Abstract
KRAS mutational status has been shown to be a predictive biomarker of resistance to anti-EGFR monoclonal antibody (mAb) therapy in patients with metastatic colorectal cancer. We report the spectrum of KRAS mutation in 1506 patients with colorectal cancer and the identification and characterization of rare insertion mutations within the functional domain of KRAS. KRAS mutations are found in 44.5% (670/1506) of the patients. Two cases are found to harbor double mutations involving both codons 12 and 13. The frequencies of KRAS mutations at its codons 12, 13, 61, and 146 are 75.1%, 19.3%, 2.5%, and 2.7%, respectively. The most abundant mutation of codon 12 is G12D, followed by G12V and G12C while G13D is the predominant mutation in codon 13. Mutations in other codons are rare. The KRAS mutation rate is significantly higher in women (48%, 296/617) than in men (42.1%, 374/889, P = 0.023). Tumors on the right colon have a higher frequency of KRAS mutations than those on the left (57.3% vs. 40.4%, P < 0.0001). Two in-frame insertion mutations affect the phosphate-binding loop (codon 10–16) of KRAS are identified. One of them has never been reported before. Compared with wild-type protein, the insertion variants enhance the cellular accumulation of active RAS (RAS-GTP) and constitutively activate the downstream signaling pathway. NIH3T3 cells transfected with the insertion variants show enhanced anchorage-independent growth and in vivo tumorigenicity. Potentially these mutations contribute to primary resistance to anti-EGFR mAb therapy but the clinical implication requires further validation.
Keywords: colorectal cancer, KRAS, targeted therapy
Introduction
Colorectal cancer (CRC) is one of the most common lethal cancers worldwide. In 2008, more than 1.2 million new cases were diagnosed, with approximately 608 700 deaths estimated to have occurred.1 Epidermal growth factor receptor (EGFR), a critical molecule in CRC initiation and progression, is frequently overexpressed in metastatic CRC (mCRC) tumors.2,3 The phenomena lead to the development of molecular targeting therapy to inhibit the EGFR signaling pathway. Using anti-EGFR monoclonal antibodies (mAbs) such as cetuximab and panitumumab, have been approved in treating mCRC to inhibit EGFR activity and hence switching off downstream pathways.2,3
However, anti-EGFR therapy does not work on all CRCs, largely due to the resistance to the anti-EGFR mAbs.4 Different studies have reported the response and outcome of CRCs to the anti-EGFR mAbs was poor with KRAS mutation which accounting for 30–40% of non-responsive cases.4-7 KRAS mutation status is now considered to be a predictive biomarker of resistance to anti-EGFR mAbs treatment for mCRC patients. KRAS is one of the RAS superfamily of proto-oncoproteins which is small signal switch molecule called GTPase, cycling between inactive GDP-bound (RAS-GDP) and active GTP-bound (RAS-GTP) forms, to regulate cellular growth and differentiation.8 Activating mutations of RAS proto-oncogenes continuously elevate the cytoplasmic RAS-GTP level. Oncogenic signaling pathways, such as Raf-MEK-ERK and PI3K/AKT cascades, are then constitutively activated in an EGFR activation-independent manner and therefore promote cell cycle progression.6,8 KRAS mutation is found in 40% of CRCs and missense point mutation is the most common mutation. The majority of the point mutation sites of KRAS in CRC patients are located at codons 12 and 13 (~80% and ~17%, respectively), together with rare mutations at codons 61 and 146 (~1–4%).3,9-11
Most clinical studies of KRAS mutation in CRC were conducted in western countries. However, KRAS mutation rate or spectrum in CRCs may partially depends on the population studied.12 It has been reported that KRAS mutations were identified in CRC patients from the UK, Switzerland, and Spain, for 27.4%, 38%, and 41% respectively.12 This epidemiological variation indicates the essence of establishment of a local CRC KRAS mutation data in different populations. There has been a dramatic increase in reported incidence of colorectal cancer in Asian.12 It is of paramount importance to investigate the KRAS mutation spectrum in our locality in view of the implication in using anti-EGFR targeting therapy. We aim to analyze the KRAS mutation status and the clinical correlation in Chinese patients with CRC in Hong Kong. Here we report the spectrum of KRAS mutation in a large cohort of colorectal cancer and the identification and characterization of a novel insertion mutation within the function domain of KRAS.
Results
Clinical characteristics of the patients
We tested a total of 1506 patients with colorectal cancer. Of them 889 (59%) were males and 617 (41%) were females. The median age at presentation was 61 ± 11.3 y (range 21–89 y). The clinical characteristics were in keeping with other reported populations of colorectal cancer.11 The age of female patients were slightly younger than males (59 ± 12.1 vs 61 ± 11.2, P = 0.014). There was significantly higher frequency of left colon tumor (75.8%) than the right side (24.2%, P < 0.0001). However, the right side tumors were more common in females (28.7%) compared with males (21.1%, P = 0.001). When rectal tumor was considered a separate entity, female patients had a higher frequency of right side tumor whereas the rectal tumors were more commonly found in male patients (P < 0.0001). The clinical characteristics of the patients tested were summarized in Table 1.
Table 1. Clinical characteristics of 1506 patients tested for KRAS status.
| Total | Female | Male | P value | |
|---|---|---|---|---|
| n = | 1506 | 617 (41%) | 889 (59%) | |
| Age | 61 ± 11.3 | 59 ± 12.1 | 61 ± 11.2 | 0.014 |
| Tumor site (right vs left) | 0.001 | |||
| Right | 365 (24.2%) | 177 (28.7%) | 188 (21.1%) | |
| Left | 1141 (75.8%) | 440 (71.3%) | 701 (78.9%) | |
| Tumor site (right vs left vs rectum) | < 0.0001 | |||
| Right | 365 (24.2%) | 177 (28.7%) | 188 (21.1%) | |
| Left | 538 (35.7%) | 228 (40.0%) | 310 (34.9%) | |
| Rectum | 603 (40.1%) | 212 (34.3%) | 391 (44.0%) | |
Status of KRAS mutation
KRAS mutations on codons 12, 13, 61 and 146 were analyzed by PCR-direct sequencing using microdissected FFPE tumor tissues from 1506 patients. A total of 672 KRAS mutations were identified from 670 patients (44.5%, 670 out of 1506, Table 2). Two cases were found to harbor double mutations. Both cases involved codon 12 and codon 13 of KRAS gene. One case harbored concomitant G12C and G13D, while the other had both G12V and G13D. Within 672 KRAS mutations identified, the frequencies of mutations at codons 12, 13, 61, and 146 were 75.1%, 19.3%, 2.5%, and 2.7%, respectively. Majority of the mutations occurred at codons 12 and 13 which accounted for more than 94% of all mutations identified. The most common mutation was glycine to aspartate on codon 12 (G12D), which accounted for 37.5% of all mutations (252 out of 672). Mutation from glycine to valine (G12V) was the second most common of all specified mutations (20.1%; 135 of 672). Mutation from glycine to aspartate on codon 13 (G13D) accounted for 19.0% (128 of 672) of specified mutations.
Table 2. KRAS mutations spectrum in 670 colorectal cancers.
| Mutation | Frequency | Percentage |
|---|---|---|
| Codon 12 | 505 | 75.1% |
| G12D | 252 | 37.5% |
| G12V | 135 | 20.1% |
| G12C | 46 | 6.8% |
| G12S | 33 | 4.9% |
| G12A | 29 | 4.3% |
| G12R | 10 | 1.5% |
| Codon13 | 130 | 19.3% |
| G13D | 128 | 19.0% |
| G13C | 2 | 0.3% |
| Codon 61 | 17 | 2.5% |
| Q61H | 9 | 1.3% |
| Q61L | 5 | 0.7% |
| Q61K | 1 | 0.1% |
| Q61R | 2 | 0.3% |
| Codon 146 | 18 | 2.7% |
| A146T | 18 | 2.7% |
| Others | 2 | 0.3% |
| c.30_31insGGA, p.G10_A11insG | 1 | 0.1% |
| c.33_34insGGAGCT:p.A11_G12insGA | 1 | 0.1% |
| Total | 672a | 100% |
aA total of 672 KRAS mutations were detected from 670 colorectal tumors. Two tumors harbored double mutations.
The KRAS mutation rate was significantly higher in women (48%, 296 of 617, Table 3) than in men (42.1%, 374 of 889, P = 0.023). The mutation rate did not differ according to the primary tumor site if the tumor location was classified as either ascending, hepatic flexure, transverse, splenic flexure, descending, sigmoid, or rectum. If the tumors on the right side of the colon (ascending and transverse colon) were group together and compared with those on the left (splenic flexure to rectum), the frequency of KRAS mutations were significantly higher in the right colon (57.3% vs. 40.4%, P < 0.0001). The KRAS mutation was not associated with the age of the patient. In comparison of the most frequently mutated codons between left and right colon, codon 12 mutations were significantly more likely to occur in rectum (right colon 28.8%, left colon 29.7%, rectum 41.6%), while codon 13 mutations were slightly more frequent in the right colon (right colon 40%, left colon 30.8%, rectum 29.2%, P = 0.013)
Table 3. Correlation of KRAS mutation status with clinical features.
| Characteristics | KRAS mutation | Total | P value | |
|---|---|---|---|---|
| + | − | |||
| n = 1506 | ||||
| No. cases | 670 (44.5%) | 836 (55.5%) | ||
| Age | 61.3 ± 11.3 | 60.5 ± 11.3 | NS | |
| Gender | 0.023 | |||
| F | 296 (48%) | 321 (52%) | 617 | |
| M | 374 (42.1%) | 515 (57.9%) | 889 | |
| Age | 62.1 ± 10.1 | 60.4 ± 9.0 | NS | |
| Tumor site (right vs left) | ||||
| Right | 209 (57.3%) | 156 (42.7%) | 365 | < 0.0001 |
| Left | 461 (40.4%) | 680 (59.6%) | 1141 | |
| Total | 670 | 836 | 1506 | |
| Tumor site (right vs left vs rectum) | < 0.0001 | |||
| Right | 209 (57.3%) | 156 (42.7%) | 365 | |
| Left | 198 (36.8%) | 340 (63.2%) | 538 | |
| Rectum | 263 (43.6%) | 340 (56.4%) | 603 | |
| Total | 670 | 836 | 1506 | |
Identification of rare KRAS insertion mutations
In the pool of CRC cases, we identified two rare KRAS mutations which were defined as in-frame insertion mutations. The Insertion mutations in KRAS exon 2 of patient #286 and #833 were further validated by direct sequencing of the cloned PCR products (Fig. 1). In patient #286, an in-flame insertion of 3-nucleotide (GGA) between codons 10 and 11 was observed (c.30_31insGGA: p.G10_A11insG). This rare mutation, which suggested the insertion of a glycine residue between glycine (amino acid 10) and alanine (amino acid 11), was reported once in the patient with myeloid leukemia.8 In patient #833, a tandem repeat sequences of codon 10 and 11 (GGA GCT) was in-flame inserted after codons 11 and introduced extra glycine and alanine residues between alanine (codon 11) and glycine (codon 12). This insertion mutation (c.33_34insGGAGCT:p.A11_G12insGA) has never been reported before. These two mutations are named 10G11 and 11GA12 respectively.
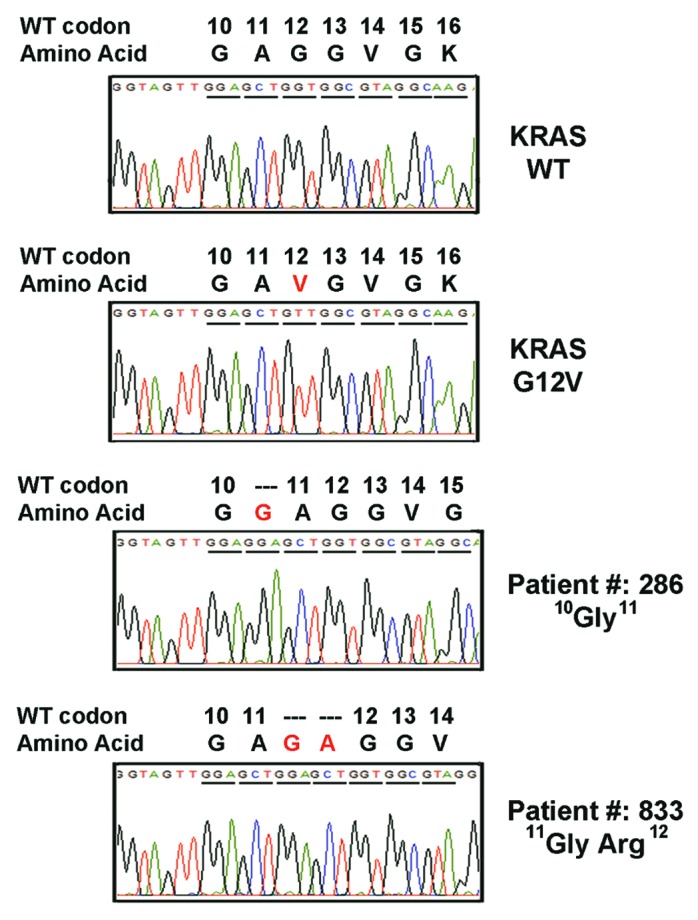
Figure 1. Electropherogram for KRAS mutants. Tissue DNA from the patient with colorectal cancer were amplified and cloned for sequencing analysis. Two novel in-flame insertions (10G11 and 11GA12) in exon 2 of KRAS gene were identified.
KRAS exon 2 insertions activate RAS signaling pathway and enhance NIH3T3 cells transformation
To investigate whether the newly found 10G11 and 11GA12 KRAS mutation activate RAS activity, we constructed expression plasmids and transiently transfected into 293FT and NIH3T3 cells. As a control, expression plasmids carrying wild-type KRAS (KRAS-WT) and a well-known active KRAS mutant (KRAS-G12V) were used for comparison during the basic functional assay.
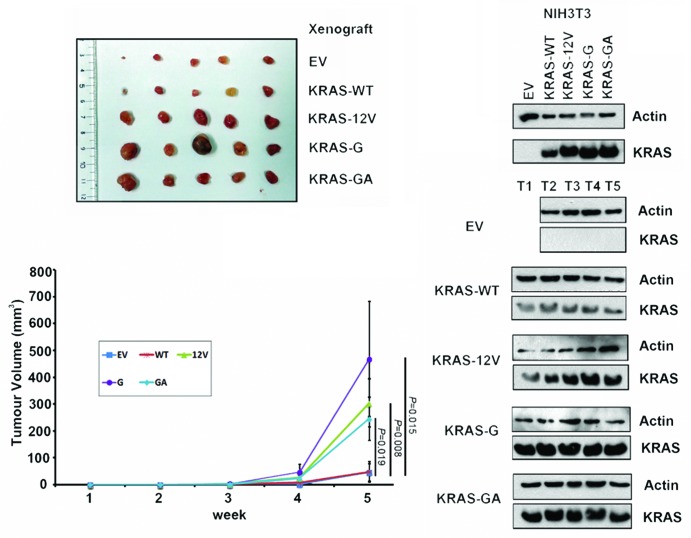
Compared with the cells transfected with WT expression plasmid, overexpression of 10G11 and 11GA12 KRAS mutants in cell lines resulted in elevated protein levels of both active RAS (Ras-GTP) and its downstream signaling molecule, phosphorylated extracellular signal-regulated kinase (p-ERK). The elevated levels of these two proteins are similar to the cells transfected with the KRAS-G12V mutant construct (Fig. 2). To further demonstrate the biological effect of 10G11 and 11GA12 KRAS mutants, NIH3T3 cells which stably transfected with empty vector, KRAS-WT, KRAS-G12V, 10G11 or 11GA12 mutant were prepared. Although NIH3T3 stable transfectants showed similar proliferation rate in MTT assay (data not shown), they have apparent differences in anchorage-independent growth property. We demonstrated in soft-agar colony formation assay that only a few number of colonies of the cells transfected with either empty vector or KRAS-wild type expression vector were observed. In contrast, more colonies were counted in all three transfectants with mutant KRAS and the differences were statistically significant compared with cells transfected with KRAS-wild type (Fig. 3). Furthermore, the colony sizes of the mutant KRAS transfectants were, in general, bigger than that in KRAS-wild type transfectant. To assess the in vivo tumorigenicity of novel KRAS variants, NIH3T3 transfectants containing empty vector or different KRAS mutants were injected subcutaneously into the dorsal flank of Balb/c nude mice. Compared with KRAS wild type and empty vector controls, KRAS 10G11 and 11GA12 significantly enhanced in vivo tumor growth as showed in Figure 4. Collectively, these observations suggested that both newly identified KRAS mutants could activate the Raf-MEK-ERK pathway by elevating RAS-GTP level and contribute in vitro and in vivo cell transformation.
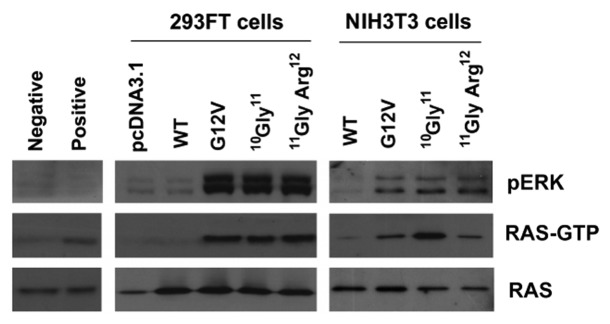
Figure 2.KRAS insertion mutants activated RAS signaling by enhancing cellular accumulation of active RAS (RAS-GTP) and activating p-ERK. NIH3T3 and 293FT cells were transfected with KRAS mutants, and RAS-GTP protein in the cell extract were immunoprecipitated with agarose beads containing Ras binding domain of Raf-1. Protein levels in both whole cell extracts (pan-RAS and pERK) and precipitated samples (RAS-GTP) were analyzed by western blot analysis as indicated. Representative results from 3 independent experiments were shown.
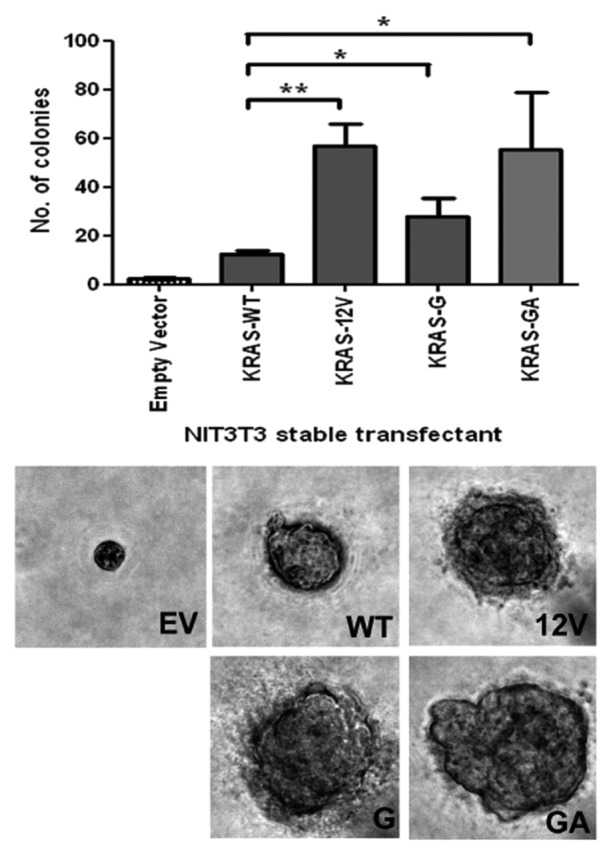
Figure 3.KRAS insertion mutants promoted anchorage-independent growth in soft agar. NIH3T3 cells stably transfected with pcDNA3.1 empty vector (EV), wild-type KRAS (WT), G12V KRAS mutant (G12V), 10G11 and 11GA12 mutants were cultured in soft agar for analysis. Representative microscopic pictures of colony from each transfectant were taken (Magnification, 400×). The number of colony in each transfectant was plot in the bar chart and the results shown were mean and standard deviation from three independent experiments. The P value of < 0.05 and < 0.001 were denoted as * and ** respectively.
Figure 4.KRAS insertion mutants promoted in vivo growth of NIH3T3 cells. In vivo tumorgenic assay in nude mice showed that tumors formed in the sites implanted with NIH3T3 cells expressing KRAS mutants (G12V, 10G11, or 11GA12) were consistently larger than that implanted with wild-type KRAS (WT) and empty vector (EV) controls. By western blotting, the expression of KRAS protein in the NIH3T3 transfectants and tumors dissected from the xenografts (T1–T5) was detected.
Discussion
In the current study, we report the KRAS mutation frequency in a large cohort of patients with colorectal cancer in Hong Kong. KRAS mutation is found in 44.5% (670 out of 1506) of colorectal cancers. The mutation rate is similar to KRAS studies previously reported.13-20 Table 4 summarized the KRAS mutation rates and the distribution of mutants in representative studies. Codon 12 is the most common KRAS mutation and the most frequently found mutation is G12D (35% of all mutations found). Our data demonstrate the predominance of KRAS-mutant carcinoma in right colon and in female patients. This is in keeping with some previous reports although other studies might not have demonstrated such relationship.21,22 The preference of site of KRAS mutation might be correlate with the different molecular pathways involved in right and left side colon CRCs. The right and left side colon cancers have been considered as distinct tumor entities because of their epidemiological, clinicopathologic, and molecular biologic features. Right side colon cancer was found to be associated with female, older age, advanced stage, and poorly differentiated mucinous histology.23-26 Higher rates of microsatellite instability and KRAS mutations were common molecular events found in right side colon cancer.27,28 Whereas the left side tumor were more common to be chromosomal instable and harbor more TP53 mutation.27-30 The reason for the observed differences between left and right side colon adenocarcinoma remains unclear. It is likely to be multifactorial and complex including embryologic origin, and the effect of chemical and bacterial luminal microenvironments. Moreover, we have reported the predominant KRAS mutations in left colon are located in codon 12 and right colon in codon 13. This finding is different from a large population-based study which found significantly more codon-12 mutation cases in proximal (right colon) than distal (left colon) tumors (29.1% vs 20.5%; P < 0.01).21 Another study also showed rectosigmoid tumor (left colon) had the highest frequency of codon 13 mutations.31 There is no consistent trend, further study is necessary.
Table 4. Comparison of KRAS mutation distribution in reported series.
| Studies | Current study | COSMIC database | Rosty 201323 | Imamura 201241 | De Roock 201042 | Chang 200943 | Karapetis 200844 | Amado 20087 | Brink 200331 | Samowitz 200021 | Andreyev 199811 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| n = | 1506 | 17316 | 776 | 1261 | 747 | 228 | 394 | 427 | 737 | 1416 | 2214 |
| Mutation rate % | 44.5 | 34.9 | 28 | 35.8 | 36.3 | 36.4 | 41.6 | 43.1 | 36.8 | 31.8 | 37.7 |
| Relative mutation distribution (%) by codon | |||||||||||
| Codon 12 | 75.1 | 79.3 | 87 | 74.6 | 69.3 | 69.9 | 63.8 | 84.2 | 70 | 77.9 | 54 |
| Codon 13 | 19.1 | 17.6 | 13 | 25.4 | 20.1 | 25.3 | 11.7 | 15.8 | 21.6 | 22.1 | 16.7 |
| Codon 61 | 2.5 | 0.58 | 5.3 | 1.2 | |||||||
| Codon 146 | 2.7 | 0.19 | 5 | 2.4 | |||||||
| Relative mutation distribution (%) by nucleotide substitution | |||||||||||
| G12D | 37.6 | 35 | 161 | 35.2 | 27.4 | 35.7 | 38 | 26.1 | 31.1 | 30.6 | |
| G12V | 20.0 | 21.5 | 95 | 20.8 | 19.8 | 28.1 | 21.7 | 24.4 | 21.4 | 23.4 | |
| G12C | 6.7 | 8.3 | 44 | 9.6 | 7.3 | 7.6 | 5.9 | 9.5 | |||
| G12S | 4.9 | 6.3 | 12 | 2.6 | 6.3 | 7.6 | 5.6 | 6.8 | |||
| G12A | 4.3 | 6.7 | 20 | 4.4 | 6.9 | 8.2 | 5.6 | 3.5 | |||
| G12R | 1.5 | 1.1 | 8 | 1.8 | 1.7 | 1.6 | 2.4 | 0.7 | |||
| G13D | 18.8 | 17.4 | 110 | 24.1 | 20.1 | 11.7 | 15.8 | 20.2 | 20.8 | 16.7 | |
| G13C | 0.3 | 3 | 0.7 | 0.3 | 0.4 | ||||||
| Q61H | 1.3 | 0.3 | 2.3 | ||||||||
| Q61L | 0.7 | 0.2 | 1 | ||||||||
| Q61R | 0.3 | 0.1 | 1.3 | ||||||||
| A146T | 2.7 | 0.2 | 5 | ||||||||
We report two rare in-frame insertion mutations in this study, c.30_31insGGA: p.G10_A11insG (duplication of codon 10) and c.33_34insGGAGCT:p.A11_G12insGA (duplication of codon 10–11). In-frame Insertion mutations in KRAS are rarely reported. Almost all reported KRAS in-frame insertions are tandem duplications. Three-nucleotide insertions resulting in codon 9, codon 10, and codon 12 duplications have been reported in colorectal cancer and leukemia.8,32-34 A Netherland cohort study found a duplication of six nucleotides in a colorectal tumor, leading to two additional amino acids added in codon 9 of KRAS.31 A 15-bp insertion in exon 3 that resulted in tandem duplication of codons 62–66 has been found in a case of primary lung adenocarcinoma.35 Another study also reported the identical 15-bp in-frame insertion mutation in a colorectal carcinoma.36
Wild-type KRAS regulate cellular growth and differentiation by cycling between inactive GDP-bound form (Ras-GDP) and active GTP-bound form (Ras-GTP). Mutant KRAS is defective in intrinsic GTP hydrolysis. Therefore, it is accumulated in cells in active GTP-bound form, resulting in constitutive activation of downstream signaling through effector proteins. Both insertion mutations found in the current study (10G11 and 11GA12) affect the phosphate-binding loop (codon 10–16) of KRAS. Our in vitro functional analyses have confirmed that similar to the KRAS mutant G12V, both rare mutants enhance the cellular accumulation of active RAS (Ras-GTP), and activate the Raf-MEK-ERK pathway. Using soft agar assays, we demonstrate the ability of both insertion variants in driving in vitro cell transformation. We also show that both insertion mutants demonstrate enhanced tumorigenicity in nude mice. Our finding is concordant with previous in vivo analysis of KRAS 10Gly11 mutation in acute leukemia which showed duplication of amino acid residue in codon 12 could lead to the activation of KRAS.8 In addition, another RAS protein member, HRAS with an insertion mutation in codon 12 was reported to gain the ability in cell transformation.8 These results suggest that both point mutation and insertion mutation within codon 12 and sites nearby could activate RAS protein through interrupting the GTP binding site of RAS family protein.
In summary, this study has provided a KRAS mutation database in colorectal cancer of local Chinese population and the correlation between KRAS status with gender and primary site in the colon. Furthermore, we report the identification and characterization of two rare KRAS insertion mutations. In vitro and in vivo functional studies confirm the oncogenic properties of these insertion mutations. KRAS mutations beyond the “hotspots” can be oncogenic by conveying selective growth advantage to the cells. These mutations might potentially contribute to primary resistance for anti-EGFR mAb targeted therapy. The clinical implication for these mutations requires further validation.
Materials and Methods
Patient sample
A total of 1506 consecutive colorectal adenocarcinoma specimens sent for KRAS mutational analysis in Prince of Wales Hospital, Hong Kong between 2008 and 2012 were included in this study. The study protocol was approved by the Joint CUHK-NTE Clinical Research Ethics Committee, Hong Kong.
Tumor DNA extraction
The location of tumor cells in the formalin-fixed, paraffin-embedded (FFPE) tissue were first marked on the standard H&E-stained histological slides. Subsequently, the corresponding tumor tissues on the unstained glass slide were microdissected manually for DNA extraction using QIAamp DNA tissue mini kit with standard procedure (Qiagen).
Sequencing analysis
Mutational hot spots including KRAS codons 12, 13, 61, and 146 were investigated by PCR-direct sequencing. PCR reactions were performed using primers listed in Table 5. Cycling sequencing reaction of the PCR fragments was performed with BigDye Terminator system (Applied Biosystems) using primers from both directions. The sequencing results were analyzed with the ABI PRISM® 3130XL Genetic Analyzer (Applied Biosystems). The data was collected and analyzed using Applied Biosystems sequencing analysis software.
Table 5. The sequences of oligonucleotides used in this study.
| PCR primers | Forward sequence | Reverse sequence |
|---|---|---|
| KRAS codon 12/13 | GTATTAACCT TATGTGTGAC A | GTCCTGCACC AGTAATATGC |
| KRAS codon 61 | TGCACTGTAA TAATCCAGAC TGTG | TGCACTGTAA TAATCCAGAC TGTG |
| KRAS codon 146 | TCTGAAGATG TACCTATGGT CCTAGT | AAGAAGCAAT GCCCTCTCAA |
| Mutagenesis primers | ||
| KRAS-WT | 5′-GGTAGTTGGA GCTGGTGGCG TAGGCAAGA-3′ | 5′- TCTTGCCTAC GCCACCAGCT CCAACTACC-3′ |
| KRAS-10G11 | 5′-GTGGTAGTTG GAGGAGCTGG TGGCGTAGGC AAG-3′ | 5′-CTTGCCTACG CCACCAGCTC CTCCAACTAC CAC-3′ |
| KRAS-11GA12 | 5′-GGTAGTTGGA GCTGGAGCTG GTGGCGTAGG CAAG-3′ | 5′-CTTGCCTACG CCACCAGCTC CAGCTCCAAC TACC-3′ |
Detection of the precise sequence of the rare mutation
PCR product corresponding to KRAS exon 1 was amplified from the patient genomic DNA and subsequently cloned using the TOPO-TA Cloning kit (Invitrogen). Ten colonies of each transformation were randomly selected for sequencing analysis.
Cell culture and transfection
Human embryonic kidney cells (293FT) and mouse embryonic fibroblast cells (NIH3T3) were obtained from Invitrogen and American Type Culture Collection (ATCC) respectively. Both cell lines were cultured in Dulbecco modified Eagle medium plus 10% FBS (Gibco, Invitrogen). Transfection of 293FT and NIH3T3 cells were performed using LipofectamineTM LTX reagent (Invitrogen) following the manufacturer’s protocol.
Site-direct mutagenesis and active RAS measurement
Full-length of KRAS cDNA was cut from pBabe K-Ras 12V vector (Addgene plasmid 12544)37 and cloned into pcDNA3.1 (+) expression vector (Invitrogen) via BamH1 and Xba1 restriction sites. Corresponding KRAS mutations were introduced into the expression vector using QuickChange® II Site-Directed Mutagenesis Kit according to the manufacturer’s recommendations (Stratagene). The desired mutations in each construct were finally confirmed by direct sequencing. The primer sequences for mutagenesis were listed in Table 5. Ras Activation Assay Kit (Millipore) was used to measure the level of active RAS (RAS-GTP) after transient transfection of corresponding plasmid into the cell lines. In brief, 0.5 mg of cell extract was immunoprecipitated with agarose beads containing human Ras Binding Domain (RBD, residues 1–149) of Raf-1. After washing, the beads were mixed with protein loading buffer and 10% of the mixture was electrophoresed by 12% SDS-PAGE for western blot analysis as previously described.38,39 The primary antibodies used were pan-RAS (RAS10, Millipore; 1:2000) and p-ERK1/2 (9102, Cell Signaling; 1:1000). HRP conjugated anti-mouse secondary antibody used was purchased from DAKO (1:20000 dilution).
Soft agar colony formation assay
NIH3T3 cells transfected with corresponding KRAS expression plasmids were selected in culture medium containing 400 μg/mL of G418 (Invitrogen) for one month before preparing colony formation assay. In the assay, culture medium containing 0.7% agarose was set as a bottom layer in 6-well dishes. A total of 3000 cells, which mixed with culture medium containing 0.35% agarose, were added over the bottom layer. After 25 d of incubation, colonies were stained with 0.005% crystal violet overnight and were counted under dissection microscope. Each experiment was performed in triplicate.
In vivo tumorigenicity
NIH3T3 transfectants (1 × 106 cells suspended in 0.1 mL phosphate-buffered saline), containing empty vector or different KRAS mutant, were injected subcutaneously into the dorsal flank of five 5-wk-old male Balb/c nude mice. The tumor volume was determined as previously described.40 All experimental procedures were approved by the Animal Ethics Committee of the Chinese University of Hong Kong.
Statistical analysis
Statistical analysis of two times two contingency tables of categorical variables was performed using the Chi-square test or Fisher exact test, as appropriate. The t test was performed to compare continuous variables between two groups. All statistical analyses were performed by using statistical program SPSS version 16.0. A two-tailed P value of <0.05 was regarded as statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Glossary
Abbreviations:
- mCRC
metastatic colorectal carcinoma
- mAbs
monoclonal antibodies
- EGFR
epidermal growth factor receptor
- FFPE
formalin-fixed, paraffin-embedded
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010;51:71–119. doi: 10.1016/S0065-2423(10)51004-7. [DOI] [PubMed] [Google Scholar]
- 3.Brand TM, Wheeler DL. KRAS mutant colorectal tumors: past and present. Small GTPases. 2012;3:34–9. doi: 10.4161/sgtp.18751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–8. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 5.Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–9. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 6.Jimeno A, Messersmith WA, Hirsch FR, Franklin WA, Eckhardt SG. KRAS mutations and sensitivity to epidermal growth factor receptor inhibitors in colorectal cancer: practical application of patient selection. J Clin Oncol. 2009;27:1130–6. doi: 10.1200/JCO.2008.19.8168. [DOI] [PubMed] [Google Scholar]
- 7.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 8.Bollag G, Adler F, elMasry N, McCabe PC, Conner E, Jr., Thompson P, McCormick F, Shannon K. Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem. 1996;271:32491–4. doi: 10.1074/jbc.271.51.32491. [DOI] [PubMed] [Google Scholar]
- 9.Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol. 2012;30:3570–7. doi: 10.1200/JCO.2012.42.2592. [DOI] [PubMed] [Google Scholar]
- 10.Edkins S, O’Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5:928–32. doi: 10.4161/cbt.5.8.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90:675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 12.Zulhabri O, Rahman J, Ismail S, Isa MR, Wan Zurinah WN. Predominance of G to A codon 12 mutation K-ras gene in Dukes’ B colorectal cancer. Singapore Med J. 2012;53:26–31. [PubMed] [Google Scholar]
- 13.Russo A, Bazan V, Agnese V, Rodolico V, Gebbia N. Prognostic and predictive factors in colorectal cancer: Kirsten Ras in CRC (RASCAL) and TP53CRC collaborative studies. Ann Oncol. 2005;16(Suppl 4):iv44–9. doi: 10.1093/annonc/mdi907. [DOI] [PubMed] [Google Scholar]
- 14.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–7. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 15.Kim ST, Park KH, Kim JS, Shin SW, Kim YH. Impact of KRAS Mutation Status on Outcomes in Metastatic Colon Cancer Patients without Anti-Epidermal Growth Factor Receptor Therapy. Cancer Res Treat. 2013;45:55–62. doi: 10.4143/crt.2013.45.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrelli F, Coinu A, Cabiddu M, Ghilardi M, Barni S. KRAS as prognostic biomarker in metastatic colorectal cancer patients treated with bevacizumab: a pooled analysis of 12 published trials. Med Oncol. 2013;30:650. doi: 10.1007/s12032-013-0650-4. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe T, Yoshino T, Uetake H, Yamazaki K, Ishiguro M, Kurokawa T, Saijo N, Ohashi Y, Sugihara K. KRAS mutational status in Japanese patients with colorectal cancer: results from a nationwide, multicenter, cross-sectional study. Jpn J Clin Oncol. 2013;43:706–12. doi: 10.1093/jjco/hyt062. [DOI] [PubMed] [Google Scholar]
- 18.Adelstein BA, Dobbins TA, Harris CA, Marschner IC, Ward RL. A systematic review and meta-analysis of KRAS status as the determinant of response to anti-EGFR antibodies and the impact of partner chemotherapy in metastatic colorectal cancer. Eur J Cancer. 2011;47:1343–54. doi: 10.1016/j.ejca.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 19.Mao C, Zhou J, Yang Z, Huang Y, Wu X, Shen H, Tang J, Chen Q. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One. 2012;7:e36653. doi: 10.1371/journal.pone.0036653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yunxia Z, Jun C, Guanshan Z, Yachao L, Xueke Z, Jin L. Mutations in epidermal growth factor receptor and K-ras in Chinese patients with colorectal cancer. BMC Med Genet. 2010;11:34. doi: 10.1186/1471-2350-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9:1193–7. [PubMed] [Google Scholar]
- 22.Elnatan J, Goh HS, Smith DR. C-KI-RAS activation and the biological behaviour of proximal and distal colonic adenocarcinomas. Eur J Cancer. 1996;32A:491–7. doi: 10.1016/0959-8049(95)00567-6. [DOI] [PubMed] [Google Scholar]
- 23.Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, Pavluk E, Nagler B, Pakenas D, Jass JR, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26:825–34. doi: 10.1038/modpathol.2012.240. [DOI] [PubMed] [Google Scholar]
- 24.Lin JK, Chang SC, Wang HS, Yang SH, Jiang JK, Chen WC, Lin TC, Li AF. Distinctive clinicopathological features of Ki-ras mutated colorectal cancers. J Surg Oncol. 2006;94:234–41. doi: 10.1002/jso.20438. [DOI] [PubMed] [Google Scholar]
- 25.Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, Arber DA, Balise RR, Tubbs RR, Shadrach B, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36:744–52. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benedix F, Kube R, Meyer F, Schmidt U, Gastinger I, Lippert H, Colon/Rectum Carcinomas (Primary Tumor) Study Group Comparison of 17,641 patients with right- and left-sided colon cancer: differences in epidemiology, perioperative course, histology, and survival. Dis Colon Rectum. 2010;53:57–64. doi: 10.1007/DCR.0b013e3181c703a4. [DOI] [PubMed] [Google Scholar]
- 27.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–88. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 28.Sugai T, Habano W, Jiao YF, Tsukahara M, Takeda Y, Otsuka K, Nakamura S. Analysis of molecular alterations in left- and right-sided colorectal carcinomas reveals distinct pathways of carcinogenesis: proposal for new molecular profile of colorectal carcinomas. J Mol Diagn. 2006;8:193–201. doi: 10.2353/jmoldx.2006.050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soong R, Powell B, Elsaleh H, Gnanasampanthan G, Smith DR, Goh HS, Joseph D, Iacopetta B. Prognostic significance of TP53 gene mutation in 995 cases of colorectal carcinoma. Influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer. 2000;36:2053–60. doi: 10.1016/S0959-8049(00)00285-9. [DOI] [PubMed] [Google Scholar]
- 30.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–8. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 31.Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruïne AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–10. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 32.Reimann C, Arola M, Bierings M, Karow A, van den Heuvel-Eibrink MM, Hasle H, Niemeyer CM, Kratz CP. A novel somatic K-Ras mutation in juvenile myelomonocytic leukemia. Leukemia. 2006;20:1637–8. doi: 10.1038/sj.leu.2404303. [DOI] [PubMed] [Google Scholar]
- 33.Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, Palmi C, Carta C, Pession A, Aricò M, et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104:307–13. doi: 10.1182/blood-2003-11-3876. [DOI] [PubMed] [Google Scholar]
- 34.Servomaa K, Kiuru A, Kosma VM, Hirvikoski P, Rytömaa T. p53 and K-ras gene mutations in carcinoma of the rectum among Finnish women. Mol Pathol. 2000;53:24–30. doi: 10.1136/mp.53.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmid K, Oehl N, Wrba F, Pirker R, Pirker C, Filipits M. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–60. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 36.Wójcik P, Kulig J, Okoń K, Zazula M, Moździoch I, Niepsuj A, Stachura J. KRAS mutation profile in colorectal carcinoma and novel mutation--internal tandem duplication in KRAS. Pol J Pathol. 2008;59:93–6. [PubMed] [Google Scholar]
- 37.Khosravi-Far R, White MA, Westwick JK, Solski PA, Chrzanowska-Wodnicka M, Van Aelst L, Wigler MH, Der CJ. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–33. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong JH, Ng DC, Chau SL, So KK, Leung PP, Lee TL, Lung RW, Chan MW, Chan AW, Lo KW, et al. Putative tumour-suppressor gene DAB2 is frequently down regulated by promoter hypermethylation in nasopharyngeal carcinoma. BMC Cancer. 2010;10:253. doi: 10.1186/1471-2407-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lung RW, Tong JH, Sung YM, Leung PS, Ng DC, Chau SL, Chan AW, Ng EK, Lo KW, To KF. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174–84. doi: 10.1593/neo.09888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kang W, Tong JH, Chan AW, Lee TL, Lung RW, Leung PP, So KK, Wu K, Fan D, Yu J, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17:2130–9. doi: 10.1158/1078-0432.CCR-10-2467. [DOI] [PubMed] [Google Scholar]
- 41.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, Qian ZR, Nishihara R, Meyerhardt JA, Haigis KM, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18:4753–63. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 43.Chang YS, Yeh KT, Chang TJ, Chai C, Lu HC, Hsu NC, Chang JG. Fast simultaneous detection of K-RAS mutations in colorectal cancer. BMC Cancer. 2009;9:179. doi: 10.1186/1471-2407-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]