Abstract
Salivary adenoid cystic carcinoma (SACC) is a common type of salivary gland cancer. The poor long-term prognosis of patients with SACC is primarily due to local recurrence, distant metastasis and perineural invasion. MicroRNAs (miRNAs) have been identified as important post-transcriptional regulators, which are involved in various biological processes. The aim of the present study was to identify the miRNA expression profiles that are involved in the metastatic progression of SACC. Therefore, microarray technology was employed to identify miRNA expression profiles in an SACC cell line, ACC-2, and a highly metastatic SACC cell line, ACC-M, which was screened from ACC-2 by a combination of in vivo selection and cloning in vitro. Differences in miRNA expression were assessed by quantitative polymerase chain reaction (qPCR) assay. In addition, the potential target genes that are regulated by selected miRNAs were analyzed by various target prediction tools. The microarray data revealed that the levels of 38 miRNAs significantly differed between the ACC-M cells and the control ACC-2 cells. Six miRNAs (miR-4487, -4430, -486-3p, -5191, -3131 and -211-3p) were selected to validate the microarray data via qPCR. The expression of two miRNAs (miR-4487 and -4430) was significantly upregulated in the ACC-M cells, while the expression of two other miRNAs (miR-5191 and -3131) was significantly downregulated in the ACC-M cells. The potential target genes that were identified to be controlled by the six selected miRNAs were divided into four groups according to function, as follows: Apoptosis and proliferation (46 genes), cell cycle (30 genes), DNA damage and repair (24 genes) and signaling pathway (30 genes). The identification of microRNA expression profiles in highly metastatic SACC cells may provide an improved understanding of the mechanisms involved in metastatic progression, which would aid in the development of novel strategies for the treatment of SACC.
Keywords: microRNA profile, adenoid cystic carcinoma, metastasis
Introduction
Salivary adenoid cystic carcinoma (SACC) is a frequent subtype of salivary gland malignancy accounting for 25% of malignant tumors in the major salivary glands (1) and 50% in the minor glands (2). The neoplasm is characterized by heterogeneous phenotypic features and persistently progressive biological behavior. Although various treatment options have been extensively investigated, the poor long-term prognosis for patients with SACC is primarily due to local recurrence associated with perineural invasion (PNI) and delayed onset of distant metastasis, particularly to the lungs (3,4).
Therefore, it is necessary to identify and understand the diverse mechanisms behind metastasis so as to improve the treatment strategies for SACC patients. Genomic and proteomic studies have yielded various novel insights into molecular targets and the mechanisms of SACC metastasis (5,6). A number of mechanisms underlying the distant metastasis of SACC have been described, including the Notch gene family, matrix metalloproteinases, nerve growth factor, and vascular endothelial growth factor (7–12). However, the mechanisms of distant metastasis are complicated and remain obscure; therefore, further elucidation is required.
The changes in gene expression that accompany metastasis include DNA, mRNA and protein levels, however, the changes in the levels of mRNA and the encoded proteins are often disproportionate. This may be due to a number of reasons, among which post-transcriptional regulation by microRNAs (miRNAs) is particularly important. miRNAs are small non-coding RNAs of 21–25 nt that negatively modulate protein expression (13). miRNAs are involved in various biological processes, including development, proliferation, apoptosis and differentiation (14,15). In addition, studies have demonstrated that aberrant microRNA expression is correlated with malignant transformation and tumor development (16,17).
Thus, it was hypothesized that miRNAs may be significant in mediating metastatic progression in SACC. In order to investigate this hypothesis, a microRNA array was employed to detect a distinctive miRNA expression pattern between an SACC cell line, ACC-2, and a highly metastatic SACC cell line, ACC-M, which was screened from ACC-2 by a combination of in vivo selection and cloning in vitro (18). Since ACC-2 and ACC-M cells have an identical genetic background (with the exception of different metastatic behavior), it was presumed that differentially expressed miRNAs were metastasis-related miRNAs, which are involved directly or indirectly in the progression of metastasis. The results of the miRNA microarray analysis were further verified by quantitative polymerase chain reaction (qPCR). In addition, bioinformatic methods were employed in the analysis of the miRNA expression arrays to identify the target genes, which are regulated by miRNAs, and analyze the gene functions that are associated with the metastasis of tumors. The miRNA signature of metastatic progression may aid with the development of novel therapeutic strategies for SACC patients.
Materials and methods
Cell lines and cell culture condition
The cell lines, ACC-2 and ACC-M, were provided by the Department of Oral Biology, School of Stomatology, Fourth Military Medical University (Xi’an, China). The two types of cells were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Life Technologies), 2.05 mM L-glutamine, 100 μg/ml penicillin and 100 μg/ml streptomycin at 37°C with 5% CO2.
miRNA microarray
Total RNA from the SACC cell line, ACC-2 and the highly metastatic SACC cell line, ACC-M was isolated using TRIzol reagent (Life Technologies). The quality and quantity of the RNA samples was assessed by standard electrophoresis and ultraviolet (UV) spectrophotometry methods (19). The miRNA microarray analysis was performed using the Human miRNA OneArray v4 (Phalanx Biotech Group, Hsinchu, Taiwan), which contains 1,884 unique microRNA probes that are complementary to human microRNA sequences (Sanger miRBase Release 18.0). The data from the microarray were collected and analyzed in accordance with the Minimum Information About a Microarray Experiment guidelines (20). The differential miRNA expression was determined using a two-tailed t-test on a single miRNA basis. The signals were considered to be different when P<0.01 and these miRNAs were subsequently selected for cluster analysis. The miRNA microarray analyses were performed in duplicate and the miRNAs that exhibited common differential expression levels were selected.
qPCR analysis of miRNA expression
qPCR assays were performed on two samples. Using the TaqMan MicroRNA assay kit (Life Technologies), which utilizes miRNA-specific primers, each RNA sample (10 μg) was reverse transcribed according to the manufacturer’s instructions. The resulting cDNA was semi-quantitatively amplified in 45 cycles on an ABI 7500 Real-Time PCR system, using TaqMan® Universal PCR Master Mix, No AmpErase® UNG and TaqMan MicroRNA assays for hsa-miR-4487, hsa-miR-4430, hsa-miR-486-3p, hsa-miR-5191, hsa-miR-3131, hsa-miR-211-3p and U6 snRNA (all Applied Biosystems, Carlsbad, CA, USA). These miRNAs were selected as they exhibited the greatest fold change (hsa-miR-4487 and hsa-miR-4430 were upregulated, and hsa-miR-5191 and hsa-miR-3131 were downregulated), or were of potential importance as indicated by previous studies (hsa-miR-486-3p was upregulated and hsa-miR-211-3p was downregulated). Each qPCR assay was performed at least three times (21,22).
miRNA target prediction
Candidate miRNAs that were selected for the qPCR analysis were utilized for target prediction. The potential miRNA targets among the genes, which were negatively correlated with miRNA expression, were determined using the publicly available TargetScan (http://www.targetscan.org), miRBase (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5) and PicTar (http://pictar.mdc-berlin.de) algorithms. The genes that were predicted as candidate miRNA targets and those that were selected on the basis of Gene Ontology (http://www.geneontology.org) were compared, and the genes that appeared in the two lists were included in the present study.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA). All numerical data from the qPCR assays were analyzed following the derivation of standard curves for each miRNA of interest. Statistical analysis was performed with Student’s t-test and P<0.05 was considered to indicate a statistically significant difference. Each experiment was performed in triplicate and each individual sample was run in triplicate.
Results
Analysis of the quality of RNA isolated from ACC-2 and ACC-M cells
The quality of total RNA was analyzed via denaturing agarose gel electrophoresis and UV spectrophotometry (UV-2600; Shimadzu Corporation, Kyoto, Japan). RNA that was isolated from the ACC-2 and ACC-M cells exhibited clear bands of 28S rRNA and 18S rRNA (Fig. 1). The OD260/OD280 values of each sample were 2.05 and 2.06, respectively, and the OD260/OD230 values of each sample were 2.05 and 2.17, respectively, as determined by the UV spectrophotometer. The RNA quality was confirmed by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) prior to further hybridization. All these results indicated that the obtained high quality RNA was suitable for microRNA microarray analysis and the following qPCR analysis to verify the results of the microarray.
Figure 1.
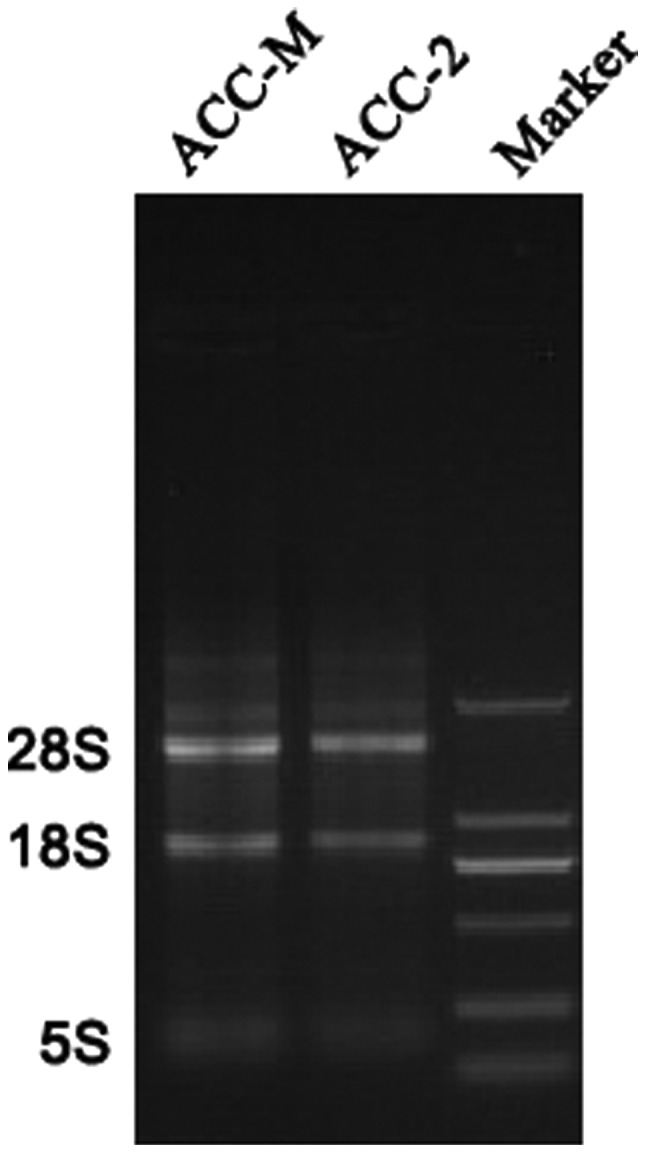
Denaturing agarose gel electrophoresis of RNA isolated from the cell samples. There are three clear bands (28S, 18S and 5S). The brightness of the 28S band was approximately twice that of the 18S bank, suggestive of high quality RNA. ACC-M was screened from ACC-2 by a combination of in vivo selection and in vitro cloning. The size of marker bands are 100, 250, 500, 750, 1,000 and 2,000bp from the bottom to top.
miRNA expression profiles in the SACC cell line, ACC-2 and the highly metastatic SACC cell line, ACC-M
To identify the changes in the miRNA expression profiles between the SACC cell line, ACC-2 and the highly metastatic SACC cell line, ACC-M, miRNA microarray analysis was conducted. The miRNA expression profile of the two SACC cell lines revealed that 38 out of 1,884 human miRNAs were differentially expressed between the ACC-2 and ACC-M cell lines. Of the 38 miRNAs identified as differentially expressed, 20 were upregulated (Table I) and 18 were downregulated (Table II) in the ACC-M cell line compared with the control ACC-2 cell line. In the average fold-change analysis, 17 of the 38 flagged miRNAs exhibited >2-fold change in expression level, while there were 21 miRNAs that exhibited <2-fold change in expression level (Table I and II).
Table I.
miRNAs upregulated in the ACC-M cell line compared with the control ACC-2 cell line.
| Phalanx ID | Name | Average fold change |
|---|---|---|
| PH_mr_0004751 | hsa-miR-4487a | 32.24 |
| PH_mr_0004716 | hsa-miR-4430a | 4.05 |
| PH_mr_0004876 | hsa-miR-5096a | 3.61 |
| PH_mr_0000840 | hsa-miR-1285-3pa | 2.85 |
| PH_mr_0008030 | hsa-miR-1273g-3pa | 2.60 |
| PH_mr_0004900 | hsa-miR-3150b-3pa | 2.59 |
| PH_mr_0004874 | hsa-miR-1273fa | 2.46 |
| PH_mr_0001931 | hsa-miR-1273aa | 2.43 |
| PH_mr_0004658 | hsa-miR-1273ea | 2.25 |
| PH_mr_0000379 | hsa-miR-574-5p | 1.98 |
| PH_mr_0004873 | hsa-miR-5095 | 1.88 |
| PH_mr_0004549 | hsa-miR-4638-5p | 1.88 |
| PH_mr_0004565 | hsa-miR-4688 | 1.87 |
| PH_mr_0001461 | hsa-miR-877-5p | 1.86 |
| PH_mr_0004257 | hsa-miR-3154 | 1.82 |
| PH_mr_0003162 | hsa-miR-766-3p | 1.80 |
| PH_mr_0004018 | hsa-miR-1972 | 1.78 |
| PH_mr_0000953 | hsa-miR-486-3p | 1.78 |
| PH_mr_0004577 | hsa-miR-4728-5p | 1.74 |
| PH_mr_0004187 | hsa-miR-1976 | 1.74 |
miRNAs exhibiting >2-fold expression change compared with ACC-2.
miRNA, microRNA.
Table II.
miRNAs downregulated in the ACC-M cell line compared with the control ACC-2 cell line.
| Phalanx ID | Name | Average fold change |
|---|---|---|
| PH_mr_0008120 | hsa-miR-5191a | 0.028 |
| PH_mr_0004035 | hsa-miR-3131a | 0.22 |
| PH_mr_0004331 | hsa-miR-4278a | 0.26 |
| PH_mr_0004534 | hsa-miR-4498a | 0.39 |
| PH_mr_0008014 | hsa-miR-211-3pa | 0.40 |
| PH_mr_0004731 | hsa-miR-4450a | 0.41 |
| PH_mr_0000102 | hsa-miR-373-5pa | 0.44 |
| PH_mr_0000010 | hsa-miR-7-5pa | 0.48 |
| PH_mr_0008044 | hsa-miR-5010-5p | 0.52 |
| PH_mr_0004714 | hsa-miR-4428 | 0.53 |
| PH_mr_0001673 | hsa-miR-18b-5p | 0.53 |
| PH_mr_0001728 | hsa-miR-18a-5p | 0.53 |
| PH_mr_0004543 | hsa-miR-1587 | 0.54 |
| PH_mr_0001696 | hsa-miR-20a-5p | 0.55 |
| PH_mr_0000136 | hsa-miR-1265 | 0.55 |
| PH_mr_0000436 | hsa-miR-92a-3p | 0.56 |
| PH_mr_0000747 | hsa-miR-186-5p | 0.57 |
| PH_mr_0000812 | hsa-miR-92b-3p | 0.57 |
miRNAs exhibiting less than half of the expression levels of ACC-2 cells.
miRNA, microRNA.
Confirmation of miRNA microarray data by qPCR
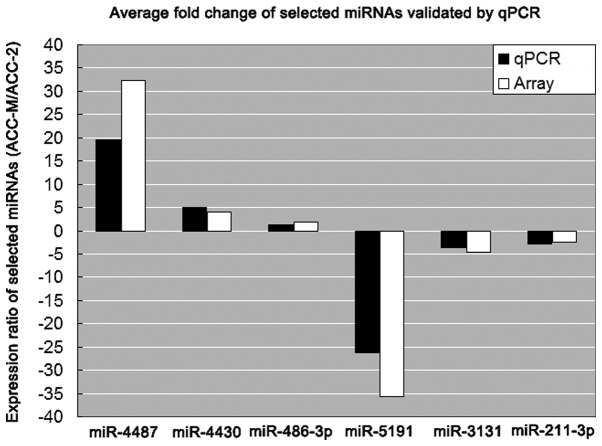
To confirm the miRNA microarray data, a TaqMan MicroRNA assay kit was used to perform qPCR analyses of the expression levels of miR-4487, -4430, -486-3p, -5191, -3131 and -211-3p in the ACC-2 and ACC-M cell lines. It is generally accepted that gene expression levels should be normalized by a carefully selected, stable internal control gene. To validate the assumption of stable expression of a given control gene, prior knowledge of a reliable measure to normalize this gene, in order to eliminate any non-specific variation, is required. For each sample, the expression values were normalized to the U6 snRNA gene (a stable housekeeping gene) and the expression levels relative to the ACC-2 cell line were calculated. The expression pattern that was identified in the arrays was confirmed by this additional analysis. Compared with the ACC-2 cells, three miRNAs (miR-4487, -4430 and -486-3p) were significantly upregulated, while three miRNAs (miR-5191, -3131 and -211-3p) were significantly downregulated in the highly metastatic SACC cell line, ACC-M. For these candidate miRNAs, the qPCR analysis revealed similar patterns of up- or downregulation to those revealed by the results of the microarray analysis (Table III; Fig. 2), although the magnitude of the reported changes in expression differed. Thus, miRNA expression profiles clearly exhibit significant differences between ACC-2 and ACC-M, indicating that miRNAs are significant during the metastatic progression of SACC.
Table III.
Validation of microarray analysis data by qPCR for candidate microRNAs.
| Average expression ratio | |||
|---|---|---|---|
|
|
|||
| Phalanx ID | Name | qPCR | Array |
| PH_mr_0004751 | hsa-miR-4487 | 19.560 | 32.240 |
| PH_mr_0004716 | hsa-miR-4430 | 4.960 | 4.050 |
| PH_mr_0000953 | hsa-miR-486-3p | 1.210 | 1.780 |
| PH_mr_0008120 | hsa-miR-5191 | 0.038 | 0.028 |
| PH_mr_0004035 | hsa-miR-3131 | 0.280 | 0.220 |
| PH_mr_0008014 | hsa-miR-211-3p | 0.340 | 0.400 |
All data were confirmed by qPCR. qPCR, quantitative polymerase chain reaction.
Figure 2.
Validation of microarray analysis data by qPCR for candidate miRNAs associated with metastatic progression of ACC-M cells. The expression levels of six miRNAs (miR-4487, -4430, -486-3p, -5191, -3131 and -211-3p) were determined by qPCR. Each qPCR assay was performed at least three times. miRNA, microRNA; qPCR, quantitative polymerase chain reaction.
Prediction of miRNA targets
The identification of potential target genes of miRNAs that are associated with the metastatic progression of SACC is essential to investigate their biological functions. Previous studies have shown that miRNAs are able to regulate expression of their target genes by decreasing mRNA stability in addition to translational inhibition (23). The candidate miRNAs, which were used for qPCR, were selected for target prediction and analyzed with the following three target prediction tools: TargetScan 6.2 (Whitehead Institute for Biomedical Research website; http://www.targetscan.org); miRBase Targets Release v5 (the Enright Group; http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5); and PicTar (http://pictar.mdc-berlin.de). By analysis of these databases, human genes, which were known to be involved in cell proliferation, apoptosis, cell cycle, DNA damage, DNA repair and signaling pathways were selected from the Gene Ontology website (http://www.ebi.ac.uk/GOA). Those genes that were predicted to be targets of the candidate miRNAs were selected and are listed in Table IV. These data may provide the foundation for further analysis of the mechanisms of metastatic progression in SACC.
Table IV.
Predicted target genes of miRNAs.
| Putative targets of miRNAs according to function | ||||
|---|---|---|---|---|
|
|
||||
| miRNA name | Apoptosis and proliferation | Cell cycle | DNA damage and repair | Signaling pathways |
| hsa-miR-4487 | GIPC3, URGCP, ITGA5, VASH1, FGD1, HECA, FLT1 | GALNTL2, NAP1L5, ZNF740, TBC1D4, CDKN1A | PSMA5, HIPK2, USH1G, BOLL | EFNB3, SORT1, RORC, FMOD, FAM123C, CUL3, CXCR3, WNT4 |
| hsa-miR-4430 | MAP3K9, MAP2K7, SHROOM4, KRT85, MNT, GLI2, IREB2 | SEZ6, GRIN3A, MOCS1, PARP16 | POMT2, RPP14, ZDHHC22 | POP4, EIF2C1, POFUT1, EFNB3, CD72, LPHN2, EIF2C3 |
| hsa-miR-486-3p | NAT15, GDI1, DMBX1, CNP, PAK6, CD276, MAP4, VASH1, PLA2G6, SPTBN4, SAP30BP, COL4A2, HS6ST1, DCAF7, TANC2 | SF3A1, ATXN7L3, SDC3, TTYH3, STX1B, GATAD2B | FYCO1, STIM1, CACNA2D2 | FLOT2, CTNNBIP1, MARK2, MAPKBP1, LOC388630 |
| hsa-miR-5191 | FOXL1, AHI1, PERP | MAP1B, CDH12, ABCA9, WWP2, HOXC6, EIF4E, CD1D | PPEF2, ZNF547, RANBP10, FLG2, NIPA2, PARP9, ZNF32, ZFP28 | RBPJL, GNB1, GNB4, SOX9, YWHAZ, NOTCH2 |
| hsa-miR-3131 | SEMA6C, MEF2D, TMEM119, TRAF3, EIF4G1 | RIMBP2, SYN1, FBXO22 | NFIX | PPARGC1B |
| hsa-miR-211-3p | NFASC, SEMA5A, DMC1, MAFG, PITPNA, ERLIN2, PDXK, SMARCD1, SOCS5, TNFAIP1 | MAU2, PDS5A, NOS1, ATP11A, XPO4 | SMCR7L, ZNF217, OBFC1, RBFOX2, ATG10 | PIK3CG, LONP2, PPP2R5E |
miRNA, microRNA.
Discussion
SACC is a common malignant tumor that arises from the secretory epithelial cells in the salivary glands of the head and neck. SACC accounts for 1% of all head and neck malignancies and 10% of all salivary neoplasms (24). Although SACC tend to grow slowly, this neoplasm has a poor prognosis owing to its insidious invasion into adjacent tissues and hematogenous spread to distant organs (lungs, bone and liver) (24,25). The five-year disease-free survival rate is ≤90%, however, this reduces to 40% after 15 years (27). Advanced tumor stage, distant metastasis to the lungs, PNI, and solid subtype are among the clinicopathological parameters that have been proposed as useful predictors of the clinical course of SACC (28). However, accurate prediction is often challenging, and in-depth studies on invasion and metastatic mechanisms are of great significance for prognosis and evaluation, and the selection of appropriate treatment protocols.
miRNAs are short non-coding RNAs of 21–25 nt that regulate gene expression in multicellular organisms. Numerous studies have indicated that miRNAs are significant in carcinogenesis and tumor progression in certain types of cancer, including SACC (29–31). However, to the best of our knowledge, the altered expression of miRNAs, which is associated with metastatic progression in SACC, has not been investigated. Therefore, in the present study, high-throughput miRNA microarray technology, with 1,884 miRNAs selected from the miRBase, was used to detect miRNA expression in parental ACC-2 cells and highly metastatic ACC-M cells. The microarray data revealed that 38 miRNAs exhibited significant differences in expression between ACC-2 and ACC-M cell lines; 20 were upregulated and 18 were downregulated in the ACC-M cells. In the present analysis, 17 miRNAs in the ACC-M cells exhibited >2-fold changes compared with the control cells (ACC-2). Although not all miRNAs in ACC-M cells exhibit similar regulation patterns, numerous miRNAs with ≥2-fold expression changes were identified in the present study for further investigation. Therefore, to confirm the expression of the six miRNAs that were identified in the microarray analysis, a qPCR assay was performed. The results of the qPCR analysis of the upregulated (miR-4487, -4430 and -486-3p) and downregulated (miR-5191, -3131 and -211-3p) miRNAs are demonstrated in Fig. 2. For all six miRNAs, the qPCR experiment confirmed their down- or upregulation, however, the magnitude of the changes in the expression levels was found to differ between the microarray and qPCR analyses.
Thus far, to the best of our knowledge, there have been no reports of a correlation between the six miRNAs discussed in the present study and the metastatic progression of SACC. To elucidate the regulatory mechanisms of those miRNAs, which are associated with the metastatic progression of SACC, various bioinformatic methods were employed to identify potential target genes of those miRNAs. The target genes, which have been reported to be associated with the metastatic progression of human tumors, were divided into four groups according to function (as described by the Gene Ontology website, http://www.geneontology.org): Apoptosis and proliferation (46 genes); cell cycle (30 genes); DNA damage and repair (24 genes); and signaling pathway (30 genes; Table IV).
Computational predictions indicate that one miRNA is able to target numerous mRNAs and each mRNA can also be targeted by numerous miRNAs. Thus, a one-to-one association between miRNAs and target genes should not be expected, rather a one-to-many or many-to-one association. Previous studies have demonstrated that functional miRNA binding sites can lie in the coding or 5′-untranslated regions of endogenous mRNAs (31). Therefore, the number of target genes that are regulated by miRNAs may be larger than what was identified by the bioinformatic methods used in the present study. Although the expression levels of miRNAs may demonstrate inverse correlations with the protein expression levels of their potential target genes, further studies are required to confirm a regulatory association between miRNAs and the predicted target genes.
In conclusion, SACC is a common subtype of salivary gland malignancy, which exhibits biological behavior that facilitates hematogenous spread to lungs. In the present study, the experimental data demonstrated that alteration of the miRNA expression profile in the ACC-M cell line may be associated with metastatic progression. These findings may aid with elucidating the potential mechanisms that underlie the metastatic progression of SACC and provide a novel molecular therapeutic target for the treatment of SACC.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81102051) and the Natural Science Foundation of Jiangsu Province (grant no. BK2011659).
Abbreviations
- SACC
salivary adenoid cystic carcinoma
- miRNA
microRNA
- PNI
perineural invasion
- qPCR
quantitative PCR
References
- 1.Renehan A, Gleave EN, Hancock BD, Smith P, McGurk M. Long-term follow-up of over 1000 patients with salivary gland tumours treated in a single centre. Br J Surg. 1996;83:1750–1754. doi: 10.1002/bjs.1800831228. [DOI] [PubMed] [Google Scholar]
- 2.Anderson JN, Jr, Beenken SW, Crowe R, Soong SJ, Peters G, Maddox WA, Urist MM. Prognostic factors in minor salivary gland cancer. Head Neck. 1995;17:480–486. doi: 10.1002/hed.2880170605. [DOI] [PubMed] [Google Scholar]
- 3.van der Wal JE, Becking AG, Snow GB, van der Waal I. Distant metastases of adenoid cystic carcinoma of the salivary glands and the value of diagnostic examinations during follow-up. Head Neck. 2002;24:779–783. doi: 10.1002/hed.10126. [DOI] [PubMed] [Google Scholar]
- 4.Ramer N, Wu H, Sabo E, Ramer Y, Emanuel P, Orta L, Burstein DE. Prognostic value of quantitative p63 immunostaining in adenoid cystic carcinoma of salivary gland assessed by computerized image analysis. Cancer. 2010;116:77–83. doi: 10.1002/cncr.24657. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Zhang HL, Shao XJ, Jiang YG, Zhao XG, Gao X, Li JH, Yang J, Zhang YF, Liu BL, Sun MY. Gene expression profile of salivary adenoid cystic carcinoma associated with perineural invasion. Tohoku J Exp Med. 2007;212:319–334. doi: 10.1620/tjem.212.319. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima D, Uzawa K, Kasamatsu A, Koike H, Endo Y, Saito K, Hashitani S, Numata T, Urade M, Tanzawa H. Protein expression profiling identifies maspin and stathmin as potential biomarkers of adenoid cystic carcinoma of the salivary glands. Int J Cancer. 2006;118:704–713. doi: 10.1002/ijc.21318. [DOI] [PubMed] [Google Scholar]
- 7.Chen W, Zhang H, Wang J, Cao G, Dong Z, Su H, Zhou X, Zhang S. Lentiviral-mediated gene silencing of Notch-4 inhibits in vitro proliferation and perineural invasion of ACC-M cells. Oncol Rep. 2013;29:1794–1804. doi: 10.3892/or.2013.2317. [DOI] [PubMed] [Google Scholar]
- 8.Ding LC, She L, Zheng DL, Huang QL, Wang JF, Zheng FF, Lu YG. Notch-4 contributes to the metastasis of salivary adenoid cystic carcinoma. Oncol Rep. 2010;24:363–368. doi: 10.3892/or_00000868. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Zhang P, Ma Q, Kong L, Li Y, Liu B, Lei D. EMMPRIN silencing inhibits proliferation and perineural invasion of human salivary adenoid cystic carcinoma cells in vitro and in vivo. Cancer Biol Ther. 2012;13:85–91. doi: 10.4161/cbt.13.2.18455. [DOI] [PubMed] [Google Scholar]
- 10.Luukkaa H, Klemi P, Hirsimäki P, Vahlberg T, Kivisaari A, Kähäri VM, Grénman R. Matrix metalloproteinase (MMP)-1, -9 and -13 as prognostic factors in salivary gland cancer. Acta Otolaryngol. 2008;128:482–490. doi: 10.1080/00016480801922895. [DOI] [PubMed] [Google Scholar]
- 11.Hao L, Xiao-lin N, Qi C, Yi-ping Y, Jia-quan L, Yan-ning L. Nerve growth factor and vascular endothelial growth factor: retrospective analysis of 63 patients with salivary adenoid cystic carcinoma. Int J Oral Sci. 2010;2:35–44. doi: 10.4248/IJOS10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa AF, Tasso MG, Mariano FV, Soares AB, Chone CT, Crespo AN, Fresno MF, Llorente JL, Suárez C, de Araújo VC, Hermsen M, Altemani A. Levels and patterns of expression of hypoxia-inducible factor-1α, vascular endothelial growth factor, glucose transporter-1 and CD105 in adenoid cystic carcinomas with high-grade transformation. Histopathology. 2012;60:816–825. doi: 10.1111/j.1365-2559.2011.04128.x. [DOI] [PubMed] [Google Scholar]
- 13.Chekanova JA, Belostotsky DA. MicroRNAs and messenger RNA turnover. Methods Mol Biol. 2006;342:73–85. doi: 10.1385/1-59745-123-1:73. [DOI] [PubMed] [Google Scholar]
- 14.Lages E, Ipas H, Guttin A, Nesr H, Berger F, Issartel JP. MicroRNAs: molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–2540. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Hashimi SM, Good DA, Cao S, Duan W, Plummer PN, Mellick AS, Wei MQ. Apoptosis and microRNA aberrations in cancer. Clin Exp Pharmacol Physiol. 2012;39:739–46. doi: 10.1111/j.1440-1681.2012.05700.x. [DOI] [PubMed] [Google Scholar]
- 16.Nishizawa T, Suzuki H. The role of microRNA in gastric malignancy. Int J Mol Sci. 2013;14:9487–9496. doi: 10.3390/ijms14059487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761–5771. doi: 10.1038/onc.2010.352. [DOI] [PubMed] [Google Scholar]
- 18.Guan XF, Qiu WL, He RG, Zhou XJ. Selection of adenoid cystic carcinoma cell clone highly metastatic to the lung: an experimental study. Int J Oral Maxillofac Surg. 1997;26:116–119. doi: 10.1016/s0901-5027(05)80830-6. [DOI] [PubMed] [Google Scholar]
- 19.Vela J, Vitorica J, Ruano D. Rapid PCR-mediated synthesis of competitor molecules for accurate quantification of beta(2) GABA(A) receptor subunit mRNA. Brain Res Brain Res Protoc. 2001;8:184–190. doi: 10.1016/s1385-299x(01)00109-x. [DOI] [PubMed] [Google Scholar]
- 20.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 21.Liu SG, Qin XG, Zhao BS, Qi B, Yao WJ, Wang TY, Li HC, Wu XN. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett. 2013;5:1639–1642. doi: 10.3892/ol.2013.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, Kreis S. New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion. PLoS One. 2013;8:e73473. doi: 10.1371/journal.pone.0073473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikitina EG, Urazova LN, Stegny VN. MicroRNAs and human cancer. Exp Oncol. 2012;34:2–8. [PubMed] [Google Scholar]
- 24.Matsuba HM, Spector GJ, Thawley SE, Simpson JR, Mauney M, Pikul FJ. Adenoid cystic salivary gland carcinoma. A histopathologic review of treatment failure patterns. Cancer. 1986;57:519–524. doi: 10.1002/1097-0142(19860201)57:3<519::aid-cncr2820570319>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 25.Kim KH, Sung MW, Chung PS, Rhee CS, Park CI, Kim WH. Adenoid cystic carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 1994;120:721–726. doi: 10.1001/archotol.1994.01880310027006. [DOI] [PubMed] [Google Scholar]
- 26.Wiseman SM, Popat SR, Rigual NR, Hicks WL, Jr, Orner JB, Wein RO, McGary CT, Loree TR. Adenoid cystic carcinoma of the paranasal sinuses or nasal cavity: a 40-year review of 35 cases. Ear Nose Throat J. 2002;81:510–517. [PubMed] [Google Scholar]
- 27.Fordice J, Kershaw C, El-Naggar A, Goepfert H. Adenoid cystic carcinoma of the head and neck: predictors of morbidity and mortality. Arch Otolaryngol Head Neck Surg. 1999;125:149–152. doi: 10.1001/archotol.125.2.149. [DOI] [PubMed] [Google Scholar]
- 28.Sequeiros-Santiago G, García-Carracedo D, Fresno MF, Suarez C, Rodrigo JP, Gonzalez MV. Oncogene amplification pattern in adenoid cystic carcinoma of the salivary glands. Oncol Rep. 2009;21:1215–1222. doi: 10.3892/or_00000344. [DOI] [PubMed] [Google Scholar]
- 29.He Q, Zhou X, Li S, Jin Y, Chen Z, Chen D, Cai Y, Liu Z, Zhao T, Wang A. MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK-Snai2 pathway. Biochim Biophys Acta. 2013;1830:5258–5266. doi: 10.1016/j.bbagen.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 30.Mitani Y, Roberts DB, Fatani H, Weber RS, Kies MS, Lippman SM, El-Naggar AK. MicroRNA profiling of salivary adenoid cystic carcinoma: association of miR-17–92 upregulation with poor outcome. PLoS One. 2013;8:e66778. doi: 10.1371/journal.pone.0066778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Hu Y, Fu J, Yang X, Zhang Z. MicroRNA155 in the growth and invasion of salivary adenoid cystic carcinoma. J Oral Pathol Med. 2013;42:140–147. doi: 10.1111/j.1600-0714.2012.01189.x. [DOI] [PubMed] [Google Scholar]
- 32.Pinós T, Barbosa-Desongles A, Hurtado A, Santamaria-Martínez A, de Torres I, Reventós J, Munell F. Human SHBG mRNA translation is modulated by alternative 5′-non-coding exons 1A and 1B. PLoS One. 2010;5:e13844. doi: 10.1371/journal.pone.0013844. [DOI] [PMC free article] [PubMed] [Google Scholar]