Abstract
The diagnosis of glioblastoma is still based on tumor histology, but emerging molecular diagnosis is becoming an important part of glioblastoma classification.
Besides the well-known cell cycle-related circuitries that are associated with glioblastoma onset and development, new insights may be derived by looking at pathways involved in regulation of epigenetic phenomena and cellular metabolism, which may both be highly deregulated in cancer cells.
We evaluated if in glioblastoma patients the high grade of malignancy could be associated with aberrant expression of some genes involved in regulation of epigenetic phenomena and lipid metabolism. We measured the mRNA levels of ZFP57, TRIM28, CPT1A, CPT1B, and CPT1C in a cohort of 80 patients divided in two groups: grade II and grade IV. We evidenced that high grade glioblastoma is associated with increased level of ZFP57, a protein involved in gene imprinting, and aberrant expression of CPT1A and CPT1C, regulators of fatty acid oxidation.
Our study may pave the way to identify new markers that could be potentially useful for diagnosis and/or prognosis of glioblastoma.
Keywords: glioblastoma, mitochondria, lipid metabolism, imprinting
Introduction
Gliomas of astrocytic, oligodendroglial and ependymal origin account for more than 70% of all brain tumors. The most frequent (65%) and most malignant histological type is the glioblastoma multiforme (GBM).1 The median survival of people diagnosed with these cancers is between 12 and 15 mo. Currently there are no definitive treatments for malignant gliomas and for this reason researchers are highly involved in dissecting molecular pathways that are altered in this cancer. Among studies aiming to investigate glioblastoma-related pathways, the Cancer Genome Atlas studied several glioblastoma samples using microarray technology and the analyses identified three core signal pathways implicated in glioblastoma, including receptor tyrosine kinase (RTK) signaling, and the P53 and RB tumor suppressor pathways.2,3
Besides these well-known pathways involved in glioblastoma onset and development, new insights may derive by looking at pathways involved in regulation of epigenetic phenomena and cellular metabolism, which may be both highly deregulated in cancer cells.
Epigenetic alterations alone or in combination with genetic mechanisms play a key role in brain tumorigenesis. In GBM the presence of epigenetic lesions has been widely described. In each tumor, hundreds of genes are subject to DNA hypermethylation at their CpG island promoters. A subset of GBMs is also characterized by locus-specific and genome-wide decrease in DNA methylation, or DNA hypomethylation. Moreover, other epigenetic alterations, such as aberrant imprinting of maternally or paternally expressed genes, changes in the position of histone variants and changes in histone modifications are also likely important in the molecular pathology of GBM.4-6
Zinc finger transcription factor ZFP57 is one of a group of genes expressed in very early embryogenesis and downregulated upon differentiation of embryonic stem cells.7 The monoallelic expression of several imprinted genes are regulated by DNA methylation of CpG-rich sequences known as imprinting control regions (ICRs). ZFP57 recognizes the methylated CpG within the TGCCGC element that are found in most ICRs. In this way, ZFP57 contributes to the maintenance of both maternally and paternally imprinted loci and is also involved in imprint establishment.8,9 ZFP57 and its co-factor KAP1(TRIM28) bind to H3K9me3 histones in ICRs and may contribute to regulation of chromatin status. Indeed, KAP1 deletion induces a loss of heterochromatin marks at ICRs, whereas deleting ZFP57 leads to ICR DNA demethylation.9
Impairment of ZFP57 has significant consequences on embryo development and may also have pathological effects. Loss of ZFP57 function in mouse zygote induces partial neonatal lethality, whereas eliminating both the maternal and zygotic function of ZFP57 results in embryonic lethality. ZFP57 mutations were found in patients with transient neonatal diabetes. Altered function of ZFP57/KAP1 axes has been found also in some cancers. Fitzgerald and coworkers evidenced a relationship between epithelial and stromal KAP1 (TRIM28) expression and survival in colorectal cancer patients. They suggest that a high TRIM28 expression ratio between stromal and epithelial compartments in colorectal cancer tissue is an independent predictor of poor prognosis.10 KAP1 could be considered also a prognostic marker for non-small cell lung cancer.11
Altered metabolism is one of the hallmarks of cancer cells. The best-known metabolic abnormality in cancer cells is the Warburg effect, which is an increased glycolysis even in the presence of oxygen.12 Besides alteration in glucose metabolism, there are compelling evidences showing that that cancer cells have specific alterations in different aspects of lipid metabolism. These alterations can affect the availability of structural lipids for the synthesis of membranes, the synthesis and degradation of lipids that contribute to energy homeostasis and the abundance of lipids with signaling functions.13 Indeed, we demonstrated that an inhibitor of carnitine-palmitoyl transferase 1A (CPT1A), the rate-limiting enzyme for fatty acid (FA) import into mitochondria can prevent myc-induced lymphomagenesis.14
The degradation of long-chain fatty acids occurs in mitochondria and is catalyzed by several carnitine acyl transferases, including carnitine palmitoyltransferase I (CPT1), which is located in the outer membrane, and carnitine palmitoyltransferase II (CPTII), which is located in the inner membrane, together with a carnitine-acylcarnitine translocase (CAT).15
Mammals have three paralog genes (CPT1A, B, and C). CPT1A is expressed mainly in liver and CPT1B in muscle. The central nervous system, notably the hypothalamus, expresses a low level of CPT1A and no CPT1B. However nervous system cells express a brain-specific CPT1, i.e., CPT1C. All the three CPT enzymes are negatively regulated by malonyl-CoA, which derives from acetyl-CoA through the activity acetyl-CoA carboxylase that is the primary regulatory step in fatty acid synthesis. Unlike CPT1A and CPT1B, CPT1C does not catalyze the acyltransferase activity by using the prototypic substrates, i.e., fatty acyl-CoA derivatives and carnitine. Differences in the activities of CTP1 enzymes are related to tissue-specific needs in fatty acid metabolism and energy expenditure.16,17
Several findings suggest that chemotherapy resistant cancer cells may represent a small subpopulation of malignant cells with stem-like properties. Glioblastoma cells, exhibiting stem-like properties, show unique energy metabolic characteristics including low mitochondrial respiration, increased glycolysis for ATP generation, and preference for hypoxia to maintain their stemness and tumor forming capacity.18
On these premises, we decided to investigate if in glioblastoma patients the high grade of malignancy could be associated with aberrant expression of some genes involved in regulation of epigenetic phenomena and lipid metabolism. We evaluated the mRNA levels of ZFP57, TRIM28, CPT1A, CPT1B, and CPT1C in a cohort of 80 patients divided in two groups: grade II and grade IV. Our data suggest that increased level of ZFP57 and deregulated expression of CPT1A an CPT1C are associated with high grade glioblastoma.
Results
We analyzed a cohort of 80 patients with grade II (46 patients) or grade IV (34 patients) gliomas, hereafter indicated as GII and GIV, respectively. The main clinical and pathological characteristics of patients are listed in Table 1.
Table 1. Main clinical and pathological characteristics of patients.
| Grade II gliomas | Grade IV gliomas | |
|---|---|---|
| Number of patients | 46 | 34 |
| Age >50 y | 9 | 14 |
| Age <50 y | 37 | 20 |
| Males | 30 | 14 |
| Females | 16 | 20 |
| Presence of seizure | 16 | 7 |
| Increased intracranial pressure | 14 | 18 |
| Mean tumor diameter (>5 cm) | 20 | 20 |
| Mean tumor diameter (<5 cm) | 26 | 14 |
| Radiotherapy | 25 | 21 |
| Chemotherapy | 22 | 22 |
| Median follow up (mo) | 80 | 15 |
| Recurrence | 20 | 30 |
We performed quantitative RT-PCR on samples from these patients to detect the expression level of several genes involved in pathways that are altered in most gliomas.
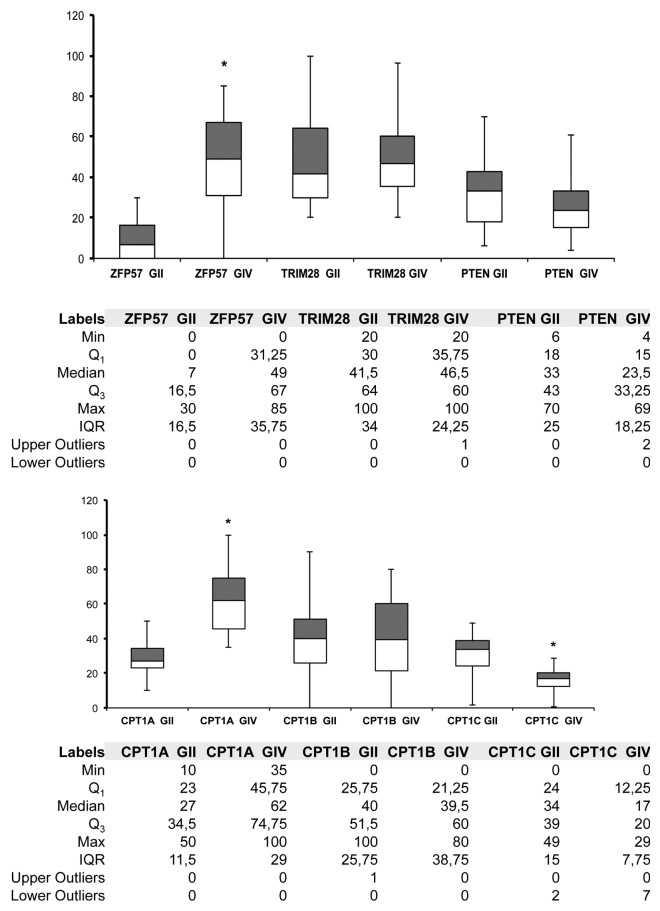
We analyzed the expression of ZFP57 and TRIM28, which are involved in epigenetic regulation of imprinted genes. The expression level of ZFP57 is significantly higher in GIV patients compared with GII group (P < 0.05), whereas no significant differences were detected in the expression of TRIM28 (Fig. 1).
Figure 1. mRNA expression level box plot charts. The upper chart shows the expression levels of ZFP57, TRIM28 and PTEN in the grade II (GII) and grade IV (GIV) cohorts. The lower chart shows the levels of CPT1A, CPT1B, and CPT1C in the same group of patients. The gray boxes represent Q3–Q2, whites boxes are Q2–Q1. The ends of the whisker are set at 1.5× Interquartile (IQR) above the third quartile (Q3) and 1.5× IQR below the first quartile (Q1). For each data set the number of outliers are included in the tables next to the charts. *P < 0.05.
These data suggested that progression of disease is associated with aberrant expression of genes involved in the regulation of the expression of imprinted genes.
We then analyzed the expression of three isoform of CPT1 enzymes, which regulate fatty acid degradation.
Of interest, we detected the expression of all the three isoforms in the great majority of analyzed samples (Fig. 1). In particular, to our knowledge, this is the first time that CPT1B expression was detected in brain samples. The expression of CPT1A was significant higher (P < 0.05) in GIV compared with GII, whereas the levels of CPT1C were higher in GII group (Fig. 1). No significant changes were observed in the CPT1B mRNA levels.
These results indicated that important changes in lipid metabolism may occur during glioblastoma progression.
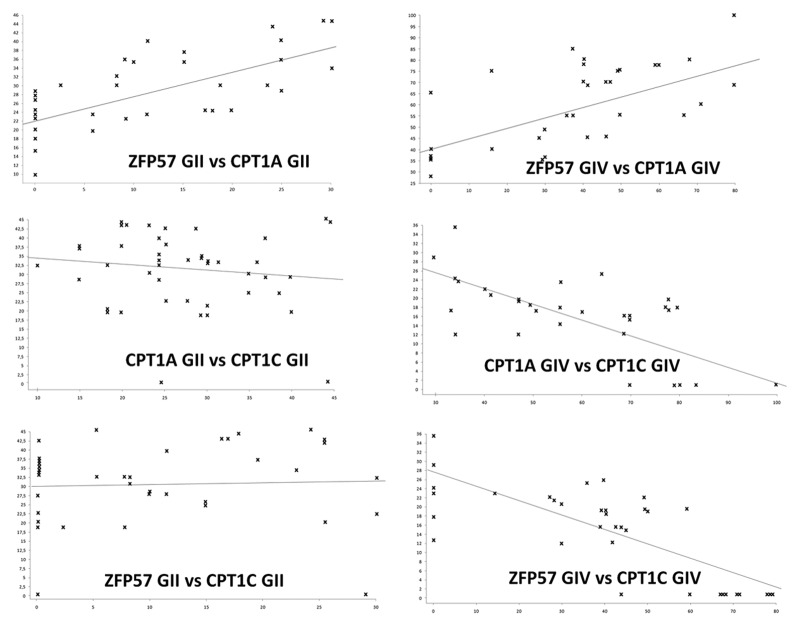
We hypothesized that the expression of these genes may globally distinguish GIV from GII group. To this end, we applied Pearson Correlation Coefficient (PCC) to identify disease-specific biomarkers. Most striking results were obtained comparing the expression of ZFP57 and CPT1A and those of CPT1A and CPT1C.
The Pearson correlation indicated that in GIV and GII groups the increase in the expression of ZFP57 is related to corresponding upregulation of CPT1A level (Fig. 2; Table 2). A negative correlation between ZFP57 and CPT1C was observed only in GIV group (Fig. 2; Table 2). Finally, in group IV we detected also negative correlation between CPT1A and CPT1C (Fig. 2; Table 2).
Figure 2. Regression curves of gene expression analysis. Each graph shows the correlation between two genes in GII and GIV cohorts of patients. Data on equations are in Table 2.
Table 2. Gene expression correlation analysis.
| ZFP57 GII vs CPT1A GII | Sample size | 46 |
| r coefficient | 0.7 | |
| Mean x (xˉ) | 9.13 | |
| Mean y (yˉ) | 27.52 | |
| Intercept (a) | 22.16 | |
| Slope (b) | 0.59 | |
| Regression line equation | y = 22.16 + 0.59x | |
| ZFP57 GIV vs CPT1A GIV | Sample size | 34 |
| r coefficient | 0.65 | |
| Mean x (xˉ) | 37.64 | |
| Mean y (yˉ) | 59.65 | |
| Intercept (a) | 40.50 | |
| Slope (b) | 0.51 | |
| Regression line equation | y = 40.50 + 0.51x | |
| CPT1A GII vs CPT1C GII | Sample size | 46 |
| r coefficient | −0.16 | |
| Mean x (xˉ) | 27.52 | |
| Mean y (yˉ) | 30.67 | |
| Intercept (a) | 36.18 | |
| Slope (b) | −0.2 | |
| Regression line equation | y = 36.18 − 0.20x | |
| CPT1A GIV vs CPT1C GIV | Sample size | 34 |
| r coefficient | −0.71 | |
| Mean x (xˉ) | 59.65 | |
| Mean y (yˉ) | 15.24 | |
| Intercept (a) | 36.72 | |
| Slope (b) | −0.36 | |
| Regression line equation | y = 36.72 − 0.36x | |
| ZFP57 GII vs CPT1C GII | Sample size | 46 |
| r coefficient | 0.05 | |
| Mean x (xˉ) | 9.13 | |
| Mean y (yˉ) | 30.67 | |
| Intercept (a) | 30.1 | |
| Slope (b) | 0.01 | |
| Regression line equation | y = 30.10 + 0.01x | |
| ZFP57 GIV vs CPT1C GIV | Sample size | 34 |
| r coefficient | −0.75 | |
| Mean x (xˉ) | 37.64 | |
| Mean y (yˉ) | 15.24 | |
| Intercept (a) | 26.31 | |
| Slope (b) | −0.29 | |
| Regression line equation | y = 26.31 − 0.29x | |
| ZFP57 GII vs PTEN GII | Sample size | 46 |
| r coefficient | 0.24 | |
| Mean x (xˉ) | 9.13 | |
| Mean y (yˉ) | 31.35 | |
| Intercept (a) | 27.78 | |
| Slope (b) | 0.38 | |
| Regression line equation | y = 27.78 + 0.38x | |
| ZFP57 GIV vs PTEN GIV | Sample size | 34 |
| r coefficient | −0.68 | |
| Mean x (xˉ) | 37.64 | |
| Mean y (yˉ) | 26.08 | |
| Intercept (a) | 44.16 | |
| Slope (b) | −0.48 | |
| Regression line equation | y = 44.15 – 0.48x | |
| CPT1A GII vs PTEN GII | Sample size | 46 |
| r coefficient | 0.31 | |
| Mean x (xˉ) | 27.52 | |
| Mean y (yˉ) | 31.35 | |
| Intercept (a) | 14.99 | |
| Slope (b) | 0.60 | |
| Regression line equation | y = 14.99 + 0.60x | |
| CPT1A GIV vs PTEN GIV | Sample size | 34 |
| r coefficient | −0.71 | |
| Mean x (xˉ) | 59.65 | |
| Mean y (yˉ) | 26.08 | |
| Intercept (a) | 64.64 | |
| Slope (b) | −0.64 | |
| Regression line equation | y = 64.64 – 0.64x |
PTEN is a tumor suppressor gene involved in several signaling pathways and the loss of its activity, through deletion, mutation or methylation, has been associated with several forms of cancer, including glioblastoma.19-22 Alteration of PTEN pathway is very frequent in glioblastoma, with mutations in 5–40% of all GBM cases, and loss of heterozygosity (LOH) in 60–80% of all cases.19,23 For many years PTEN mutations have been considered a powerful prognostic marker for glioblastoma tumors.21 Recently, some findings suggest a more complex and nonlinear molecular relationship between PTEN, its regulators and effectors in the tumorigenesis of glioblastoma.19 Besides association studies between PTEN mutations and glioblastoma prognosis, some authors reported that GBM and lower grades of gliomas may be classified according differences in PTEN expression.20 On this premise, we evaluated PTEN expression in our cohort of patients and evidenced a difference in PTEN mRNA levels between GII and GIV group (Fig. 1; Table 2). In this latter the mRNA expression was higher than in low-grade group. Of great interest, in GIV group the expression of PTEN was negatively correlated both with CPT1A and ZFP57 (Table 2). No significant correlation was detected in GII group between PTEN and CPT1A or ZFP57. Finally, we did not evidence any correlation between PTEN and CTP1C or TRIM28 in both the groups of patients (data not shown).
Discussion
Glioblastoma is the most common and most malignant tumor of the central nervous system and is currently non-curable. The diagnosis of glioblastoma is still based on tumor histology, but emerging molecular diagnostics of key genetic and epigenetic changes is becoming an important part of glioblastoma classification.
Our study aimed to identify new markers that could be potentially useful for diagnosis and/or prognosis of glioblastoma.
We evidenced that grade IV glioma (GBM) have specific expression pattern for ZFP57, CPT1A, and CPT1C. Our data do not allow us to determine if these specific changes are the cause or a consequence of tumor progression. Moreover, the small cohort of patients did not allow us to carry out a Kaplan and Meyer survival analysis to evaluate if the changes we identified may be associated with reduced patients’ survival. Nevertheless our data could be useful to follow glioma progression and may pave the way for further investigations on large cohorts of patients.
Altered expression of ZFP57 suggests that changes in DNA methylation status occur not only before onset of glioblastoma but also during its development to high-grade malignancy.
The reciprocal changes in CPT1A and CPT1C expression indicate that in glioma at advanced stages there is a complete changes in cell metabolism, since CPT1A and CPT1C have different preferences for fatty acid substrates and are differently regulated by malonyl-CoA.
Globally speaking our data suggest that grade II and grade IV glioma show significant metabolic and epigenetic differences that could be exploited for a proper diagnosis/prognosis and may also indicate which are the pathways that therapeutic tools should target.
Correlation analysis of ZFP57 and CPT1A with the expression of PTEN, which is well-known glioblastoma prognostic marker, reinforces the hypothesis that the expression level of these genes may be useful for diagnosis and/or prognosis of glioblastoma. Of course, further studies on larger cohorts of patients are needed to confirm this preliminary finding.
Material and Methods
Patients and tumor samples
Between 1998 and 2011, clinical tumor samples were collected from 80 patients. The samples were either frozen tissues stored at –80 °C or formalin-fixed paraffin embedded tissues.
RNA extraction and RT-PCR
Total RNA was extracted from cell cultures using TRI REAGENT (Molecular Research Center Inc.) according to the manufacturer’s protocol. RNA from paraffin-embedded tissues was extracted with RNeasy FFPE kit (Qiagen Italia).
The mRNA levels of the genes analyzed were measured by quantitative reverse-transcription PCR. These assays were run on an Opticon 4 machine (MJ Research). Reactions were performed according to the manufacturer’s instructions by using SYBR green PCR Master mix. Primer sequences were designed with Primer express software and are listed in Table S1.
Statistical analysis
Statistical analyses (Fisher exact test; ANOVA test followed by the Student t and Bonferroni tests; Pearson Correlation) were evaluated using Prism software (Graphpad Software).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was partially supported by Progetto PON—“Ricerca e Competitivitá 2007 2013” —PON01_00802 entitled, “Sviluppo di molecole capaci di modulare vie metaboliche intracellulari redox sensibili per la prevenzione e la cura di patologie infettive, tumorali, neurodegenerative e loro delivery mediante piattaforme nano tecnologiche” to G.P.
References
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Galderisi U, Cipollaro M, Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–6. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- 3.Galderisi U, Cipollaro M, Giordano A. Stem cells and brain cancer. Cell Death Differ. 2006;13:5–11. doi: 10.1038/sj.cdd.4401757. [DOI] [PubMed] [Google Scholar]
- 4.Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–64. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- 5.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19:188–97. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Otsuka S, Maegawa S, Takamura A, Kamitani H, Watanabe T, Oshimura M, Nanba E. Aberrant promoter methylation and expression of the imprinted PEG3 gene in glioma. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:157–65. doi: 10.2183/pjab.85.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, Toh H, Sasaki H, Zhang X, Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26:2374–9. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–72. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fitzgerald S, Sheehan KM, O’Grady A, Kenny D, O’Kennedy R, Kay EW, Kijanka GS. Relationship between epithelial and stromal TRIM28 expression predicts survival in colorectal cancer patients. J Gastroenterol Hepatol. 2013;28:967–74. doi: 10.1111/jgh.12157. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Zhao E, Li C, Huang L, Xiao L, Cheng L, Huang X, Song Y, Xu D. TRIM28, a new molecular marker predicting metastasis and survival in early-stage non-small cell lung cancer. Cancer Epidemiol. 2013;37:71–8. doi: 10.1016/j.canep.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 12.Bensinger SJ, Christofk HR. New aspects of the Warburg effect in cancer cell biology. Semin Cell Dev Biol. 2012;23:352–61. doi: 10.1016/j.semcdb.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–23. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 14.Pacilli A, Calienni M, Margarucci S, D’Apolito M, Petillo O, Rocchi L, Pasquinelli G, Nicolai R, Koverech A, Calvani M, et al. Carnitine-acyltransferase system inhibition, cancer cell death, and prevention of myc-induced lymphomagenesis. J Natl Cancer Inst. 2013;105:489–98. doi: 10.1093/jnci/djt030. [DOI] [PubMed] [Google Scholar]
- 15.Bennett MJ. Pathophysiology of fatty acid oxidation disorders. J Inherit Metab Dis. 2010;33:533–7. doi: 10.1007/s10545-010-9170-y. [DOI] [PubMed] [Google Scholar]
- 16.Lee J, Wolfgang MJ. Metabolomic profiling reveals a role for CPT1c in neuronal oxidative metabolism. BMC Biochem. 2012;13:23. doi: 10.1186/1471-2091-13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolfgang MJ, Kurama T, Dai Y, Suwa A, Asaumi M, Matsumoto S, Cha SH, Shimokawa T, Lane MD. The brain-specific carnitine palmitoyltransferase-1c regulates energy homeostasis. Proc Natl Acad Sci U S A. 2006;103:7282–7. doi: 10.1073/pnas.0602205103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y, Zhou Y, Shingu T, Feng L, Chen Z, Ogasawara M, Keating MJ, Kondo S, Huang P. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem. 2011;286:32843–53. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carico C, Nuño M, Mukherjee D, Elramsisy A, Dantis J, Hu J, Rudnick J, Yu JS, Black KL, Bannykh SI, et al. Loss of PTEN is not associated with poor survival in newly diagnosed glioblastoma patients of the temozolomide era. PLoS One. 2012;7:e33684. doi: 10.1371/journal.pone.0033684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sano T, Lin H, Chen X, Langford LA, Koul D, Bondy ML, Hess KR, Myers JN, Hong YK, Yung WK, et al. Differential expression of MMAC/PTEN in glioblastoma multiforme: relationship to localization and prognosis. Cancer Res. 1999;59:1820–4. [PubMed] [Google Scholar]
- 21.Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, et al. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–56. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 22.Wang SI, Puc J, Li J, Bruce JN, Cairns P, Sidransky D, Parsons R. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res. 1997;57:4183–6. [PubMed] [Google Scholar]
- 23.Srividya MR, Thota B, Shailaja BC, Arivazhagan A, Thennarasu K, Chandramouli BA, Hegde AS, Santosh V. Homozygous 10q23/PTEN deletion and its impact on outcome in glioblastoma: a prospective translational study on a uniformly treated cohort of adult patients. Neuropathology. 2011;31:376–83. doi: 10.1111/j.1440-1789.2010.01178.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.