Abstract
Heterochromatin is the enigmatic eukaryotic genome compartment found mostly at telomeres and centromeres. Conventional approaches to sequence assembly and genetic manipulation fail in this highly repetitive, gene-sparse, and recombinationally silent DNA. In contrast, genetic and molecular analyses of euchromatin-encoded proteins that bind, remodel, and propagate heterochromatin have revealed its vital role in numerous cellular and evolutionary processes. Utilizing the 12 sequenced Drosophila genomes, Levine et al1 took a phylogenomic approach to discover new such protein “surrogates” of heterochromatin function and evolution. This paper reported over 20 new members of what was traditionally believed to be a small and static Heterochromatin Protein 1 (HP1) gene family. The newly identified HP1 proteins are structurally diverse, lineage-restricted, and expressed primarily in the male germline. The birth and death of HP1 genes follows a “revolving door” pattern, where new HP1s appear to replace old HP1s. Here, we address alternative evolutionary models that drive this constant innovation.
Keywords: Heterochromatin Protein 1, gene duplication, germline, phylogenomics, heterochromatin, chromodomain, chromoshadow domain, pseudogenization, Drosophila, HP1
HP1: A Flashlight for the Dark Matter of Eukaryotic Genomes
The human genome sequence is not complete.2 Neither are the Mus musculus,3 Drosophila melanogaster4 nor Arabidopsis thaliana5 genomes. Indeed, “complete genome sequence” assemblies from many eukaryotes may be missing up to 30% of nuclear-encoded DNA. The unassembled genome compartment is mostly comprised of heterochromatin—packed with satellites and transposable elements, but relatively sparse in protein-coding genes and recombination events. The challenges of assembling repetitive DNA and genetic mapping to regions of low recombination, combined with a mistaken perception that heterochromatin harbors few functional elements or no genes, has contributed to decades of scientific neglect. With the advent of new sequencing technologies and painstaking efforts of large consortia of researchers (e.g., the Drosophila Heterochromatin Genome Project4), this slight is being slowly corrected with significant advances in heterochromatin function and evolution.
Despite renewed interest in the study of heterochromatin DNA sequence, most insights into heterochromatin function have emerged from genetic and molecular studies of euchromatin-encoded proteins that affect heterochromatin properties, especially the expression of proximal genes in the phenomenon of position-effect-variegation (PEV).6 These and subsequent studies led to the awareness that heterochromatin participates in many essential cellular processes, including chromosome segregation,7 genome defense,8 and gene regulation,9 transforming the scientific community from disinterest to broad appreciation of heterochromatin’s biological significance. The resulting picture revealed that rather than representing a sea of functionally uninteresting homogeneous repeats, heterochromatin contains many disparate elements of varied functions (e.g., piwi-associated RNA, or piRNA, clusters for genome defense8).
Our understanding of heterochromatin function was transformed by a pivotal 1986 publication describing D. melanogaster’s Heterochromatin Protein 1 (HP1).10 Using monoclonal antibodies against a protein fraction tightly bound to DNA, James and Elgin uncovered a chromosomal protein that localizes primarily to pericentric heterochromatin and was encoded by Su(var)2–511, one of the genes previously shown to suppress PEV. Subsequent analysis of HP1 (now called “HP1A”) and its numerous interacting partners illuminated an unprecedented number of heterochromatin-dependent essential processes. Centromere maintenance,12 RNA interference,13 and telomere protection,14 for example, all rely on HP1-dependent heterochromatin integrity. This progress highlights the power of heterochromatin-bound proteins as molecular tools to reveal new roles for this ubiquitous but cryptic genome compartment.
More recently, these heterochromatin-bound proteins have been used to reveal the evolutionary forces that may act on the rapidly evolving but unassembled, sometimes undefined heterochromatic sequence to which they bind.15 For example, the finding that a female germline-restricted HP1 protein, Rhino/HP1D, had evolved under positive selection (faster than expected amino acid divergence) predicted its engagement in a molecular arms-race with transposable elements.16 This prediction was supported by the finding that Rhino binds heterochromatin-embedded, rapidly evolving piRNA clusters,17 which themselves likely evolve under positive selection18 to “immunize” genomes against new invasions of transposable elements. Thus, rigorous population genetic and molecular evolution analyses on heterochromatin protein “surrogates” could reveal evolutionary dynamics at the repetitive heterochromatin sequence for which such tests are undeveloped.
HP1 Phylogenomics: Expanding the Family Business
We set out to discover new “heterochromatin surrogates” for both functional and evolutionary analysis. We focused our analysis on the gene family founded by HP1A.a At first glance, the HP1 gene family seems like a poor target for phylogenomics. For many years, it was considered small, static, and structurally homogenous.19 HP1 family members are traditionally defined by a single, common domain structure—a chromodomain, hinge, and chromoshadow domain20 (Fig. 1). Using this definition, HP1B and HP1C were discovered in the newly sequenced D. melanogaster genome in 2000,21 bringing the family size up to three. Additional HP1s in non-Drosophila genomes were also identified, including three in humans (HP1α, HP1β, HP1γ),22-24 which are not orthologous to any of the Drosophila HP1 genes25 and instead likely derived from an HP1B-like ancestor.25
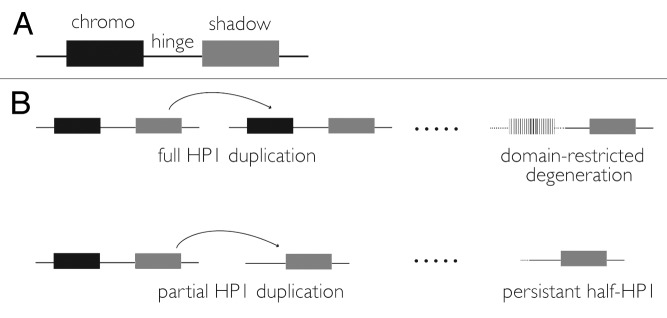
Figure 1. (A) Canonical HP1 domain structure. The chromodomain (”chromo”) recognizes H3K9me, the hinge binds DNA and/or RNA, and the chromoshadow (”shadow”) homodimerizes and heterodimerizes. (B) Alternative paths underlying half-HP1 birth. Drift or selection drives the degeneration of the chromodomain (in this example) following a full HP1 duplication event. Alternatively, the duplication itself is restricted to a single domain. A 3′ retrotransposition bias43 may underlie the enrichment of chromoshadow domain-only HP1s observed in our data set.1
At this time, a family size of three was the maximum number across any eukaryote, including yeast, worms, mouse, and Arabidopsis.19 Furthermore, all three Drosophila HP1s and all three mammalian HP1s are highly conserved across broad evolutionary distances.26 Remarkably, the human HP1α can rescue D. melanogaster HP1A-dependent loss of silencing27 despite paralogy. These observations supported the idea that functional and evolutionary stasis is a defining feature of this small family. However, the fortuitous discovery of a new D. melanogaster HP1 in a female sterility screen,28 called “rhino,” together with its signature of strong positive selection,16 suggested that HP1s are potentially more numerous and more plastic than originally thought. The subsequent discovery of a fifth HP116 in D. melanogaster, HP1E, supported this prediction and motivated our comprehensive phylogenomic analysis.
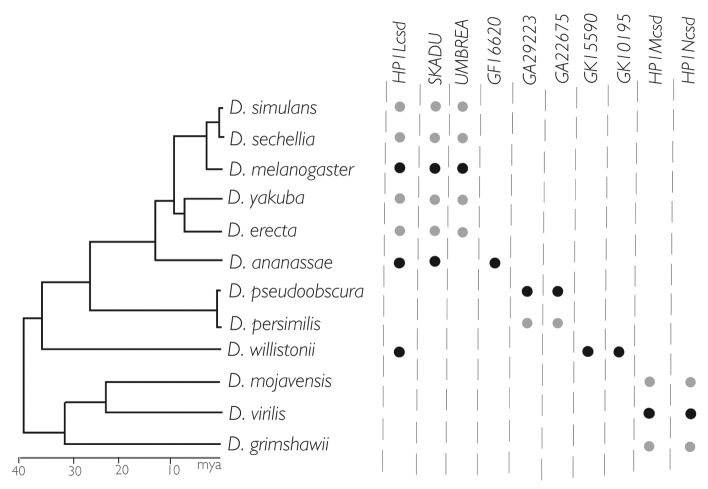
HP1A through HP1E served as entry points into characterizing the HP1 gene family in the recently sequenced 12 Drosophila genomes, which represent over 40 million years of Drosophila evolution.29 Using all five chromodomains and chromoshadow domains as queries, we computationally searched the 12 genomes for significant tBLASTn hits. Our iterative search strategy, in which any significant domain hit becomes a search query itself, returned over 100 hits across this 40 million year snapshot. Using the evolutionary definition of “gene family,” we conditioned membership of these hits into the HP1 family based on phylogenetic relationships. Combined with information about synteny, we identified orthologous, paralogous, and anciently diverged members relative to known HP1s. Orthologs have diverged through speciation events; thus, they are both syntenic (in the same genomic location) and more closely related to each other than to other HP1 genes. For example, the HP1A alleles from all 12 genomes form a single monophyletic (single evolutionary origin) clade. Paralogs represent sister clades; for example, the HP1G and HP1A gene clades share a common ancestor that is more than 40 million years old. We also identified younger paralogs that cluster within older HP1s, like the relatively young, HP1D/Rhino-derived Oxpecker genes. These represent cases of more recent gene duplication events within pre-existing HP1 clades. Finally, a few paralogs defied groupings within old or new HP1 gene clades, representing either ancient origins or very rapid divergence (they still share stronger identity to HP1 chromo or shadow domains than to any other Drosophila proteins).
We found that HP1A, HP1B, HP1C and HP1D orthologs occurred in all 12 genomes in the syntenic locations. However, HP1E orthologs had clearly degenerated in several species, implicating recurrent HP1E gene loss. This seemingly unique species-specificity of HP1E proved to be the rule for all new HP1 paralogs we discovered. Indeed, none of the other HP1 genes discovered are present in all 12 genomes. Virtually all of these newly identified HP1s evolved within the last 40 million years and so appear in only a restricted set of lineages.
Even more unexpectedly, while the canonical HP1 domain structure is defined by the presence of both a chromodomain and a chromoshadow domain (Fig. 1A), the majority of new HP1 family members encoded only one of these domains, having lost or degenerated either the original chromo or chromoshadow domain during or after duplication (Fig. 1B). We initially disregarded these “half-HP1s” as duplicate genes caught in the act of pseudogenization. However, upon examining the syntenic locations of these half-HP1s, we confirmed that many had been retained for many millions of years. Long-term retention is consistent with function, particularly in Drosophila where the half-life of pseudogenes is remarkably short.30 Further supporting this prediction, we found evidence of transcription for 18 of the 19 genes in our list. Intriguingly, virtually all are transcribed primarily in the male germline. Four genes are encoded in the well-annotated genome of D. melanogaster—one chromodomain-only HP1, and three chromoshadow-only HP1s (one of which we already know is essential31).
Our data are consistent with a minimum HP1 gene family size of 26 in the 12 Drosophila species sampled. This represents a 4-fold increase in HP1 gene number. Furthermore, the unprecedented structural diversity of the new HP1 family members offers a dramatically expanded toolkit for discovering new heterochromatin functions.b The pervasive lineage-restriction is consistent with species- or clade- specific adaptations that rely on young HP1 genes (below), and may offer insights into the evolutionary significance of the rampant between-species divergence observed at the heterochromatin sequence itself. Moreover, the predominance of male germline expression is consistent with currently uncharacterized male-specific chromosome biology driving this lineage-specific adaptation.
A ‘Revolving Door’ of HP1 Proteins in the Drosophila Male Germline
The male germline has recurrently emerged as a venue enriched for signatures of positive selection across many gene classes.32 These DNA signatures include statistical enrichment for new amino acid changes and retention of gene duplicates, the latter of which results in expansion of gene families. An agnostic analysis of Drosophila gene family evolution across the 12 genomes33 demonstrated that significant family expansions (and contractions) are enriched for male reproduction-related functions. Intriguingly, the abundant male germline-expressed HP1 paralogs are not part of gene family expansion in the strict sense. Despite rampant gene birth and death over the 40 million years, the number of HP1 genes in a given Drosophila species varies only modestly (for the chromoshadow-only class, see Figure 2). This gene number stasis, despite prolific gene birth, is consistent with functional gene replacements over time. Borrowing a term from Demuth and Hahn,34 we refer to this pattern as an HP1 gene family “revolving door.”
Figure 2. Revolving door dynamics: gene number stasis, recurrent birth, and recurrent death. The 10 chromoshadow-only genes represented are all expressed primarily in testis. Each lineage harbors either two or three HP1s of this domain class, but these genes are rarely shared across distant lineages. ● Expression assayed directly by tissue-restricted RTPCR. ● Expression inferred.
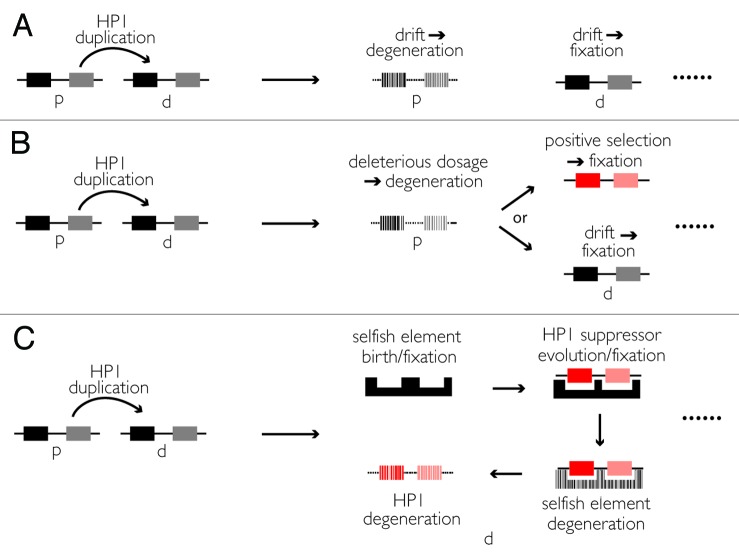
At a genome-wide scale, recurrent gene turnover is consistent with a neutral model of gene family evolution.35 Averaged across all genes, a steady-state birth/death process (that assumes an equilibrium genome size) readily accounts for gene turnover. Under this model, a gene duplication event generates a daughter copy that is ultimately retained while the parent copy accumulates mutations under genetic drift (Fig. 3A). At the level of a single gene family with elevated birth/death rates, however, this model is less satisfying; specifically, under neutrality the gene death rate varies independently of gene copy number. Chromatin proteins, and specifically HP1s, however, are typically dosage-sensitive.36 An extra gene copy can, for example, suppress or enhance heterochromatin spreading along a chromosome.37 We speculate that gene duplications of some chromatin-protein encoding loci are instantly visible to natural selection. Consequently, an HP1 gene death rate parameter may vary positively with gene copy number, which at least partially explains the gene family-wide revolving door pattern (Fig. 3B). This slight variation on Birchler’s “gene balance hypothesis”38 may also explain our observation that half-HP1s evolve exclusively from full HP1s. A mutation that breaks a chromodomain or chromoshadow domain in the full HP1’s daughter copy instantly relieves deleterious dosage-effects. Relief from deleterious dosage effects may free up the daughter copy to evolve along its own evolutionary trajectory.
Figure 3. Alternative forces driving gene replacements. (A) Birth and then fixation/death under neutral forces. (B) Birth and then fixation under neutral forces or positive selection, death driven by negative selection to relieve dosage effects, such as heterochromatin expansion/contraction or positive/negative transcriptional regulation.44-46 (C) Birth and then fixation under positive selection to suppress recurrently evolving selfish elements, death under neutral forces. “p” = parent gene, “d” = daughter gene.
In addition to this negative selection, positive selection may also explain the recurrent gene birth and death across the HP1 family (Fig. 3C). Heterochromatin is riddled with selfish elements.39,40 These genomic parasites gain a fitness advantage upon self-replication or drive in the germline where they have direct access to the next generation. Germline-restricted HP1s like the numerous Rhino/HP1D-derived chromodomain-only Oxpecker genes may suppress this selfish activity. Once successfully silenced, the selfish element and its suppressor degenerate. Recurrent bouts of selfish element birth and degeneration41 may explain at least some of this HP1 turnover in the male germline.
Just like the discovery and study of histone variants have greatly transformed our understanding of chromatin functions and states,42 analysis of this diverse toolkit of heterochromatin surrogates promises to reveal both currently unknown cellular roles for heterochromatin as well as the evolutionary forces that act on this still understudied genome compartment. Moreover, this kind of phylogenomic approach is gene family- and taxon- independent. As more and more complete genome sequencing data sets become available, we anticipate many more analyses that overturn false perceptions of stasis at gene families that encode essential proteins.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank B. Ross and S. Zanders for comments on the manuscript. Our work is supported by an NRSA fellowship F32-GM097897–02 (MTL) and NIH grant R01-GM74108 (HSM). HSM is an Early Career Scientist of the Howard Hughes Medical Institute.
Footnotes
We refer to the Su(var)2–5 gene as HP1A here purely for ease of referral and comparison to the other paralogous HP1 genes in Drosophila.
The HP1 family designation is an evolutionary classification; significant functional work needs to be done to ascertain whether any or all the new genes indeed encode heterochromatin-binding proteins
References
- 1.Levine MT, McCoy C, Vermaak D, Lee YC, Hiatt MA, Matsen FA, et al. Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family. PLoS Genet. 2012;8:e1002729. doi: 10.1371/journal.pgen.1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichler EE, Clark RA, She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat Rev Genet. 2004;5:345–54. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 3.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, et al. Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Smith CD, Shu S, Mungall CJ, Karpen GH. The Release 5.1 annotation of Drosophila melanogaster heterochromatin. Science. 2007;316:1586–91. doi: 10.1126/science.1139815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wortman JR, Haas BJ, Hannick LI, Smith RK, Jr., Maiti R, Ronning CM, et al. Annotation of the Arabidopsis genome. Plant Physiol. 2003;132:461–8. doi: 10.1104/pp.103.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Girton JR, Johansen KM. Chapter 1 Chromatin Structure and the Regulation of Gene Expression: The Lessons of PEV in Drosophila. In: Veronica van H, Robert EH, eds. Advances in genetics: Academic Press, 2008:1-43. [DOI] [PubMed] [Google Scholar]
- 7.Le HD, Donaldson KM, Cook KR, Karpen GH. A high proportion of genes involved in position effect variegation also affect chromosome inheritance. Chromosoma. 2004;112:269–76. doi: 10.1007/s00412-003-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–35. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elgin SC. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/S0959-437X(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 10.James TC, Elgin SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol Cell Biol. 1986;6:3862–72. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eissenberg JC, James TC, Foster-Hartnett DM, Hartnett T, Ngan V, Elgin SC. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1990;87:9923–7. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kellum R. HP1 complexes and heterochromatin assembly. Curr Top Microbiol Immunol. 2003;274:53–77. doi: 10.1007/978-3-642-55747-7_3. [DOI] [PubMed] [Google Scholar]
- 13.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–11. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capkova Frydrychova R, Biessmann H, Mason JM. Regulation of telomere length in Drosophila. Cytogenet Genome Res. 2008;122:356–64. doi: 10.1159/000167823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermaak D, Bayes JJ, Malik HS. A surrogate approach to study the evolution of noncoding DNA elements that organize eukaryotic genomes. J Hered. 2009;100:624–36. doi: 10.1093/jhered/esp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermaak D, Henikoff S, Malik HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–49. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Assis R, Kondrashov AS. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proc Natl Acad Sci U S A. 2009;106:7079–82. doi: 10.1073/pnas.0900523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissenberg JC, Elgin SC. The HP1 protein family: getting a grip on chromatin. Curr Opin Genet Dev. 2000;10:204–10. doi: 10.1016/S0959-437X(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 20.Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–73. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 22.Ye Q, Worman HJ. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem. 1996;271:14653–6. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- 23.Saunders WS, Chue C, Goebl M, Craig C, Clark RF, Powers JA, et al. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J Cell Sci. 1993;104:573–82. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 24.Singh PB, Miller JR, Pearce J, Kothary R, Burton RD, Paro R, et al. A sequence motif found in a Drosophila heterochromatin protein is conserved in animals and plants. Nucleic Acids Res. 1991;19:789–94. doi: 10.1093/nar/19.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vermaak D, Malik HS. Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet. 2009;43:467–92. doi: 10.1146/annurev-genet-102108-134802. [DOI] [PubMed] [Google Scholar]
- 26.Lomberk G, Wallrath L, Urrutia R. The Heterochromatin Protein 1 family. Genome Biol. 2006;7:228. doi: 10.1186/gb-2006-7-7-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norwood LE, Grade SK, Cryderman DE, Hines KA, Furiasse N, Toro R, et al. Conserved properties of HP1(Hsalpha) Gene. 2004;336:37–46. doi: 10.1016/j.gene.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA. Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics. 2001;159:1117–34. doi: 10.1093/genetics/159.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, et al. Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–18. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 30.Lozovskaya ER, Nurminsky DI, Petrov DA, Hartl DL. Genome size as a mutation-selection-drift process. Genes Genet Syst. 1999;74:201–7. doi: 10.1266/ggs.74.201. [DOI] [PubMed] [Google Scholar]
- 31.Chen S, Zhang YE, Long M. New genes in Drosophila quickly become essential. Science. 2010;330:1682–5. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–44. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 33.Hahn MW, Han MV, Han SG. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 2007;3:e197. doi: 10.1371/journal.pgen.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS One. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch M. The Origins of Genome Architecture. Sunderland: Sinauer Associates, 2007. [Google Scholar]
- 36.Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol. 2003;14:67–75. doi: 10.1016/S1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- 37.Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–52. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birchler JA, Veitia RA. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A. 2012;109:14746–53. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, et al. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci U S A. 1995;92:3804–8. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–6. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 41.Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A. 2001;98:13183–8. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 43.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 44.Cryderman DE, Grade SK, Li Y, Fanti L, Pimpinelli S, Wallrath LL. Role of Drosophila HP1 in euchromatic gene expression. Developmental dynamics: an official publication of the American Association of Anatomists 2005; 232:767-74. [DOI] [PubMed]
- 45.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, et al. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15:467–76. doi: 10.1016/j.molcel.2004.06.036. [DOI] [PubMed] [Google Scholar]
- 46.Riddle NC, Jung YL, Gu T, Alekseyenko AA, Asker D, Gui H, et al. Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet. 2012;8:e1002954. doi: 10.1371/journal.pgen.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]