Abstract
ARID1A has emerged as a tumor suppressor gene, which is mutated in a broad spectrum of cancers, especially in those arising from ectopic or eutopic endometrium. As a subunit of SWI/SNF chromatin remodeler, ARID1A facilitates target-specific binding of SWI/SNF complexes to chromatin, thereby altering the accessibility of chromatin to a variety of nuclear factors. In human cancer, ARID1A possesses not only features of a gatekeeper, regulating cell cycle progression, but also features of a caretaker, preventing genomic instability. An increasing body of evidence suggests crosstalk between ARID1A and PI3K/Akt pathways, and between ARID1A and p53. In this review, we discuss the spectrum of ARID1A alterations in cancers, tumor suppression mechanisms of ARID1A, oncogenic pathways cooperating with ARID1A, and clinical implications of ARID1A mutation.
Keywords: ARID1A, PI3K, SWI/SNF, chromatin remodeling complex, clear cell carcinoma, endometrial cancer, endometrioid carcinoma, gastric cancer, ovarian cancer, tumor suppressor gene
Introduction
Dysregulation of ATP-dependent chromatin remodeling complexes has been implicated in a variety of cancers in an increasing number of studies.1,2 These complexes utilize the energy of ATP hydroxylation to reposition, eject, or exchange nucleosomes, thereby modulating DNA accessibility to other cellular machineries involved in transcription, DNA replication, methylation, and repair.3 Among different ATP-dependent chromatin remodelers, SWitch/Sucrose NonFermentable (SWI/SNF) is the most commonly dysregulated in cancer, and ARID1A (AT-rich interactive domain-containing protein 1A) is the most frequently mutated among all genes encoding subunits of SWI/SNF complexes.4
Recently, ARID1A has been established as a tumor suppressor gene through the discovery of recurrent inactivating ARID1A mutations in a broad spectrum of cancers, and its tumor suppressor role has been supported by functional studies. In this review, we discuss the spectrum of ARID1A alterations in cancers, known biological functions of ARID1A, the possible mechanism how ARID1A suppresses tumor development, and the potential of using ARID1A as a prognostic biomarker.
ARID1A is a Subunit of SWI/SNF Chromatin Remodeler
ARID1A, also known as BAF250a, SMARCF1, or p270, belongs to a family of proteins containing a highly conserved, approximately 100-amino acid DNA binding domain called ARID (AT-rich interacting domain). Although the ARID domains in general preferentially bind AT-rich DNA sequences, the ARID domain of mammalian ARID1A exhibits general DNA binding character without sequence specificity.5,6 The C-terminal domain of ARID1A, presumably important for protein–protein interaction, contains multiple copies of the sequence motif, LXXLL, which has been shown to facilitate the interaction of various proteins with nuclear hormone receptors.7
ARID1A, as well as its paralog ARID1B, associates in a mutually exclusive fashion with several other proteins to form the BRG1-associated factor (BAF) complexes, a subfamily of mammalian SWI/SNF chromatin remodelers. BAF complexes are composed of one of two mutually exclusive ATPase subunits (BRM and BRG1), a set of core subunits (BAF47, BAF155, and BAF170) that augment catalytic activity, and variant subunits that were thought to provide target specificity.8 Mammalian SWI/SNF complexes are essential in regulating gene expression, and have been implicated in several cellular functions, including proliferation, cell fate determination, self-renewal in stem cells, DNA methylation, and damage repair.1,8-10 Like ARID1A, several subunits of SWI/SNF complexes have multiple paralogs, making it possible to form hundreds of different SWI/SNF complex combinations by incorporating different paralogous subunits.11 This combinatorial assembly of SWI/SNF complexes is thought to provide the target and lineage specificity exhibited by different forms of SWI/SNF complexes.12 Indeed, distribution of ARID1A and ARID1B varies in mouse embryonic tissues, and they demonstrate different kinetic patterns during cell cycle progression; ARID1A accumulates in G0/G1 phase and is downregulated in S and G2/M phases, whereas ARID1B expression remains constant throughout all phases.13 Therefore, differential incorporation of ARID1 proteins is likely responsible for directing BAF complexes to various targets, thus contributing to cell cycle regulation in different biological contexts.14
It is thought that ARID1A contributes to targeting of BAF complexes in two ways. First, ARID1A may mediate recruitment of BAF complexes to target DNA regulatory elements through interacting with other transcription (co-)factors. ARID1A has been shown to interact with ligand-bound nuclear hormone receptors and p53 through its C-terminal domain and stimulate transcriptional activity of these transcription factors.15-17 Second, the ARID domain, despite showing no sequence specificity in vitro, may be crucial for improving BAF affinity to chromatin for certain targets in vivo. Chandler et al. demonstrated that ARID–DNA interactions contribute to SWI/SNF activity in mouse embryos in which missense mutation of the ARID domain disrupted ARID1A–DNA interactions. This results in a decrease in promoter occupancy by SWI/SNF, and defects in cardiovascular development.18 Since missense mutations involving the ARID domain are present in human cancers, it raises a possibility that ARID–DNA interactions are essential for the tumor suppressor function of ARID1A and loss of such interaction (due to mutations) abolishes its tumor suppression function.
ARID1A acts as a nucleocytoplasmic protein whose stability depends on its subcellular localization. Nuclear ARID1A is less stable than cytoplasmic ARID1A because the protein is rapidly degraded by the ubiquitin–proteasome system present in the nucleus.19 A naturally occurring in-frame deletion that disrupts the consensus nuclear export signal results in reduced steady-state protein levels of ARID1A due to its retention in the nuclei and subsequent degradation.19 These findings delineate the basic biological mechanism regulating ARID1A subcellular distribution and protein stability.
ARID1A Mutations in Human Cancers
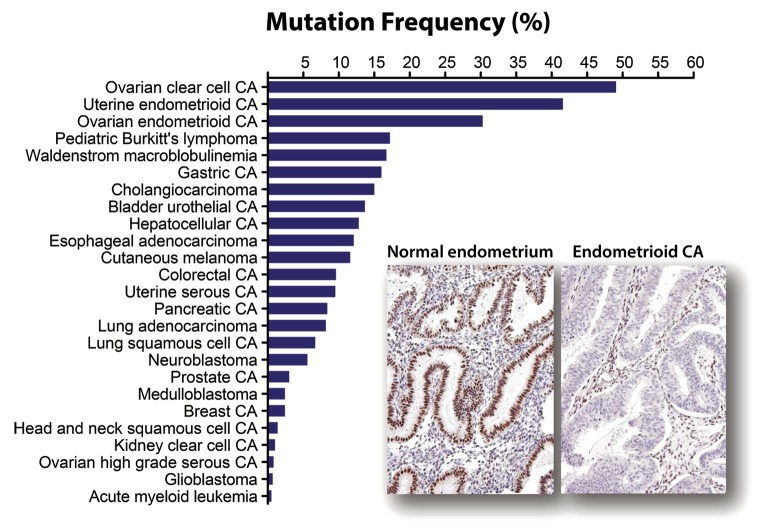
The ARID1A gene maps to chromosome 1p36.11, a region frequently deleted in cancer.20 Initial clues that ARID1A was a tumor suppressor came from expression analyses that showed decreased ARID1A expression in 30% of renal cancer and 10% of breast cancer,21 as well as discovery of ARID1A deletions and rearrangement in a few cancer cell lines.22 Not until the advent of next-generation sequencing did the first concrete evidence that ARID1A was a tumor suppressor gene emerge. Two genome-wide sequencing studies in 2010 discovered frequent loss-of-function somatic mutations in endometriosis-associated ovarian cancers, including ovarian clear cell carcinoma and ovarian endometrioid carcinoma, which harbored ARID1A somatic mutation in 46–57% and 30%, respectively.23,24 Subsequent comprehensive sequencing efforts have reported frequent somatic mutations in ARID1A in other types of cancers in addition to endometriosis-associated neoplasms (Table 1), including uterine endometrioid carcinoma (39~44%),30-32 gastric carcinoma (8~29%),20,43-45 esophageal adenocarcinoma (9~19%),21,40-42 Waldenstrom macroglobulinemia (17%),22,63 pediatric Burkitt lymphoma (17%),23,62,67 hepatocellular carcinoma (10~16%),48-50,67 cholangiocarcinoma (14~15%),32,51,52 and urothelial carcinoma of bladder (12~15%).57-59 These mutations, most of which are deletion or nonsense mutations, are distributed throughout the ARID1A gene. Figure 1 summarizes their mutation frequencies in reported human neoplastic diseases. In addition, studies applying immunohistochemistry have also identified frequent loss of ARID1A expression in several additional tumor types (Table 1) including ovarian endocervical-type mucinous borderline tumor (33%),29 cervical adenocarcinoma (24~31%),38 endometrial clear cell carcinoma (21~26%),33,35-37 endometrial carcinosarcoma (14%),33 and anaplastic thyroid carcinoma (14%).33 Comparative genomic hybridization studies have also detected frequent heterozygous deletions involving ARID1A in pancreatic cancer (36~47%),53,68 breast cancer (13~35%),55,69 and clear cell renal cell carcinoma (16%).70 Overall, the frequency and pattern of ARID1A mutations indicates that ARID1A is altered in a broad spectrum of human cancers.
Table 1. Recurrent mutation and protein loss of ARID1A in cancersa.
| Cancer types | Mutation (%) | Protein loss (%) | References |
|---|---|---|---|
| Ovarian clear cell carcinoma | 24/42 (57%) | N/A | 23 |
| 55/119 (46%) | 55/132 (42%) | 24 | |
| N/A | 88/149 (59%) | 25 | |
| N/A | 35/68 (52%) | 26 | |
| N/A | 40/90 (44%) | 27 | |
| N/A | 34/82 (41%) | 28 | |
| Ovarian endometrioid carcinoma | 10/33 (30%) | 39/125 (31%) | 24 |
| N/A | 62/130 (48%) | 28 | |
| Ovarian endocervical-type mucinous borderline tumor | N/A | 8/24 (33%) | 29 |
| Endometrial endometrioid carcinoma | 82/186 (44%) | N/A | 30 |
| 10/25 (40%) | 15/58 (26%) | 31 | |
| 73/186 (39%) | N/A | 32 | |
| N/A | 73/214 (34%) | 33 | |
| N/A | 27/111 (24%) | 34 | |
| N/A | 84/436 (19%) | 35 | |
| Endometrial serous carcinoma | 4/42 (10%) | N/A | 32 |
| N/A | 17/95 (18%) | 33 | |
| N/A | 1/44 (3%) | 35 | |
| N/A | 0/17 (0%) | 31 | |
| Endometrial clear cell carcinoma | N/A | 6/23 (26%) | 33 |
| N/A | 5/22 (23%) | 36 | |
| N/A | 4/19 (21%) | 35 | |
| N/A | 10/50 (20%) | 37 | |
| Endometrial carcinosarcoma | N/A | 18/127 (14%) | 33 |
| Cervical adenocarcinoma | N/A | 14/45 (31%) | 38 |
| N/A | 6/25 (24%) | 39 | |
| Cervical squamous cell carcinoma | N/A | 19/116 (16%) | 39 |
| N/A | 3/46 (7%) | 38 | |
| Esophageal adenocarcinoma | 10/54 (19%) | N/A | 40 |
| 3/20 (15%) | 12/98 (12%) | 41 | |
| 14/149 (9%) | N/A | 42 | |
| Gastric carcinoma | 32/109 (29%) | 22/109 (20%) | 43 |
| 10/100 (10%) | N/A | 44 | |
| 9/110 (8%) | N/A | 45 | |
| N/A | 26/180 (14%) | 33 | |
| N/A | 5/45 (11%) | 31 | |
| N/A | 94/857 (11%) | 46 | |
| Colorectal carcinoma | 12/119 (10%) | N/A | 44 |
| 21/224 (9%) | N/A | 47 | |
| N/A | 2/49 (4%) | 31 | |
| N/A | 2/250 (1%) | 33 | |
| Hepatocellular carcinoma | 20/125 (16%) | N/A | 48 |
| 14/110 (13%) | N/A | 49 | |
| 15/147 (10%) | N/A | 50 | |
| N/A | 0/41 (0%) | 31 | |
| Cholangiocarcinoma | 32/209 (15%) | N/A | 51 |
| 9/64 (14%) | N/A | 52 | |
| N/A | 2/27 (7%) | 31 | |
| Pancreatic adenocarcinoma | 10/119 (8%) | N/A | 44 |
| 3/36 (8%) | N/A | 53 | |
| Breast carcinoma | 4/114 (4%) | N/A | 44 |
| 11/507 (2%) | N/A | 54 | |
| 0/11 (0%) | 64% (n = 236)b | 55 | |
| N/A | 63/112 (56%)b | 56 | |
| N/A | 11/315 (3%) | 33 | |
| N/A | 1/91 (1%) | 31 | |
| Urothelial carcinoma of bladder | 15/99 (15%) | N/A | 57 |
| 13/97 (13%) | N/A | 58 | |
| 6/52 (12%) | N/A | 59 | |
| Anaplastic thyroid carcinoma | N/A | 5/35 (14%) | 33 |
| Cutaneous melanoma | 14/121 (12%) | N/A | 60 |
| 3/25 (12%) | N/A | 61 | |
| Pediatric Burkitt lymphoma | 5/29 (17%) | N/A | 62 |
| Waldenstrom macroglobulinemia | 5/30 (17%) | N/A | 63 |
| Lung adenocarcinoma | 15/183 (8%) | N/A | 64 |
| Lung squamous cell carcinoma | 12/178 (7%) | N/A | 65 |
| Neuroblastoma | 4/71 (6%) | N/A | 66 |
aOnly cancer types with >5% mutation or protein loss were shown. bLess stringent criteria were used for IHC interpretation.
Figure 1. Mutation frequency of ARID1A among human cancers. A total of 594 mutated samples from 5160 tumors belonging to 25 different tumor types have been reported (as of January 2014). Among all neoplastic diseases analyzed, ARID1A mutation is most prevalent in ovarian clear cell carcinoma, uterine endometrioid carcinoma, and ovarian endometrioid carcinoma, all of which are derived from either ectopic or eutopic endometrial epithelium. ARID1A mutations are associated with loss of ARID1A expression as shown in an example of uterine endometrioid carcinoma while normal endometrium expresses ARID1A protein.
ARID1A Mutations in Precancerous Lesions
Loss of ARID1A protein expression is a surrogate marker for ARID1A loss of function mutations,24 and immunohistochemistry has been used to study ARID1A expression in tumor precursor lesions of which the small size challenges direct sequence analysis. Loss of ARID1A appears to be an early molecular event in developing ovarian and endometrial cancers. Yamamoto et al. showed a strikingly high prevalence of ARID1A loss in the precursor lesions adjacent to ovarian clear cell carcinomas, including 86% of non-atypical and 100% of atypical endometriosis and clear-cell adenofibroma components.71 In a study by Ayhan et al., all 31 ARID1A-negative ovarian carcinomas arising in ovarian endometriotic cysts lost ARID1A expression in the contiguous endometriotic cyst epithelium.72 In contrast, ARID1A loss is rarely found in endometriosis not associated with cancerous tissue, or in precursor lesions of ARID1A-expressing cancers.71,72 In endometrial cancer, Werner et al. found frequent ARID1A loss (16%) in complex endometrial hyperplasia with atypia, the precursor lesion of uterine endometrioid carcinoma.35 Similarly, Mao et al. reported that the percentage of complete ARID1A loss increased from 0% in complex atypical hyperplasia, to 25% in low-grade endometrioid carcinoma, to 44% in high-grade endometrioid carcinoma.73 These findings suggest that loss of ARID1A expression, presumably due to mutation, plays an important role in tumor progression of uterine endometrioid carcinoma. In addition to gynecologic cancers, ARID1A protein loss was also identified in Barrett esophagus, a precancerous lesion of esophageal adenocarcinoma, and frequency of loss was higher in lesions with more severe dysplasia.41 The occurrence of ARID1A mutations in these precancerous lesions suggests an important role for ARID1A inactivation in tumor initiation.
Mechanisms of Tumor Suppression: Gatekeeper or Caretaker?
Tumor suppressor genes can be broadly grouped into two classes—“caretakers” and “gatekeepers”.74 Gatekeepers control cellular proliferation, usually through regulating cell cycle or promoting apoptosis, whereas caretakers maintain the integrity of the genome. Functional studies of ARID1A have shown that it is a tumor suppressor with functions related to both gatekeepers and caretakers in different model systems. Thus, inactivating ARID1A through somatic mutations and other epigenetic mechanisms results in loss of both gatekeeper and caretaker functions in cells, thus promoting tumor initiation.
ARID1A as a Gatekeeper
ARID1A has been demonstrated to be capable of repressing cellular proliferation in a variety of cancers. For endometriosis-associated gynecologic cancers, restoration of ARID1A expression suppressed both in vitro cellular proliferation and tumor xenograft growth in an ovarian clear cell carcinoma cell line and a uterine endometrioid carcinoma cell line; whereas, silencing ARID1A expression in cell lines derived from normal ovarian surface epithelium increased the rate of cellular proliferation.17 In breast cancer, ARID1A restoration inhibited proliferation in culture and growth in soft agar of ARID1A-mutated, T47D cells, whereas silencing ARID1A in ARID1A wild-type MCF-7 cells led to increased proliferation.55 In gastric cancer, knockdown of wild-type ARID1A in four gastric cancer cell lines enhanced proliferation, whereas restoring ARID1A expression suppressed tumor proliferation in three ARID1A-deleted gastric cancer cell lines.45,75 In bile duct cancer, silencing of ARID1A in three cell lines with wild-type ARID1A resulted in increased proliferation, which was reversed when ARID1A was re-expressed ectopically.51 In hepatocellular carcinoma, ARID1A knockdown significantly promoted cellular proliferation of four wild-type cell lines but did not affect a cell line with ARID1A mutation.49 The results of these functional studies indicate that ARID1A functions as a gatekeeper tumor suppressor gene.
As a gatekeeper, ARID1A regulates cell cycle entry and progression. In an MC3T3-E1 pre-osteoblast cell line model, ARID1A-depleted cells failed to undergo differentiation-associated cell cycle arrest upon induction with ascorbic acid, a differentiation agent.76,77 During differentiation, ARID1A-containing BAF complexes directly targeted and repressed several E2F-responsive promoters, as well as the c-myc promoter, whose repression is critical for induction of p21 in this cellular model.76,77 Using the same cell model, Nagl et al. discovered that ARID1A- and ARID1B-containing BAF complexes had opposite effects on cell-cycle regulation. Cell cycle arrest induced by serum deprivation was dependent on ARID1A activity, whereas re-entry into cell cycle after serum stimulation required ARID1B.14 In general, ARID1A is required for proper cell cycle arrest at the G1 checkpoint in response to various cellular signals and environmental cues.
In an ovarian cell line model, physical interactions among p53, ARID1A, and BRG1 were observed, and both p53 and BRG1 co-occupied the promoters of CDKN1A (p21) and SMAD3.17 Silencing of ARID1A reduced the occupation of BRG1 at the promoters of CDKN1A and SMAD3, and reduced their transcriptional activity. Moreover, p53 was required for ARID1A-induced p21 expression. This evidence suggests that ARID1A-containing SWI/SNF complexes are recruited to the CDKN1A promoter through interaction with p53, leading to induction of p21 and subsequent cell cycle arrest.
ARID1A as a Caretaker
ARID1A, in addition to its role as a “gatekeeper”, may also function as a “caretaker”, a tumor suppressor that maintains the genomic stability by preventing sequence mutations and structural aberrations in chromosomes. In its caretaker function, ARID1A participates in mediating DNA decatenation, and probably, in facilitating DNA damage repair and mismatch repair.
Like BRCA1, ARID1A physically interacts with topoisomerase IIα (TOP2A), which decatenates newly replicated sister chromatids, ensuring proper chromosome segregation during mitosis.78,79 TOP2A chromatin binding depends on its interaction with ARID1A and the ATPase activity of SWI/SNF complexes. ARID1A knockdown, similar to Brg1 deletion and BRCA1 deficiency, phenocopies TOP2A inhibition. Failure to resolve catenated DNA by TOP2A leads to increased anaphase bridge formation during mitosis, resulting in aneuploidy as well as polyploidy.79 If this in vitro finding can be extrapolated to human tissues, one would expect to observe that tumors harboring ARID1A mutations are characterized by an increase in chromosomal instability or manifested by frequent anaphase bridge. Further studies are required to confirm the biological significance of the above finding.
SWI/SNF complexes have been shown to facilitate repair of DNA damage. SWI/SNF is recruited to DNA double strand breaks (DSB) by interacting with acetylated H3 in gamma-H2AX-containing nucleosomes,80 and promotes ATM-mediated phosphorylation of H2AX around DSB,81 forming a positive feedback loop that facilitates DSB repair. SWI/SNF is also involved in nucleotide excision repair, mediating the repair of UV-induced pyrimidine dimers82,83 and chemical-induced crosslinking of DNA.84 BRG1 interacts with BRCA185 and facilitates BRCA1 recruitment to DNA damage sites.86 Dysfunctional SWI/SNF, therefore, may compromise DNA damage repair and subsequently promote genomic instability. Although the extent to which ARID1A is involved in SWI/SNF-mediated DNA damage repair remains to be determined, given the high frequency of ARID1A mutation in a broad spectrum of cancers, it is likely that ARID1A plays a critical role in this process.
Defects in mismatch repair (MMR) caused by mutation or promoter hypermethylation of MMR genes are prevalent in colorectal, gastric, and endometrial cancers.87 MMR deficiency is responsible for slippage errors during DNA synthesis, resulting in microsatellite instability (MSI) involving short repeats of mono- or oligonucleotides. MSI thus represents the phenotype of MMR deficiency and is associated with a remarkably elevated rate of sequence mutations in cancer cells. ARID1A mutation has been associated with MSI in gastric and colorectal cancers.43,47 Similarly, loss of ARID1A protein expression is associated with loss of expression of mismatch repair proteins and/or MSI in gastric carcinoma46 and uterine endometrioid carcinoma.88,89 However, controversy remains as to whether loss of ARID1A is the cause or result of MMR deficiency. The ARID1A gene contains numerous short mononucleotide repeats that can be affected by slippage errors during DNA replication resulting from a mismatch repair defect. Wang et al. suggested that ARID1A mutation was the result rather than cause of mismatch repair defects because most (89%, 25/28) of the ARID1A mutations found in MSI gastric cancers were indels involving short mononucleotide repeats; whereas, similar indels only occurred in 1 of 17 ARID1A mutations in microsatellite stable gastric cancer.43 Similarly, in a study by Jones et al.,44 ARID1A indels involving short mononucleotide repeats were identified in 10 of 11 (91%) colorectal and gastric cancers with MSI but only 2 of 8 (25%) microsatellite stable ones. Cajuso et al. found a high prevalence (18/46, 39%) of ARID1A mutations in MSI colorectal cancer, with 17 of 25 (68%) mutations (from 18 tumors) representing frameshifts involving mononucleotide repeats.90 Hence, the high frequency of ARID1A mutations in gastrointestinal cancers with MSI is likely the consequence of MMR defects. A similar ARID1A mutation spectrum, however, is not observed in uterine endometrioid carcinoma with MSI, as none of the 26 ARID1A somatic mutations in MSI endometrioid carcinoma in a TCGA study are indels involving short mononucleotide repeats.32 Since the patterns of ARID1A mutations in MSI samples are quite different between endometrial and gastrointestinal cancers, it is possible that the causal relationship between ARID1A mutations and mismatch repair depends on tissue origins. In support of this view, Tjalling Bosse et al. found that ARID1A loss was significantly more prevalent in uterine endometrioid carcinomas with sporadic MSI as compared with uterine endometrioid carcinomas from patients with Lynch syndrome, a germline defect of MMR genes.89 This also suggests that ARID1A mutation is unlikely the consequence of mismatch repair defects in uterine endometrioid carcinoma.
ARID1A and Epstein–Barr Virus
ARID1A mutation or loss of its expression has been found to be associated with Epstein–Barr virus (EBV)-associated gastric cancers.46 Wang et al. found ARID1A mutation in 47% of EBV-infected, microsatellite stable gastric cancer samples, which was substantially higher than in microsatellite stable gastric cancers without EBV infection (10%).43 Abe et al. observed that loss of ARID1A expression was prevalent in EBV-associated (34%) and MLH1-lost (29%) gastric cancer, but rare in gastric cancer without EBV infection and MLH1 loss (5%).46 Histologically, EBV-associated gastric cancer is characterized by an intense infiltrate of lymphocytes within tumors,91 which is also a histological feature of colorectal and gastric cancers with MSI.92,93 Interestingly, an RNAi screen revealed that knockdown of ARID1A rendered Jurkat leukemia cells resistant to Fas (CD95)-mediated apoptosis,94 a killing mechanism used by T cells and NK cells.95 The association between ARID1A mutation and cancer subtypes rich in lymphocytic infiltration raised the speculation that ARID1A mutation may equip cancer cells to escape from immune surveillance. Elucidation of the role played by ARID1A in immune evasion may provide insights germane to approaches using immunotherapy.
Pathway Crosstalk
The observation that mutations in different cancer genes frequently co-occur suggests that these mutations may cooperate in tumor development. On the other hand, the occurrence of mutual exclusion of mutations between two different cancer genes suggests that these mutations may have similar effects or may act within the same functional pathway. Correlation of mutation status of different genes has discovered several pathways that potentially cooperate with ARID1A.
Collaboration between ARID1A Mutation and PI3K/Akt Pathway Activation
Accumulating evidence suggests that ARID1A mutation may cooperate with the PI3K/Akt pathway to promote development of cancer, especially ovarian endometriosis-associated cancers. In ovarian clear cell carcinoma, ARID1A mutation or loss of protein expression often co-occurs with PIK3CA mutation.23,26,71 Combining the data from Jones et al., Yamamoto et al. and Huang et al., ARID1A was found to be mutated or lost in 41 (72%) of 57 ovarian clear cell carcinomas with PIK3CA mutation but in only 41 (43%) of 95 tumors without PIK3CA mutation (P = 0.0007, Fig. 2A).23,26,71 In addition, Huang et al. also found that ARID1A was more frequently lost in tumors with ZNF217 amplification,26 a potential oncogene that was shown to augment the PI3K/Akt signaling activity by upregulating ErbB3 expression.96 In ovarian endometrioid carcinoma, ARID1A was mutated in 80% (4/5) of ovarian endometrioid carcinoma with PTEN mutation but in only 20% (5/25) of samples without PTEN mutation (P = 0.0195).97 In addition to gynecologic cancers, ARID1A mutations were also significantly associated with PIK3CA-activating mutations in gastric carcinomas.45 Viewing the totality of the evidence, it is likely that loss of ARID1A function and dysregulation of the PI3K/Akt signaling pathway have a synergistic effect on tumor development.
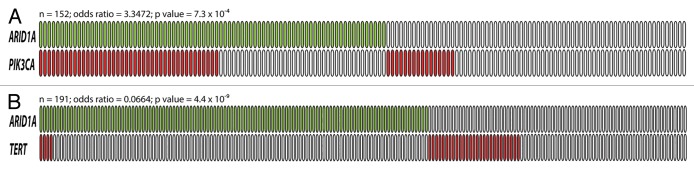
Figure 2. The relationship between ARID1A mutation and PIK3CA mutation (A), and between ARID1A mutation and TERT promoter mutation (B). Green boxes are tumors harboring somatic ARID1A mutations while red boxes are tumors with either PIK3CA mutation or TERT promoter mutation. Each box represents an individual tumor. The total number of tumors, the odds ratio (OR) and P values are shown above each diagram.
The Potential Roles of ARID1A in p53 and Telomere Biology
An inverse relationship between ARID1A and TP53 mutations has been observed in uterine endometrioid carcinoma,88,89 gastric carcinoma,43,45 and esophageal dysplasia/adenocarcinoma.41 ARID1A has been shown to directly interact with p53 and to modulate p53-mediated transcriptional regulation in ovarian cancer.17 This suggests that ARID1A and p53 suppress tumor development in a codependent fashion; therefore, loss of ARID1A may have a similar effect as p53 deficiency. In addition to TP53, Wu et al. have recently discovered a tendency toward mutual exclusivity between ARID1A loss and TERT promoter mutation in ovarian clear cell carcinoma,98 suggesting that ARID1A may also play a role in telomere biology (Fig. 2B).
Potential Synthetic Lethality with ARID1A Mutation
A recent systematic screening of genetic vulnerability across cancer cell lines identified ARID1B as the top gene required for survival of cancer cells with inactivating ARID1A mutations.99 ARID1B, a subunit of SWI/SNF complex mutually exclusive with ARID1A, was required for stable assembly of the SWI/SNF complex in ARID1A-deficient cells. Silencing ARID1B impaired cellular proliferation in cancer cells with ARID1A mutations but not in cells with wild-type ARID1A. This result suggests that ARID1B is a potential therapeutic target for cancers with ARID1A mutation.
ARID1A as a Prognostic Predictor
The prognostic significance of ARID1A mutation or loss of expression has been evaluated in ovarian, endometrial, cervical, urinary bladder, breast, and gastric cancers. These studies have revealed that in different tumor types ARID1A alterations have a different prognostic impact. In ovarian clear cell carcinoma, no significant differences in survival between ARID1A-negative and ARID1A-positive cases were observed in four different studies.25,27,28,37 In endometrial cancer, no association between ARID1A loss and disease-specific survival was observed in three studies on endometrioid carcinoma,34,35,88 or in one study on clear cell carcinoma.37 In cervical cancer, ARID1A loss was a predictor of reduced overall survival in one study,39 but not in another.38 In bladder transitional cell carcinoma, Gui et al. found no association between ARID1A mutation status and tumor grade or stage.58 In breast cancer, low ARID1A mRNA expression has been associated with aggressive features, including higher grade, higher stage, hormone receptor negativity, and ERBB2 positivity,55,56,69 although low ARID1A is not an independent prognosticator for disease-specific survival.55,56 In clear cell renal cell carcinoma, lower protein and mRNA expression levels were associated with worse prognosis.70 In gastric cancer, ARID1A loss was found to be an independent predictor for better prognosis in one study,43 but worse prognosis in another.75 These conflicting results may be due to the fact that the multivariate analyses employed did not incorporate both MSI and EBV infection, two critical confounding factors that are correlated with ARID1A loss and associated with better prognosis.100,101 Taking both MSI and EBV infection into account, Abe et al. showed that ARID1A loss was associated with poor prognosis only in gastric cancers without MSI and EBV infection.46
Conclusion
Recent cancer genome studies have established ARID1A as an important tumor suppressor, whose mechanisms in tumor suppression and interplay with other oncogenic pathways have just begun to be unraveled. Although tumor suppressor genes inherently present challenges for developing targeted therapeutics, this obstacle will likely be circumvented by addressing the following issues. First, the involvement of ARID1A in maintaining genomic stability makes tumors with ARID1A mutations potential candidates for therapeutics based on synthetic lethality—an ARID1A-deficient tumor, with genomic instability as its Achilles’ heel, may be vulnerable to therapies targeting certain pathways involving genome maintenance. Alternatively, ARID1B presents as a new promising therapeutic target for synthetic lethality in cancers with ARID1A mutation and warrants further investigation. Furthermore, with more epigenetic therapies emerging in cancer medicine,102 it is of paramount importance to delineate the landscape of ARID1A-containing SWI/SNF targets and the epigenetic alterations that follow ARID1A mutation. This analysis will serve as a springboard for future development of epigenetic therapeutics. Last but not least, due to the possible interaction between ARID1A mutation and the PI3K pathway, it is interesting to investigate the effect of PI3K inhibitors on tumors with different ARID1A mutation status. This will provide new opportunity to fine-tune personalized medicine using PI3K inhibitor. Addressing these issues will likely yield fruitful results, leading to the application of personalized medicine to ARID1A deficient tumors.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by an NIH/NCI grant CA165807.
References
- 1.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 4.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dallas PB, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol. 2000;20:3137–46. doi: 10.1128/MCB.20.9.3137-3146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilsker D, Patsialou A, Zumbrun SD, Kim S, Chen Y, Dallas PB, Moran E. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 2004;32:1345–53. doi: 10.1093/nar/gkh277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 8.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–92. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 9.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 10.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med. 2007;13:373–80. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–6. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores-Alcantar A, Gonzalez-Sandoval A, Escalante-Alcalde D, Lomelí H. Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res. 2011;345:137–48. doi: 10.1007/s00441-011-1182-x. [DOI] [PubMed] [Google Scholar]
- 14.Nagl NG, Jr., Wang X, Patsialou A, Van Scoy M, Moran E. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 2007;26:752–63. doi: 10.1038/sj.emboj.7601541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–88. doi: 10.1128/MCB.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J Biol Chem. 2002;277:41674–85. doi: 10.1074/jbc.M205961200. [DOI] [PubMed] [Google Scholar]
- 17.Guan B, Wang TL, Shih IeM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71:6718–27. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–80. doi: 10.1128/MCB.01008-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan B, Gao M, Wu C-H, Wang T-L, Shih IeM. Functional analysis of in-frame indel ARID1A mutations reveals new regulatory mechanisms of its tumor suppressor functions. Neoplasia. 2012;14:986–93. doi: 10.1593/neo.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–6. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Nagl NG, Jr., Flowers S, Zweitzig D, Dallas PB, Moran E. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer. 2004;112:636–42. doi: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Zhao Y-L, Li Y, Fletcher JA, Xiao S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosomes Cancer. 2007;46:745–50. doi: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 23.Jones S, Wang T-L, Shih IeM, Mao T-L, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr., Vogelstein B, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda D, Mao T-L, Fukayama M, Nakagawa S, Yano T, Taketani Y, Shih IeM. Clinicopathological Significance of Loss of ARID1A Immunoreactivity in Ovarian Clear Cell Carcinoma. Int J Mol Sci. 2010;11:5120–8. doi: 10.3390/ijms11125120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H-N, Lin M-C, Huang W-C, Chiang Y-C, Kuo K-T. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations and ZNF217 amplification in ovarian clear cell carcinoma. Mod Pathol. 2013 doi: 10.1038/modpathol.2013.216. forthcoming. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto S, Tsuda H, Takano M, Tamai S, Matsubara O. PIK3CA mutations and loss of ARID1A protein expression are early events in the development of cystic ovarian clear cell adenocarcinoma. Virchows Arch. 2012;460:77–87. doi: 10.1007/s00428-011-1169-8. [DOI] [PubMed] [Google Scholar]
- 28.Lowery WJ, Schildkraut JM, Akushevich L, Bentley R, Marks JR, Huntsman D, Berchuck A. Loss of ARID1A-associated protein expression is a frequent event in clear cell and endometrioid ovarian cancers. Int J Gynecol Cancer. 2012;22:9–14. doi: 10.1097/IGC.0b013e318231f140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C-H, Mao T-L, Vang R, Ayhan A, Wang T-L, Kurman RJ, Shih IeM. Endocervical-type mucinous borderline tumors are related to endometrioid tumors based on mutation and loss of expression of ARID1A. Int J Gynecol Pathol. 2012;31:297–303. doi: 10.1097/PGP.0b013e31823f8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H, Cheung LWT, Li J, Ju Z, Yu S, Stemke-Hale K, Dogruluk T, Lu Y, Liu X, Gu C, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–9. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan B, Mao T-L, Panuganti PK, Kuhn E, Kurman RJ, Maeda D, Chen E, Jeng Y-M, Wang T-L, Shih IeM. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol. 2011;35:625–32. doi: 10.1097/PAS.0b013e318212782a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al. Cancer Genome Atlas Research Network Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiegand KC, Lee AF, Al-Agha OM, Chow C, Kalloger SE, Scott DW, Steidl C, Wiseman SM, Gascoyne RD, Gilks B, et al. Loss of BAF250a (ARID1A) is frequent in high-grade endometrial carcinomas. J Pathol. 2011;224:328–33. doi: 10.1002/path.2911. [DOI] [PubMed] [Google Scholar]
- 34.Rahman M, Nakayama K, Rahman MT, Katagiri H, Katagiri A, Ishibashi T, Ishikawa M, Iida K, Miyazaki K. Clinicopathologic analysis of loss of AT-rich interactive domain 1A expression in endometrial cancer. Hum Pathol. 2013;44:103–9. doi: 10.1016/j.humpath.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 35.Werner HMJ, Berg A, Wik E, Birkeland E, Krakstad C, Kusonmano K, Petersen K, Kalland KH, Oyan AM, Akslen LA, et al. ARID1A loss is prevalent in endometrial hyperplasia with atypia and low-grade endometrioid carcinomas. Mod Pathol. 2013;26:428–34. doi: 10.1038/modpathol.2012.174. [DOI] [PubMed] [Google Scholar]
- 36.Fadare O, Renshaw IL, Liang SX. Does the Loss of ARID1A (BAF-250a) Expression in Endometrial Clear Cell Carcinomas Have Any Clinicopathologic Significance? A Pilot Assessment. J Cancer. 2012;3:129–36. doi: 10.7150/jca.4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fadare O, Gwin K, Desouki MM, Crispens MA, Jones HW, 3rd, Khabele D, Liang SX, Zheng W, Mohammed K, Hecht JL, et al. The clinicopathologic significance of p53 and BAF-250a (ARID1A) expression in clear cell carcinoma of the endometrium. Mod Pathol. 2013;26:1101–10. doi: 10.1038/modpathol.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katagiri A, Nakayama K, Rahman MT, Rahman M, Katagiri H, Ishikawa M, Ishibashi T, Iida K, Otsuki Y, Nakayama S, et al. Frequent loss of tumor suppressor ARID1A protein expression in adenocarcinomas/adenosquamous carcinomas of the uterine cervix. Int J Gynecol Cancer. 2012;22:208–12. doi: 10.1097/IGC.0b013e3182313d78. [DOI] [PubMed] [Google Scholar]
- 39.Cho H, Kim JS-Y, Chung H, Perry C, Lee H, Kim J-H. Loss of ARID1A/BAF250a expression is linked to tumor progression and adverse prognosis in cervical cancer. Hum Pathol. 2013;44:1365–74. doi: 10.1016/j.humpath.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 40.Chong IY, Cunningham D, Barber LJ, Campbell J, Chen L, Kozarewa I, Fenwick K, Assiotis I, Guettler S, Garcia-Murillas I, et al. The genomic landscape of oesophagogastric junctional adenocarcinoma. J Pathol. 2013;231:301–10. doi: 10.1002/path.4247. [DOI] [PubMed] [Google Scholar]
- 41.Streppel MM, Lata S, DelaBastide M, Montgomery EA, Wang JS, Canto MI, Macgregor-Das AM, Pai S, Morsink FHM, Offerhaus GJ, et al. Next-generation sequencing of endoscopic biopsies identifies ARID1A as a tumor-suppressor gene in Barrett’s esophagus. Oncogene. 2014;33:347–57. doi: 10.1038/onc.2012.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, Chan TL, Kan Z, Chan ASY, Tsui WY, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 44.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, Schmidt MK, Markowitz S, Yan H, Bigner D, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–3. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 46.Abe H, Maeda D, Hino R, Otake Y, Isogai M, Ushiku AS, Matsusaka K, Kunita A, Ushiku T, Uozaki H, et al. ARID1A expression loss in gastric cancer: pathway-dependent roles with and without Epstein-Barr virus infection and microsatellite instability. Virchows Arch. 2012;461:367–77. doi: 10.1007/s00428-012-1303-2. [DOI] [PubMed] [Google Scholar]
- 47.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–8. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang J, Deng Q, Wang Q, Li K-Y, Dai J-H, Li N, Zhu Z-D, Zhou B, Liu X-Y, Liu R-F, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–21. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 50.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–4. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 51.Chan-On W, Nairismägi M-L, Ong CK, Lim WK, Dima S, Pairojkul C, Lim KH, McPherson JR, Cutcutache I, Heng HL, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45:1474–8. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 52.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–3. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birnbaum DJ, Adélaïde J, Mamessier E, Finetti P, Lagarde A, Monges G, Viret F, Gonçalvès A, Turrini O, Delpero J-R, et al. Genome profiling of pancreatic adenocarcinoma. Genes Chromosomes Cancer. 2011;50:456–65. doi: 10.1002/gcc.20870. [DOI] [PubMed] [Google Scholar]
- 54.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mamo A, Cavallone L, Tuzmen S, Chabot C, Ferrario C, Hassan S, Edgren H, Kallioniemi O, Aleynikova O, Przybytkowski E, et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene. 2012;31:2090–100. doi: 10.1038/onc.2011.386. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Zhang Y, Yang Y, Niu M, Sun S, Ji H, Ma Y, Yao G, Jiang Y, Shan M, et al. Frequent low expression of chromatin remodeling gene ARID1A in breast cancer and its clinical significance. Cancer Epidemiol. 2012;36:288–93. doi: 10.1016/j.canep.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 57.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, Dean M, Huang Y, Jia W, Zhou Q, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–63. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–8. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balbás-Martínez C, Rodríguez-Pinilla M, Casanova A, Domínguez O, Pisano DG, Gómez G, Lloreta J, Lorente JA, Malats N, Real FX. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8:e62483. doi: 10.1371/journal.pone.0062483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, Palmer G, Lucena-Silva N, Pedrosa F, Pedrosa M, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–4. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao Y, Sheehy P, Manning RJ, Patterson CJ, Tripsas C, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N Engl J Med. 2012;367:826–33. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 64.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Jr., Papadopoulos N, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45:12–7. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–43. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shain AH, Giacomini CP, Matsukuma K, Karikari CA, Bashyam MD, Hidalgo M, Maitra A, Pollack JR. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109:E252–9. doi: 10.1073/pnas.1114817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cornen S, Adelaide J, Bertucci F, Finetti P, Guille A, Birnbaum DJ, Birnbaum D, Chaffanet M. Mutations and deletions of ARID1A in breast tumors. Oncogene. 2012;31:4255–6. doi: 10.1038/onc.2011.598. [DOI] [PubMed] [Google Scholar]
- 70.Lichner Z, Scorilas A, White NMA, Girgis AH, Rotstein L, Wiegand KC, Latif A, Chow C, Huntsman D, Yousef GM. The chromatin remodeling gene ARID1A is a new prognostic marker in clear cell renal cell carcinoma. Am J Pathol. 2013;182:1163–70. doi: 10.1016/j.ajpath.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto S, Tsuda H, Takano M, Tamai S, Matsubara O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod Pathol. 2012;25:615–24. doi: 10.1038/modpathol.2011.189. [DOI] [PubMed] [Google Scholar]
- 72.Ayhan A, Mao T-L, Seckin T, Wu C-H, Guan B, Ogawa H, Futagami M, Mizukami H, Yokoyama Y, Kurman RJ, et al. Loss of ARID1A expression is an early molecular event in tumor progression from ovarian endometriotic cyst to clear cell and endometrioid carcinoma. Int J Gynecol Cancer. 2012;22:1310–5. doi: 10.1097/IGC.0b013e31826b5dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mao T-L, Ardighieri L, Ayhan A, Kuo K-T, Wu C-H, Wang T-L, Shih IeM. Loss of ARID1A expression correlates with stages of tumor progression in uterine endometrioid carcinoma. Am J Surg Pathol. 2013;37:1342–8. doi: 10.1097/PAS.0b013e3182889dc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–3, 763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 75.Wang D-D, Chen Y-B, Pan K, Wang W, Chen S-P, Chen J-G, Zhao J-J, Lv L, Pan Q-Z, Li Y-Q, et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS One. 2012;7:e40364. doi: 10.1371/journal.pone.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagl NG, Jr., Patsialou A, Haines DS, Dallas PB, Beck GR, Jr., Moran E. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 2005;65:9236–44. doi: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- 77.Nagl NG, Jr., Zweitzig DR, Thimmapaya B, Beck GR, Jr., Moran E. The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 2006;66:1289–93. doi: 10.1158/0008-5472.CAN-05-3427. [DOI] [PubMed] [Google Scholar]
- 78.Lou Z, Minter-Dykhouse K, Chen J. BRCA1 participates in DNA decatenation. Nat Struct Mol Biol. 2005;12:589–93. doi: 10.1038/nsmb953. [DOI] [PubMed] [Google Scholar]
- 79.Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, Pfister S, Cho Y-J, Zhao K, Crabtree GR. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497:624–7. doi: 10.1038/nature12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–45. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park J-H, Park E-J, Lee H-S, Kim S-J, Hur S-K, Imbalzano AN, Kwon J. Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting γ-H2AX induction. EMBO J. 2006;25:3986–97. doi: 10.1038/sj.emboj.7601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ. Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle. 2008;7:1067–74. doi: 10.4161/cc.7.8.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao Q, Wang Q-E, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–32. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp Cell Res. 2012;318:1973–86. doi: 10.1016/j.yexcr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–65. doi: 10.1016/S0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 86.Zhang L, Chen H, Gong M, Gong F. The chromatin remodeling protein BRG1 modulates BRCA1 response to UV irradiation by regulating ATR/ATM activation. Front Oncol. 2013;3:7. doi: 10.3389/fonc.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–9. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 88.Allo G, Bernardini MQ, Wu R-C, Shih IeM, Kalloger S, Pollett A, Gilks CB, Clarke BA. ARID1A loss correlates with mismatch repair deficiency and intact p53 expression in high-grade endometrial carcinomas. Mod Pathol. 2014;27:255–61. doi: 10.1038/modpathol.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bosse T, ter Haar NT, Seeber LM, v Diest PJ, Hes FJ, Vasen HF, Nout RA, Creutzberg CL, Morreau H, Smit VT. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol. 2013;26:1525–35. doi: 10.1038/modpathol.2013.96. [DOI] [PubMed] [Google Scholar]
- 90.Cajuso T, Hänninen UA, Kondelin J, Gylfe AE, Tanskanen T, Katainen R, Pitkänen E, Ristolainen H, Kaasinen E, Taipale M, et al. Exome sequencing reveals frequent inactivating mutations in ARID1A,ARID1B,ARID2, and ARID4A in microsatellite unstable colorectal cancer. Int J Cancer. 2013 doi: 10.1002/ijc.28705. forthcoming. [DOI] [PubMed] [Google Scholar]
- 91.Yuen ST, Chung LP, Leung SY, Luk IS, Chan SY, Ho J. In situ detection of Epstein-Barr virus in gastric and colorectal adenocarcinomas. Am J Surg Pathol. 1994;18:1158–63. doi: 10.1097/00000478-199411000-00010. [DOI] [PubMed] [Google Scholar]
- 92.Jass JR. HNPCC and sporadic MSI-H colorectal cancer: a review of the morphological similarities and differences. Fam Cancer. 2004;3:93–100. doi: 10.1023/B:FAME.0000039849.86008.b7. [DOI] [PubMed] [Google Scholar]
- 93.dos Santos NR, Seruca R, Constância M, Seixas M, Sobrinho-Simões M. Microsatellite instability at multiple loci in gastric carcinoma: clinicopathologic implications and prognosis. Gastroenterology. 1996;110:38–44. doi: 10.1053/gast.1996.v110.pm8536886. [DOI] [PubMed] [Google Scholar]
- 94.Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:20380–5. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Igney FH, Krammer PH. Immune escape of tumors: apoptosis resistance and tumor counterattack. J Leukoc Biol. 2002;71:907–20. [PubMed] [Google Scholar]
- 96.Krig SR, Miller JK, Frietze S, Beckett LA, Neve RM, Farnham PJ, Yaswen PI, Sweeney CA. ZNF217, a candidate breast cancer oncogene amplified at 20q13, regulates expression of the ErbB3 receptor tyrosine kinase in breast cancer cells. Oncogene. 2010;29:5500–10. doi: 10.1038/onc.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McConechy MK, Ding J, Senz J, Yang W, Melnyk N, Tone AA, Prentice LM, Wiegand KC, McAlpine JN, Shah SP, et al. Ovarian and endometrial endometrioid carcinomas have distinct CTNNB1 and PTEN mutation profiles. Mod Pathol. 2014;27:128–34. doi: 10.1038/modpathol.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu R-C, Ayhan A, Maeda D, Kim K-R, Clarke BA, Shaw P, Chui MH, Rosen B, Shih IeM, Wang T-L. Frequent somatic mutations of the telomerase reverse transcriptase promoter in ovarian clear cell carcinoma but not in other major types of gynaecological malignancy. J Pathol. 2014;232:473–81. doi: 10.1002/path.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Helming KC, Wang X, Wilson BG, Vazquez F, Haswell JR, Manchester HE, Kim Y, Kryukov GV, Ghandi M, Aguirre AJ, et al. ARID1B is a specific vulnerability in ARID1A-mutant cancers. Nat Med. 2014;20:251–4. doi: 10.1038/nm.3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Beghelli S, de Manzoni G, Barbi S, Tomezzoli A, Roviello F, Di Gregorio C, Vindigni C, Bortesi L, Parisi A, Saragoni L, et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery. 2006;139:347–56. doi: 10.1016/j.surg.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 101.Camargo MC, Kim W-H, Chiaravalli AM, Kim K-M, Corvalan AH, Matsuo K, Yu J, Sung JJY, Herrera-Goepfert R, Meneses-Gonzalez F, et al. Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: an international pooled analysis. Gut. 2014;63:236–43. doi: 10.1136/gutjnl-2013-304531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Popovic R, Licht JD. Emerging epigenetic targets and therapies in cancer medicine. Cancer Discov. 2012;2:405–13. doi: 10.1158/2159-8290.CD-12-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]