more independent experiments for each cell line is shown.
(C) Craf-/- MEFs were co-transfected with the indicated constructs and then treated with PD0325901 for 1 hr. Lysates were analyzed by immunobloting to determine ERK phosphorylation. A representative example of two independent experiments for each RAS isoform is shown. WT: wild type; KD: kinase dead
See also Figure S2
CRAF knockdown enhances the antiproliferative effect of MEK inhibition in KRAS-mutant tumors
The difference in sensitivity between BRAF and KRAS mutant tumors may reflect the fact that MEK is the major substrate of mutant BRAF whereas mutant KRAS signals through multiple effectors. We hypothesized that the ability of MEK inhibitors to inhibit the proliferation of KRAS mutant cells would be augmented by concurrent targeting of other RAS-effector pathways. To address this possibility in a non-biased way, we performed an shRNA screen to identify genes whose inhibition sensitize mutant KRAS tumors to PD0325901 inhibition using a well-characterized genetic model of mutant KRAS-induced pancreatic cancer. In order to develop a system in which shRNA expression is conditionally induced by tetracycline, we engineered a “TET-ON” competent murine pancreatic cancer cell line using pancreatic progenitor cells isolated from a murine fetus harboring an endogenous KrasG12D mutation. These cells were engineered to express c-Myc and a potent p53-specific shRNA, linked to a reverse tet transactivator (Figure S1C-E). Orthotopic injection of these cells into the pancreas of a recipient mouse resulted in a tumor that displayed key histological characteristics of human pancreatic cancer (Figure S1F). A cell line derived from such tumors, referred to as KRPC, was dependent on KRAS (data not shown) and capable of inducible shRNA expression. KRPC cells also exhibited rebound ERK phosphorylation after treatment with PD0325901 (Figure S1G), similar to that in human KRAS mutant lung and pancreatic cancer cell lines.
KRPC cells were transduced with a dox-inducible shRNA library containing a panel of reporter-validated shRNAs (Fellmann et al., 2011; Zuber et al., 2011) that target 70 genes encoding various components of RAS effector pathways. Dox-induction of shRNA expression was followed by treatment with either DMSO or PD0325901 in order to identify shRNAs that were selectively depleted after 10 days of MEK inhibitor treatment (Figure S1H and 1E-F). PD0325901 was chosen because it had the highest potency of the drugs in Figure S1A. Positive control shRNAs, targeting an essential gene (Rpa3), were strongly depleted in both vehicle and drug-treated cells. Consistent with the importance of mutant KRAS in cancer maintenance, shRNAs targeting KRAS were also depleted from the population irrespective of drug treatment (Figure 1E-F). By contrast, CRAF-specific shRNAs were depleted by over 6-fold after MEK inhibitor treatment (Figure 1F), but only minimally after treatment with DMSO (Figure 1E and S1I). One of three BRAF specific shRNAs was marginally depleted with PD0325901 whereas shRNAs targeting ARAF or other RAS effectors were not selectively depleted after MEK inhibitor treatment (Figure S1J). In an independent screen with an shRNA library enriched for known drug targets, CRAF knockdown was again the most significant enhancer of MEK inhibition in KRAS mutant cells (data not shown).
To confirm these findings, we tested a series of CRAF and BRAF specific shRNAs in this pancreatic cancer model and in human KRAS mutant cancer cell lines. Four different CRAF shRNAs that effectively decreased CRAF expression (Figure 1G) were not depleted when used alone, but were selectively depleted in tumor cells exposed to PD0325901 (Figure 1H). The two effective BRAF shRNAs tested were also depleted after PD0325901 treatment, but to a lesser extent (Figure S1K-L). In human KRAS mutant pancreatic cancer cells, CRAF shRNAs were also depleted only after MEK inhibitor treatment (Figure S1M, S1N). In this experiment, BRAF-specific shRNAs were also depleted, but to a lesser extent. Similar results were observed following PD0325901 treatment of KRPC xenografts harboring a tet-responsive CRAF shRNA: when CRAF expression was knockdown in combination with PD0325901, there was a significantly greater tumor regression than that observed when PD0325901 was administered alone (Figure 1I). These results show that CRAF inhibition is required for PD0325901 to be effective against KRAS mutant cancers, and suggest that inhibitors like PD0325901 are less effective at inhibiting CRAF-activated MEK.
The rebound in ERK phosphorylation is dependent on CRAF
To directly test whether RAF family members contribute to the rebound in ERK phosphorylation observed after MEK inhibitor treatment we knocked down RAF in KRAS mutant lung cancers. CRAF siRNAs decreased pMEK levels and attenuated the rebound in ERK phosphorylation at 48 hours, whereas control siRNAs had no effect (Figure 2A and S2A-B). Knockdown of BRAF had a small effect on the rebound in cells with mutant RAS whereas knockdown of ARAF expression had no effect (Figure 2A). CRAF expression was also required for the rebound observed in KRPC cells treated with PD0325901 (Figure S2C). While these data do not exclude a potential role for wild type BRAF in mediating the rebound in ERK phosphorylation in KRAS mutant cells, they suggest that CRAF is required for the pERK rebound and are consistent with reports that CRAF is the main RAF isoform responsible for driving ERK signaling in some RAS mutant tumors (Blasco et al., 2011; Dumaz et al., 2006; Karreth et al., 2011).
The pERK rebound observed in KRAS mutant tumors was associated with induction of pMEK (Figure 1C) and induction of CRAF phosphorylation at Ser 338 (Figure S2D). The rebound in pMEK and pERK are not linearly related, probably because the level of ERK phosphorylation is due to changes in MEK kinase activity and the activity and expression of ERK phosphatases. Induction of MEK phosphorylation in cells exposed to MEK inhibitors results from inhibition of ERK-dependent inhibitory phosphorylations of CRAF, with attendant activation of CRAF kinase (Alessi et al., 1994; Dougherty et al., 2005). By contrast, and in agreement with previous data (Pratilas et al., 2009), the level of MEK phosphorylation decreased slightly after treatment in BRAFV600E tumors (Figure 1C). The selective induction of MEK phosphorylation suggests that CRAF activation occurs differentially in tumors with mutant RAS compared to those with mutant BRAFV600E.
In order to assay the induction of CRAF kinase activity, we performed in vitro kinase reactions, with CRAF that was immunoprecipitated from cells treated with a MEK inhibitor and a kinase-dead recombinant MEK1 as a substrate. In agreement with the conclusion above, PD0325901 treatment increased CRAF activity over time in KRAS mutant models, whereas only a small induction was noted in BRAFV600E cells (Figure 2B and Figure S2E). Thus, compared to cells harboring BRAFV600E, KRAS mutant cells have more pronounced reactivation of CRAF after inhibition of ERK signaling, which in turn mediates the rebound in ERK phosphorylation.
MEK is less susceptible to inhibition when activated by CRAF than by BRAFV600E
Since RAS activation and CRAF activity are both low in BRAFV600E tumors, in which MEK/ERK signaling is driven almost entirely by BRAFV600E (Wan et al., 2004), we hypothesized that the decreased MEK inhibitor sensitivity of KRAS tumors is due to ineffective inhibition of CRAF-driven MEK signaling by these drugs. To test this we undertook two experimental approaches. In the first approach, mutant KRAS was co-expressed with either wild type (WT) CRAF or kinase-dead (KD, i.e. K375M) CRAF in Craf-/- MEFs. As expected, one-hour treatment with PD0325901 inhibited ERK phosphorylation in cells expressing the CRAFKD. In those expressing CRAFWT, however, the pathway sensitivity to the drug was significantly reduced (Figure 2C). A similar effect was observed when ERK signaling was driven by active NRAS (Figure 2C). These results validate our CRAF knockdown findings and strengthen our conclusion that CRAF activation reduces the efficacy of MEK inhibitor treatment.
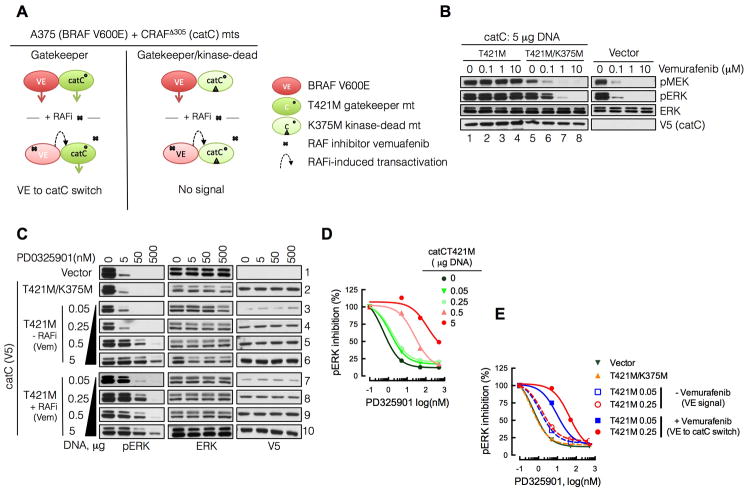
In the second approach, we developed a cellular system where ERK signaling could be selectively switched from being driven by BRAFV600E to CRAF. To accomplish this, we took advantage of the ability of RAF inhibitors to transactivate RAF dimers (Figure 3A). We utilized the BRAFV600E cell line A375, where CRAF activity is low and MEK activation is BRAFV600E-dependent (Lito et al., 2012; Pratilas et al., 2009). Vemurafenib treatment inhibits BRAFV600E and ERK signaling in these cells. An N-terminal truncated CRAF (catC) was expressed in A375 to form intracellular BRAFV600E-catC heterodimers. CatC dimerizes in a RAS-independent manner and is transactivated by RAF inhibitor vemurafenib (Poulikakos et al., 2010). CatC was engineered to harbor a gatekeeper mutation (T421M) in order to prevent its binding to vemurafenib. Thus, when vemureafenib binds to and inhibits BRAFV600E, it transactivates catCT421M in heterodimers, thereby triggering a switch from BRAFV600E- to catC-dependent MEK activation (Figure 3A). A kinase-dead catCT421M/K375M was used as a control. As shown in Figure 3B, expression of catCT421M rendered ERK signaling insensitive to vemurafenib, since this drug does not inhibit RAF dimers (lanes 1-4) whereas ERK signaling was sensitive to vemurafenib in cells expressing catCT421M/K375M (compare lanes 7-8 to 3-4). The residual signaling in lanes 3 and 4 is therefore due to transactivation of catCT421M by vemurafenib and represents activation of CRAF kinase-dependent ERK signaling. Vemurafenib treatment induced similar effects even when lower amounts of catC were expressed (Figure S3A-C).
Figure 3. MEK is less susceptible to inhibition when activated by CRAF than BRAF V600E.
(A) A schematic representation of the experimental system used. BRAFV600E mutant A375 cells have low levels of CRAF activity at baseline and MEK activation is dependent on BRAF kinase. Treatment with RAF or MEK inhibitors inhibits ERK signaling in these cells. We relied on the ability of RAF inhibitors to transactivate RAF dimers in order to switch from BRAFV600E-dependent to catC-dependent signaling. To this end, catC was expressed to form BRAFV600E-catC heterodimers in A375 cells. A gatekeeper mutation in catC (T421M) prevents RAF inhibitor binding. Thus, the RAF inhibitor vemurafenib binds to and inhibits BRAFV600E while transactivating catC in heterodimers. A kinase-dead catC (T421M/K375M) was used as a control.
(B) A375 cells expressing the indicated constructs were treated with increasing doses of vemurafenib for 1 hr and whole cell lysates were subjected to immunoblotting with the indicated antibodies.
(C) A375 cells were transfected with increasing amounts of plasmid encoding catCT421M. In the indicated rows RAF inhibitor pre-treatment (vemurafenib, 1μM, 1 hr) was used to transactivate catC in heterodimers, and switch from BRAFV600E- to catC-driven signaling. Following transfection and pretreatment as shown, the cells were treated with increasing doses of PD0325901 for 1 hr to determine its ability to inhibit ERK phosphorylation. A representative example of at least two independent experiments for each condition is shown.
(D, E) The bands in C were quantified by densitometry and normalized to the level of pERK prior to PD0325901 treatment.
See also Figure S3
We then determined whether the MEK inhibitor PD0325901 could effectively inhibit vemurafenib-induced catC-dependent ERK signaling (i.e. CRAF-dependent signaling). The inhibition of ERK phosphorylation in response to the MEK inhibitor PD0325901 was similar in BRAFV600E cells transfected with an empty vector or with catCT421M/K375M (Figure 3C: rows 1-2), situations in which ERK phosphorylation is driven by BRAFV600E. In contrast, expression of kinase active catCT421M attenuated the ability of PD0325901 to inhibit ERK phosphorylation (Figure 3C: rows 3-6) in a manner that was proportional to the level of catC expression (Figure 3D). Cells expressing low catC levels showed no change in their sensitivity to MEK inhibitors in the absence of vemurafenib (Figure 3C: rows 3-4). In these cells, however, vemurafenib treatment induced appreciable CRAF kinase signaling (Figure S3B-C), and this was sufficient to reduce the sensitivity of pERK to PD0325901 (Figure 3C: rows 7 vs 3 or 8 vs 4 and Figure 3E). In cells in which catC expression was high enough to drive CRAF kinase-dependent signaling in the absence of vemurafenib, the effects of PD0325901 were attenuated regardless of whether the cells were treated with vemurafenib (Figure 3C: rows 5-6 vs 9-10). These data show that ERK signaling activated by CRAF is less sensitive to PD0325901 than when activated by BRAF V600E.
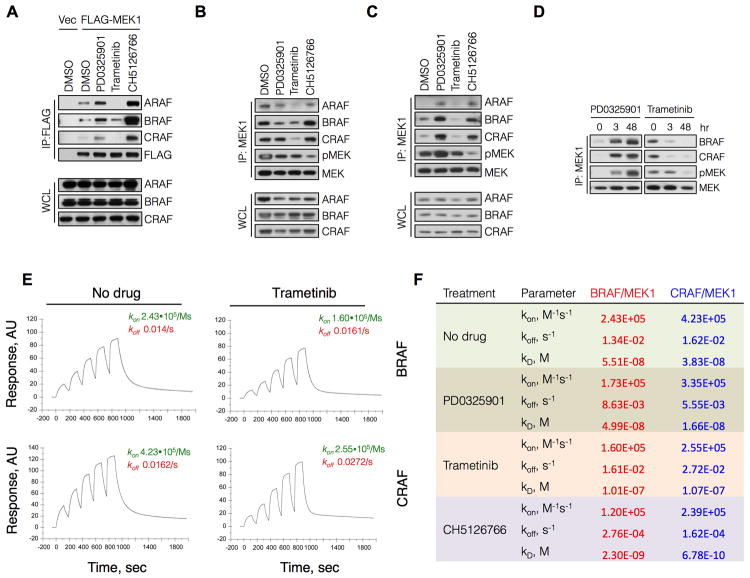
MEK inhibitors induce RAF-MEK complexes
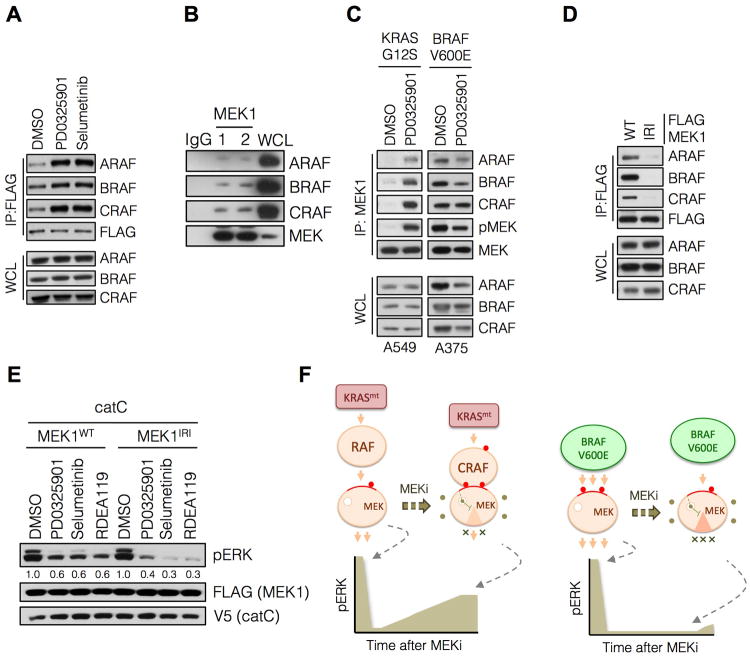
In unpublished work we observed that some MEK inhibitors induce the association of MEK with RAF. We hypothesized that MEK bound to an activated RAF kinase is less sensitive to MEK inhibitors than is unbound MEK. We first evaluated if PD0325901 or selumetinib induced RAF/MEK complexes in HEK293 cells engineered to express FLAG-MEK1. Exposure of these cells to either drug increased the interaction of each RAF kinase with FLAG-MEK1 (Figure 4A). We next tested whether induction of these complexes differed between cells with mutant BRAF or mutant KRAS. Treatment of KRAS mutant A549 cells with PD0325901 increased the association of endogenous MEK1 with all three RAF kinases (Figure 4B and 4C). In contrast, the same treatment did not induce MEK-RAF complexes in BRAFV600E A375 cells (Figure 4C). In this setting, PD0325901 reduced the level of MEK1 in complex with BRAF, as well as the level of MEK phosphorylation.
Figure 4. Allosteric MEK inhibitors induce RAF-MEK complex formation in KRAS mutant tumors.
(A) HEK293 cells were transfected with FLAG-MEK1, followed by treatment with PD0325901 for 3 hrs. MEK1 was immunoprecipitated using an anti-FLAG antibody followed by immunoblotting with the indicated antibodies to determine RAF-MEK interactions. A representative example of at least two independent experiments with each drug is shown.
(B) Whole cell lysates from A375 cells were subjected to immunoprecipitation with IgG or a MEK1 antibody and then immunoblotted for the indicated proteins. Two MEK specific IP replicates are shown.
(C) Cells were treated with PD0325901 for 3 hrs and endogenous MEK1 was immunoprecipitated to determine its interaction with RAF kinases. A representative example of three independent experiments is shown.
(D) HEK293 cells were transfected with WT MEK1 or a MEK1 mutant with impaired RAF interaction (IRI, M308A/I310A). RAF-MEK1 complexes were determined as in C.
(E) A375 cells were transfected with the indicated constructs and then treated with PD0325901 (50 nM), selumetinib (500 nM) or RDEA119 (100 nM) for 1 hr to determine the effect on ERK phosphorylation. A representative example of at least two independent experiments for PD0325901 and selumetinib are shown.
(F) A schematic diagram modeling the role of CRAF in the differential adaptation of KRAS and BRAF mutant tumors to MEK inhibitor treatment.
See also Figure S4
In order to confirm that the differences described above were related to the oncoprotein responsible for activating ERK signaling, we repeated the experiments in HEK293 cells that express FLAG-MEK1 and either KRASG12V or BRAFV600E. Induction of MEK1-RAF complexes by PD0325901 was only observed when MEK1 was co-expressed with KRASG12V (Figure S4A: lanes 1-2). In contrast, when MEK1 was co-expressed with BRAFV600E there was a decrease in MEK1-RAF complexes after 3 hours of treatment (Figure S4A: lanes 5-6). Similar findings were observed when KRAS or BRAF mutants were co-expressed with MEK2 (Figure S4B). The data suggest that binding of MEK to allosteric inhibitors cause MEK to bind to RAF in cells with mutant RAS, but not in those with BRAFV600E.
The effect of MEK inhibitors is enhanced when RAF-MEK interactions are impaired
These findings suggest the possibility that MEK bound to an activated CRAF in tumors with activated RAS is less sensitive to MEK inhibitors than is free MEK. We therefore tested whether reducing the affinity of MEK for RAF would enhance its sensitivity to drugs in cells with activated CRAF. The proline-rich region of MEK1 is thought to mediate its interaction with RAF (Dang et al., 1998). Alanine substitutions of Met 308 and Ile 310 impair this interaction (McKay et al., 2009) and, in our experiments, produced a MEK1 mutant (impaired RAF interaction, IRI) that was defective in its ability to bind RAF kinases (Figure 4D). We used the system illustrated in Figure 3 to test if altering the RAF-MEK interaction affects the inhibition of CRAF-induced ERK signaling by these drugs. In cells in which catC was co-expressed with FLAG-MEK1 there was residual pERK following treatment with PD0325901, selumetinib or RDEA119 (Figure 4E). When catC was co-expressed with MEK1IRI instead, these drugs inhibited ERK phosphorylation more effectively (Figure 4E and S4C-D). These findings suggest that MEK bound to activated CRAF is less sensitive to inhibition by MEK inhibitors like PD0325901. Together, our data support the conclusion that MEK inhibitors suppress ERK-dependent negative feedback to activate CRAF and induce complex formation between MEK and CRAF. These, in turn, cooperate to attenuate maximal pathway inhibition by these drugs in KRAS mutant tumors (Figure 4F).
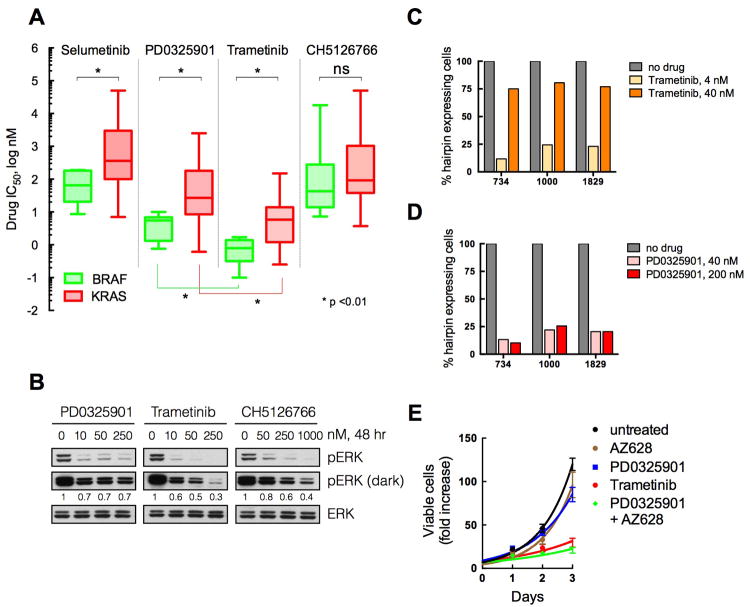
Trametinib and CH5126766 Inhibit ERK Signaling More Durably than PD0325901
Trametinib and CH5126766 are newer MEK inhibitors that have been reported to inhibit the proliferation of selected RAS-mutant xenografts better than PD0325901 does (Gilmartin et al., 2011; Ishii et al., 2013). On average, trametinib inhibited the proliferation of BRAFV600E and KRAS mutant cancer cell lines with lower IC50s than PD0325901 or selumetinib (Figure 5A and S5A), even though trametinib and PD0325901 had similar potencies against MEK1 in vitro (Figure S5B). When compared to PD0325901, CH5126766 was less potent at inhibiting MEK1 kinase activity in vitro. CH5126766, inhibited the proliferation of BRAFV600E and KRAS mutant cell lines at a similar IC50 range (Figure 5A), although CH5126766 was still less effective against some KRAS mutant cells (Figure S5A: H2030 and A549).
Figure 5. Newer allosteric MEK inhibitors inhibit ERK phosphorylation better than PD0325901.
(A) A panel of BRAFV600E (n, 15) and KRAS mutant (n, 50) tumor cell lines were subjected to proliferation assays with the indicated drugs. Cells were treated for three days with increasing amounts of the indicated compounds, in order to determine their respective IC50s. The mean (line) and range of IC50s across cell lines (n of 3 for each cell line) is shown.
(B) KRAS mutant A549 cells were treated with increasing concentrations of PD0325901, trametinib or CH5126766 for 48 hr. Lysates were evaluated by immunoblotting to determine the level of ERK phosphorylation. A representative of two independent experiments is shown.
(C, D) KRPC cells expressing dox-inducible CRAF shRNAs were treated with dox alone (no drug) or in combination with trametinib (C) or PD0325901 (D) at the indicated concentrations. The data were normalized to the number of shRNA expressing cells in the dox only controls. A representative of two independent experiments with at least two different shRNAs targeting CRAF is shown.
(E) KRAS mutant A549 cells were treated with 100 nM PD0325901, 2 μM AZ628, or 100 nM trametinib as indicated. The fold increase in viable cell number over time is shown. Means +/- SEM from a representative of two independent experiments, each performed in triplicate, are shown.
See also Figure S5
Increasing concentrations of PD0325901 did not reduce the rebound in ERK phosphorylation occurring in KRAS mutant A549 cells at 48 hours (Figure 5B). In contrast, the rebound associated with trametinib or CH5126766 decreased with increasing concentrations of these drugs (Figure 5B). As these drugs have different potencies for MEK1 in vitro (Figure S5B), we also evaluated the effects of each drug at concentrations 50 times higher than their IC50 for kinase inhibition in vitro. As compared to PD0325901, treatment with trametinib or CH5126766 for 48 hrs at these concentrations resulted in more sustained inhibition of pERK in KRAS-mutant cells (Figure S5C, H2030) and, also, in A375 cells expressing increasing amounts of active CRAF (Figure S5D).
If CRAF knockdown enhances the effect of MEK inhibitors by more durably inhibiting ERK phosphorylation, then its effect should be minimized when combined with trametinib (which inhibits ERK signaling better than PD0325901). To test this, we evaluated whether cells expressing CRAF shRNAs were depleted when trametinib was administered at two different doses, one associated with pERK rebound (4 nM), and a higher dose associated with much less rebound (40 nM). As shown in Figure 5C, cells expressing CRAF shRNAs were selectively depleted following treatment with 4 nM trametinib but not following treatment with 40 nM trametinib. By contrast, a higher dose of PD0325901 did not affect the selective depletion of cells expressing CRAF shRNA (Figure 5D). A similar effect was observed with CH5126766 (data not shown). These data support our model that PD0325901 does not sufficiently inhibit CRAF-driven ERK signaling and that CRAF knockdown enhances the effects of MEK inhibitors because the combination produces sustained ERK inhibition.
If the added benefit of trametinib occurs due to better inhibition of CRAF-driven signaling than PD0325901 then the combination of PD0325901 with a CRAF inhibitor ought to yield a similar response to that of trametinib alone. To test this, we used AZ628, which has been characterized as an irreversible CRAF inhibitor, although it also targets other RAF kinases. As shown in Figure 5E, treatment with PD0325901 or AZ628 alone did not cause significant growth inhibition in KRAS mutant A549 cells. Combined PD0325901 and AZ628 treatment, however, was not only much more effective than either drug alone but also produced a similar growth inhibition as observed with trametinib treatment alone (Figure 5E).
Trametinib decreases the interaction of MEK1 with RAF Kinases
We then asked whether the newer MEK inhibitors have a different effect on the formation of RAF-MEK complexes or MEK phosphorylation. In contrast to PD0325901, trametinib did not induce a significant interaction between MEK1 and RAF kinases in HEK293 cells, regardless of whether MEK1 was expressed alone (Figure 6A), or co-expressed with mutant KRAS or BRAFV600E (Figure S4A). Similar effects were observed when MEK2 was expressed with KRAS or BRAF (Figure S4B). Moreover, trametinib treatment diminished the ability of endogenous MEK1 to interact with RAF kinases in both BRAFV600E (Figure 6B) and KRAS mutant cells (Figure 6C). In KRAS mutant cells, this effect persisted up to 48 hr after drug treatment (Figure 6D).
Figure 6. Trametinib binding weakens the interaction of MEK1 with RAF.
(A) HEK293 cells were transfected with FLAG-MEK1 and then treated with PD0325901 (50 nM), trametinib (10 nM) or CH5126766 (250 nM) for 3 hr. MEK1 was immunoprecipitated with a FLAG antibody and RAF-MEK complexes were determined by immunoblotting.
(B, C) BRAFV600E (A375, B) and KRASG12S (A549, C) cells were treated with the indicated MEK inhibitors as in (A). Lysates were subjected to immunoprecipitations and immunoblotting to determine the interaction between endogenous MEK1 and RAF kinases.
(D) A549 cells were treated with PD0325901 (50 nM) or trametinib (10 nM) for the indicated times. RAF-MEK complexes were determined as in (C).
(E) Recombinant GST-BRAF or GST-CRAF was immobilized on a sensor chip followed by sequential injections of increasing amounts of His-MEK1 in the presence or absence of trametinib at a saturating concentration (3 μM) to determine the association and dissociation rates of MEK-RAF complexes. A representative of at least two independent experiments is shown in each panel of this figure.
(F) Effect of MEK inhibitors on the binding constants for the interaction between MEK1 and RAF.
See also Figure S6
Surface plasmon resonance (SPR) was used to determine if the change in complex formation reflected a change in the affinity of MEK for RAF. Purified BRAF or CRAF were immobilized on a sensor chip followed by sequential injections of increasing amounts of MEK1. These experiments were performed in the presence of 500 μM ATP and before or after addition of trametinib at a saturating concentration of 3 μM (Figure 6E). When bound to trametinib, the dissociation constants for the interaction of MEK1 with either BRAF or CRAF kinase were 0.101 μM and 0.107 μM, respectively. By comparison, the dissociation constants for MEK1 unbound to drug were 0.055 μM and 0.038 μM, respectively. Thus, trametinib is a MEK inhibitor that uniquely reduces the affinity of MEK from RAF, thereby reducing steady state levels of RAF-MEK complexes. Trametinib inhibits the catalytic activity of MEK by the same mechanism as PD0325901, yet it has the opposite effect on its interaction with the RAF kinases. In contrast to the effect observed with PD0325901, trametinib decreased the rebound in ERK phosphorylation in KRAS tumors cells (Figure 5B). This supports our above finding that CRAF-MEK complexes reduce the ability of PD0325901-like inhibitors to inhibit ERK signaling.
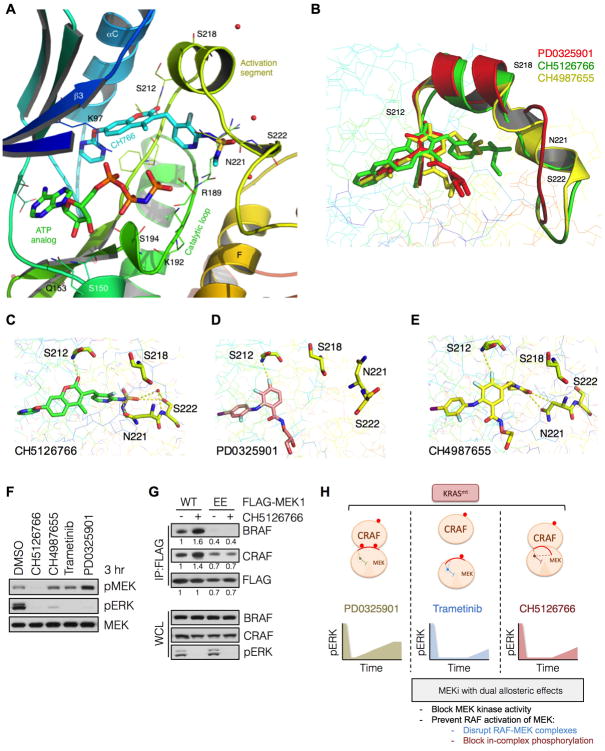
CH5126766 prevents phosphorylation of MEK and induces RAF-MEK complexes
In contrast to trametinib, CH5126766 promoted the formation of RAF-MEK complexes (Figure 6A-C and S4A-B). In addition, CH5126766 completely prevented the induction of MEK phosphorylation in KRAS mutant tumor cells (Figure S6). These results are consistent with our previous findings that CH5126766 blocks MEK phosphorylation, and that CH5126766-bound MEK1 has a markedly slower dissociation from RAF kinases (Ishii et al., 2013).
The ability of CH5126766 to prevent MEK phosphorylation, while inducing RAF-MEK complexes, demonstrates another mechanism for effectively inhibiting ERK signaling in KRAS mutant tumors. To investigate the structural basis for this mechanism we determined the ternary complex of MEK1 bound to CH5126766 and an ATP analogue at a resolution of 2.7Å (Figure 7A and Table S1). CH5126766 interacts with a binding pocket defined by the MEK1 activation segment, catalytic loop and ATP-binding site. MEK1 bound to CH5126766 was in a catalytically inactive conformation, with an outwardly shifted αC helix. CH5126766 formed a critical hydrogen bond with the backbone amide of Ser 212, in a similar manner as other MEK inhibitors, including PD03259021 (Figure S7A). This hydrogen bond is critical for the binding of allosteric inhibitors to MEK (Ohren et al., 2004) and thus represents a feature of this class of drugs.
Figure 7. Interaction with Ser 222 and allosteric displacement of the MEK activation segment by CH5126766 prevents RAF-mediated phosphorylation.
(A) The ternary structure of MEK1-bound to CH5126766 and an ATP analog at a resolution of 2.7 Å. Its interaction with the MEK activation segment residues Ser 212, Asn 221 and Ser 222 are shown.
(B) Superpositioning of CH5126766 (green), CH4987655 (yellow) and PD0325089 (an enantiomer of PD0325901, red) and their effect on the orientation of the MEK1 activation segment.
(C-E) Interactions of the indicated compounds with key residues in the MEK1 acitvation segment.
(F) KRAS mutant A549 cells were treated with the indicated inhibitors for 3 hrs and whole cell lysates were subjected to immunoblotting with the indicated antibodies. A representative example of three independent experiments is shown.
(G) HEK293 cells transfected with wild type MEK1 or a MEK1 mutant harboring phosphomimetic substitutions with glutamic acid at Ser 218 and Ser 222 (i.e. EE) were treated with CH5126766 for 3 hrs. Co-immunoprecipitations were used to determine the interaction between FLAG-tagged MEK1 and endogenous BRAF or CRAF. A representative example of two independent experiments is shown.
(H) Schematic diagram representing the allosteric effects induced by each MEK inhibitor.
The unique aspect of CH5126766 binding was its extension along the activation segment of MEK1 (Figures 7A and 7B) and its interaction with Asn 221 and Ser 222 (Figure 7C), with the latter interaction being coordinated by a water molecule. CH5126766-bound MEK1 displayed a lateral displacement of the activation segment, causing a minor and a major repositioning of the RAF phosphorylation residues Ser 218 and Ser 222, respectively (Figures 7B, C and S7B). This displacement was minimal in MEK1 bound to an enantiomer of PD0325901 (Figures 7B, D and S7C). Thus, it is likely that CH5126766 prevents the phosphorylation of MEK by binding to Ser 222, a key RAF phosphorylation site, and by displacing the activation segment of MEK.
The MEK inhibitor CH4987655 is similar to CH5126766 in that it also interacts with Asn 221 and laterally displaces the activation segment of MEK (Figures 7B and 7E). Compared to CH5126766, however, CH4987655 spans a shorter distance along the MEK activation segment (Figure 7B) and does not interact with Ser 222 (Figures 7E vs 7C). With these differences in mind, we tested the ability of CH4987655 to induce MEK phosphorylation following MEK inhibitor treatment in KRAS mutant A549 cells. CH4987655 caused a very minimal induction of pMEK, compared to standard MEK inhibitors like PD0325901. MEK phosphorylation in CH4987655 treated cells, however, was much greater than that in CH5126766 treated cells, suggesting that both lateral displacement of the activation segment and Ser 222 binding are required for maximal inhibition of MEK phosphorylation by allosteric compounds.
We then asked whether the increase in MEK-RAF complexes in cells treated with CH5126766 is caused by dephosphorylation of MEK. A MEK1 mutant with phosphomimetic glutamate substitutions at the RAF phosphorylation sites (i.e. S218E and S222E or EE) was used to answer this question. As expected, CH5126766 treatment induced the interaction of MEK1WT with BRAF and CRAF (Figure 7G). This induction was abolished in cells expressing MEK1EE treated with CH5126766, which suggests that the higher affinity of CH5126766-bound MEK1 from RAF kinases occurs because MEK bound to this compound cannot be phosphorylated. Consequently, CH5126766 is more effective at producing more sustained inhibition of ERK phosphorylation than PD0325901, despite inducing RAF-MEK complexes.
Discussion
Selective MEK inhibitors have marginal activity in patients whose tumors harbor KRAS mutations. Our data suggest that reactivation of CRAF limits the ability of MEK inhibitors to inhibit ERK signaling in KRAS mutant tumors. Knockdown of CRAF expression enhanced the effects of MEK inhibitors on signaling and on the proliferation of KRAS tumors. CRAF knockdown may enhance the antitumor effects of MEK inhibition by inhibiting other targets of RAF in addition to MEK (Blasco et al., 2011). Several findings presented here, however, suggest that MEK inhibitors do not effectively inhibit CRAF-driven ERK signaling: the rebound in ERK phosphorylation in mutant KRAS tumors exposed to MEK inhibitors was driven by CRAF, and CRAF expression reduced the ability of these drugs to inhibit ERK signaling. Furthermore, when the pathway was inhibited with the stronger drug trametinib, enhancement by CRAF knockdown was minimal.
Activated ERK feedback inhibits the RAF/MEK/ERK pathway in cells with activated RAS by directly phosphorylating and inhibiting CRAF kinase activity (Dougherty et al., 2005). MEK inhibitors relieve this feedback by inhibiting ERK and dephosphorylating and reactivating CRAF. This causes induction of MEK phosphorylation in most tumor cells, including those with mutant KRAS. (Alessi et al., 1994; Dougherty et al., 2005; Pratilas et al., 2009). By contrast, RAS-GTP levels are very low in tumors with BRAFV600E (Lito et al., 2012) and MEK inhibitors cause only marginal induction of CRAF kinase activity and do not induce MEK phosphorylation in these cells. As inhibition of cellular ERK signaling by PD0325901 varied inversely with the level of activated CRAF, it is likely that the decreased effectiveness of MEK inhibitors in mutant KRAS cells is due to release of feedback inhibition of CRAF kinase activity.
MEK inhibitors like PD0325901 and selumetinib also increase the association of MEK with RAF kinases. This occurs in cells expressing mutant KRAS but not in those expressing mutant BRAF. These data suggest that when MEK is bound to an activated CRAF, it is less sensitive to MEK inhibitors than is unbound MEK. In support of this model, expression of a MEK mutant that interacts more weakly with RAF resulted in better inhibition of CRAF-dependent ERK signaling.
Near complete inhibition of ERK signaling is required for RAF inhibitors to cause tumor regression in BRAFV600E tumors (Bollag et al., 2010). Such potent pathway inhibition is unlikely to be achieved with PD0325901, solumetinib or other standard MEK inhibitors in tumors driven by mutant KRAS. Our model suggests that ERK signaling in these tumors may be more effectively inhibited by compounds that not only inhibit the catalytic activity of MEK but also reduce its reactivation by CRAF. Indeed, we found two MEK inhibitors that display this property and produce a more durable inhibition ERK signaling than PD0325901. Trametinib and CH5126766 bind to a similar pocket in MEK as other MEK inhibitors and inhibit its catalytic activity allosterically. These compounds, however, also reduce the activation of MEK by RAF through two distinct mechanisms. Binding to trametinib increases the dissociation rate of MEK from RAF and reduces the intracellular level of RAF-MEK complexes. This reduces both the induction of MEK phosphorylation and the rebound in ERK phosphorylation observed in KRAS mutant tumors. CH5126766 works by a different mechanism: it interacts with Ser 222 and Asn 221 on MEK and prevents its phosphorylation by activated CRAF. Drug-bound, dephosphorylated MEK binds more tightly to RAF and the level of MEK-RAF complexes is induced. However, because CH512676-bound MEK cannot be phosphorylated and released from RAF, it becomes a dominant negative inhibitor of RAF (Ishii et al., 2013).
Based on computational modeling, others have proposed that MEK inhibitors that prevent MEK phosphorylation do so by interacting with Ser 212 (Hatzivassiliou et al., 2013). However, this in-silico modeling was based on a crystal structure with a limited definition of the carboxy terminal portion of the MEK activation segment, which does not include Asn 221 and Ser 222. Here we solved the ternary structure of CH512676-bound MEK1 and show that CH5126766 forms multiple interactions with the activation segment of MEK, binding not only to Ser 212 but also to Asn 221 and Ser 222 and displaces the activation segment of MEK1. We believe that this is the likely mechanism by which it prevents the phosphorylation of MEK by RAF.
Our results demonstrate that the ability of MEK inhibitors to inhibit MAPK activity and inhibit KRAS mutant cancer cells vary as a function of the specific biochemical properties of the drug-bound MEK complex. Differences in cellular responses to these drugs reveal subtleties in the biochemical regulation of the pathway, particularly the role of CRAF-MEK complex formation in rendering KRAS mutant cancers less sensitive to these drugs. Rational drug discovery relies on lead-optimization of compounds based on their selectivity, potency, and pharmacologic properties. Understanding the exact mechanism of drug activity in the appropriate signaling context is imperative in order to design more effective compounds. Here we identify two biochemical properties that, if optimized, will improve the efficacy of MEK inhibitors: increasing the dissociation rate of MEK from RAF and preventing MEK phosphorylation by reactivated CRAF. If coupled with clinical trial designs aimed at improving the therapeutic index of these drugs, e.g. pulsatile or intermittent dosing schedules, these efforts are likely to maximize the clinical benefit of MEK inhibitors in KRAS mutant tumors.
Experimental Procedures
Compounds and cell culture
PD0325901 was synthesized in the MSKCC Organic Synthesis Core Facility by O. Ouerfelli. Trametinib and CH5126766 were obtained from GlaxoSmithKline and Chugai Pharmaceuticals, respectively. All compounds were diluted in DMSO for in vitro experiments. Craf-/- MEFs were kindly provided by M. Baccarini; HEK293 were from Invitrogen while BRAF and KRAS mutant cell lines were obtained from American Type Culture Collection. Cell lines were maintained in DMEM or RPMI supplemented with 10% fetal bovine serum, glutamine and antibiotics.
Antibodies, immunoblotting and immunoprecipitations
Antibodies detecting pMEK, pERK, pCRAF, MEK, ERK and CRAF were obtained from Cell Signaling. V5 and FLAG antibodies were from Invitrogen and Sigma, respectively. Immunoblotting was performed as described (Poulikakos et al., 2010). Exogenously expressed FLAG-MEK1 was immunoprecipitated (IP) via a FLAG-specific antibody agarose conjugate (Sigma) according to the manufacturer's recommendations. Endogenous MEK1 was immunoprecipitated with MEK1 612B12 antibody (Cell signaling). In both cases, IPs were carried out at 4° C overnight.
Immunoprecipitation kinase assays
These were performed as described (Poulikakos et al., 2010). Briefly, cells were lysed in NP-40 lysis buffer supplemented with phosphatase and protease inhibitors (Roche). IPs were carried out at 4° C for 4 hrs and immunoprecipitated CRAF was subjected to a kinase assay with kinase dead MEK1 (Millipore). Kinase assays were conducted in the presence of 100 μM ATP at 30° C for 30 min. MEK phosphorylation was assayed by immunoblotting with phospho-specific antibodies.
Plasmids and transfections
MEK1, KRas and NRas encoding plasmids were obtained from Biomyx. CRAF or MEK constructs harboring various mutations were generated by site-directed mutagenesis (Agilent). DNA and siRNA transfections were carried out by using lipofectamine 2000 and RNAiMAX reagents according to the manufacturer's recommendations.
Animal Studies
Nu/nu athymic mice were obtained from the Harlan Laboratories and maintained in compliance with IACUC guidelines. Subcutaneous xenografts and tumor measurements were performed as described (Lito et al., 2012). All studies were performed in compliance with institutional guidelines under an IACUC approved protocol (Memorial Sloan-Kettering Cancer Center No. 09-05-009).
Surface plasmon resonance
This was performed as described (Ishii et al., 2013). Briefly N- terminal GST-tagged BRAF or CRAF was captured on the surface of a CM5 sensor chip (GE Healthcare) by anti-GST polyclonal antibodies that were pre-immobilized on the chip according to the manufacturer's instructions. His6-MEK1 solutions at concentrations of 0.0256, 0.064, 0.16, 0.4, and 1 mmol/L, were injected sequentially in order of increasing concentration over the sensor chip in the absence or presence of 3 mmol/L of the test compounds and then the dissociation constants of His6-MEK1 were calculated for the immobilized RAF for each condition in the presence of 500 μmol/L ATP. The effects of MEK inhibitors on the RAF-MEK1 complex formation were determined by using single-cycle kinetics.
Crystallization and structural determination of MEK1-CH5126766 complex
MEK1 kinase (residues 62-393, del 271-302) was expressed in E.coli with a His tag. The expressed protein was purified using standard chromatography. Crystals were obtained at 4°C from sitting drops using a reservoir solution (7.5-10% PEG 8000, 0.2M NaCl, 0.2M Ammonium sulfate, 0.1M citrate buffer, pH 4.2) by vapor diffusion. Diffraction data were collected at 100 K at the SGX-CAT beamline in APX synchrotron using a Mar CCD detector (Marresearch GmbH). The data set was processed with HKL2000 and scaled with SCALA in the space group P3121. The structure of the MEK1 and CH5126766 complex was determined by molecular replacement. The crystals contain one monomer of the protein in the asymmetric unit. The model was rebuilt manually in COOT, and refined with CNX 2005 (Accelrys Inc.) to a final resolution of 2.7 Å. B-factors were refined isotropically. The final model consisted of residues 62-380 with 2 breaks (residues 79-81, 268-307 were disordered). The resulting electron density revealed an unambiguous binding mode of CH5126766. For crystallographic data and refinement statistics, see Table S1. This structure has been deposited in Protein Data Bank (PDB) with accession number 3WIG.
Pooled negative selection screening
A miR30-based shRNA library containing 193 shRNAs targeting 70 genes encoding for various components of the Ras effector pathway was assembled. All hairpins included in this set were the top scoring candidates in a FACS based high-throughput reporter assay that functionally evaluates shRNA potency and thus leads to the identification of highly potent shRNAs (Fellmann et al., 2011). The hairpin pool was subcloned into TRMPV-Neo (Zuber et al., 2011) and combined with positive- and negative control shRNAs. Viral supernatant from triplicate transfections into PlatE cells was harvested and used to transduce Tet-on KRPC cells under conditions that led to single integrations (less that 5% initial infection rate). Triplicates were processed separately throughout the screen and at all times care was taken to maintain an shRNA representation exceeding 1000× per replicate.
Successfully transduced cells underwent G418 selection (Invitrogen, final concentration 500μg/ml) for 7 days. At this time, T0 samples were obtained for genomic DNA extraction and the remaining cells were cultured in doxycycline containing media (final concentration 1μg/ml) to induce shRNA expression. At 48 hr after dox treatment, cells of each triplicate were further propagated in doxycycline containing media in the presence of 40 nM PD0325901 or DMSO for the duration of the screen. On the final day of the screen (Tf), approximately 2 million cells per replicate and condition were FACS sorted (Venus+, dsRed+) and genomic DNA was extracted from all samples using the Gentra Puregene Cell kit (QIAGEN, Venlo, Netherlands). Deep-sequencing template libraries were generated for each replicate by PCR amplification of shRNA guide strands. To allow for multiplexed illumina sequencing, forward primers located in the loop region of the miR30 cassette were barcoded 3′ of the p7 adapter sequence (forward primer: 5′-CAAGCAGAAGACGGCATACGA - 8nt barcode - TAGTGAAGCCACAGATGTA -3′). The p5-adapter-coupled reverse primer was placed 3′ of the miR30 shRNA cassette within the TRMPV-Neo backbone to achieve exclusive amplification of shRNAs that were part of the library and avoid amplification of the miR30 based p53 hairpin present in the screen cell line (reverse primer: 5′ - AATGATACGGCGACCACCGACGGTAG AATTGCTAGAATTG - 3′). All PCR reactions per barcode were pooled and subjected to Agencourt AMPure XP bead purification (Beckman Coulter, Inc., Pasadena, CA). Libraries were analyzed on an Illumina 2000 Genome Analyzer (Illumina, San Diego, CA) using the custom primer miR30EcoRISeq 5′ - TAGCCCCTTGAATTCCAGG CAGTAGGCA - 3′. Sequence processing was performed using a custom script developed by the MSKCC Bioinformatics Core and downstream analysis was performed in R (http://cran.r-project.org/) and the fold change of shRNA representation was calculated by comparing normalized shRNA reads at Tf to normalized reads at T0. To identify synthetically lethal interactions, fold change in the presence of the MEK inhibitor was set in relation to the fold change in the absence of the MEK inhibitor.
Supplementary Material
Highlights.
KRAS mutant tumors are less sensitive to MEK inhibitors than BRAFV600E mutant tumors
CRAF knockdown enhances the effect of MEK inhibitors
MEK is less susceptible to MEK inhibitors when activated by CRAF than by BRAFV600E
MEK inhibitors have distinct biochemical properties that control their activity
Significance.
We show that the effects of MEK inhibitors in KRAS mutant tumors vary, owing to specific properties of drug-bound MEK. In these tumors, some MEK inhibitors induce CRAF-MEK complex formation, which prevents durable inhibition of ERK. Newer compounds associate with MEK differently and are affected less by reactivated CRAF. Through genetic, pharmacological, and structural approaches we identified two allosteric effects that improve the efficacy of MEK inhibitors in KRAS mutant tumors: increasing the dissociation of MEK from RAF or causing MEK to adapt a conformation in which it cannot be phosphorylated by RAF. Designing more effective compounds for treating cancer requires understanding the mechanism of drug action in the appropriate genetic context.
Acknowledgments
This work has been funded by the National Institutes of Health (P01CA129243-06 to N.R. and S.L.); Stand-up to cancer (SU2C) melanoma grant, the Experimental Therapeutics Center of Memorial Sloan-Kettering Cancer Center, Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research (to N.R.); the Conquer Cancer Foundation Young Investigator Award and the American Association of Cancer Institutes translational fellowship (to P.L.) and the German Research Foundation (#2278/1-1 to A.S.). The authors are grateful to Tona Gilmer for providing trametinib, Nick Socci, Geulah Livshits and Eusebio Manchado-Robles for help with the interpretation of the RNAi screen, to Takaaki Fukami for helping with the structural analysis and to Kiyoaki Sakata, Toshihiko Fujii and Toshihiro Aoki for their help with viability assays and synthesis of CH5126766.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alessi DR, Saito Y, Campbell DG, Cohen P, Sithanandam G, Rapp U, Ashworth A, Marshall CJ, Cowley S. Identification of the sites in MAP kinase kinase-1 phosphorylated by p74raf-1. The EMBO journal. 1994;13:1610–1619. doi: 10.1002/j.1460-2075.1994.tb06424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, Baccarini M, Barbacid M. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, Hirth P. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews Drug discovery. 2012;11:873–886. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang A, Frost JA, Cobb MH. The MEK1 proline-rich insert is required for efficient activation of the mitogen-activated protein kinases ERK1 and ERK2 in mammalian cells. The Journal of biological chemistry. 1998;273:19909–19913. doi: 10.1074/jbc.273.31.19909. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Dong C, Waters SB, Holt KH, Pessin JE. SOS phosphorylation and disassociation of the Grb2-SOS complex by the ERK and JNK signaling pathways. The Journal of biological chemistry. 1996;271:6328–6332. doi: 10.1074/jbc.271.11.6328. [DOI] [PubMed] [Google Scholar]
- Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, Conrads TP, Veenstra TD, Lu KP, Morrison DK. Regulation of Raf-1 by direct feedback phosphorylation. Molecular cell. 2005;17:215–224. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- Douville E, Downward J. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene. 1997;15:373–383. doi: 10.1038/sj.onc.1201214. [DOI] [PubMed] [Google Scholar]
- Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, Chresta C, McCormack R, Byrne N, Cockerill M, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, Bastian BC, Springer C, Marais R. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- Fellmann C, Zuber J, McJunkin K, Chang K, Malone CD, Dickins RA, Xu Q, Hengartner MO, Elledge SJ, Hannon GJ, et al. Functional identification of optimized RNAi triggers using a massively parallel sensor assay. Molecular cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. The New England journal of medicine. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, Rominger CM, Erskine S, Fisher KE, Yang J, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer cell. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, Hewitt JF, Zak M, Peck A, Orr C, et al. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF-driven cancers. Nature. 2013 doi: 10.1038/nature12441. [DOI] [PubMed] [Google Scholar]
- Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, et al. Kinase-Dead BRAF and Oncogenic RAS Cooperate to Drive Tumor Progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Harada N, Joseph EW, Ohara K, Miura T, Sakamoto H, Matsuda Y, Tomii Y, Tachibana-Kondo Y, Iikura H, et al. Enhanced Inhibition of ERK Signaling by a Novel Allosteric MEK Inhibitor, CH5126766, That Suppresses Feedback Reactivation of RAF Activity. Cancer Res. 2013;73:4050–4060. doi: 10.1158/0008-5472.CAN-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph EW, Pratilas CA, Poulikakos PI, Tadi M, Wang W, Taylor BS, Halilovic E, Persaud Y, Xing F, Viale A, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth FA, Frese KK, DeNicola GM, Baccarini M, Tuveson DA. C-Raf is required for the initiation of lung cancer by K-Ras(G12D) Cancer discovery. 2011;1:128–136. doi: 10.1158/2159-8290.CD-10-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W, Heidecker G, Lloyd P, Rapp UR. Raf-1 protein kinase is required for growth of induced NIH/3T3 cells. Nature. 1991;349:426–428. doi: 10.1038/349426a0. [DOI] [PubMed] [Google Scholar]
- Lito P, Pratilas CA, Joseph EW, Tadi M, Halilovic E, Zubrowski M, Huang A, Wong WL, Callahan MK, Merghoub T, et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer cell. 2012;22:668–682. doi: 10.1016/j.ccr.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCubrey JA, Steelman LS, Abrams SL, Chappell WH, Russo S, Ove R, Milella M, Tafuri A, Lunghi P, Bonati A, et al. Emerging MEK inhibitors. Expert opinion on emerging drugs. 2010;15:203–223. doi: 10.1517/14728210903282760. [DOI] [PubMed] [Google Scholar]
- Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nature structural & molecular biology. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Hanrahan AJ, Halilovic E, Persaud Y, Soh J, Chitale D, Shigematsu H, Yamamoto H, Sawai A, Janakiraman M, et al. Genetic predictors of MEK dependence in non-small cell lung cancer. Cancer Res. 2008;68:9375–9383. doi: 10.1158/0008-5472.CAN-08-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, Rosen N. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nature reviews Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Sebolt-Leopold JS, Dudley DT, Herrera R, Van Becelaere K, Wiland A, Gowan RC, Tecle H, Barrett SD, Bridges A, Przybranowski S, et al. Blockade of the MAP kinase pathway suppresses growth of colon tumors in vivo. Nature medicine. 1999;5:810–816. doi: 10.1038/10533. [DOI] [PubMed] [Google Scholar]
- Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nature reviews Molecular cell biology. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith JA, Thompson IR, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic acids research. 2013;41:D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nature biotechnology. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.