Abstract
Appropriate gene expression relies on the sophisticated interplay between genetic and epigenetic factors. Histone acetylation and an open chromatin configuration are key features of transcribed regions and are mainly present around active promoters. Our recent identification of the SET-domain containing protein UpSET established a new functional link between the modulation of open chromatin features and active recruitment of well-known co-repressors in metazoans. Structurally, the SET domain of UpSET resembles H3K4 and H3K36 methyltransferases; however, it is does not confer histone methyltransferase activity. Rather than methylating histones to regulate gene expression like other SET domain-containing proteins, UpSET fine-tunes transcription by modulating the chromatin structure around active promoters resulting in suppression of expression of off-target genes or nearby repetitive elements. Chromatin modulation by UpSET occurs in part through its interaction with histone deacetylases. Here, we discuss the different scenarios in which UpSET could play key roles in modulating gene expression.
Keywords: UpSET, Rpd3, Sin3A, Set3C, Mixed Lineage Leukemia protein 5, histone acetylation, chromatin accessibility
Introduction
The functional and structural distinction between euchromatin and heterochromatin domains within eukaryotic genomes is crucial to the maintenance of transcriptional programs that drive development and differentiation in higher organisms. Nucleosomes are the fundamental building blocks that organize the genome into chromatin and allow its compaction in the nucleus. Transcriptionally active regions are targeted by transcription factors that are necessary to remodel the local promoter landscape and engage the transcriptional machinery for transcription. It is long known that chromatin containing active genes is more sensitive to DNaseI1 and that chromatin regions regulating gene expression (i.e., promoters and enhancers) display hypersensitivity to this enzyme.2-5 Nucleosomes act as structural barriers that block the access of these DNA binding proteins to regulatory elements.6,7 Consequently, active promoters tend to be nucleosome-free regions with high histone turnover, whereas nucleosomes are well positioned over non-active promoter regions.8,9 Histone acetylation is also a landmark of euchromatic regions and relies on the transfer of an acetyl group from Acetyl-Coenzyme A to the ε-N-lysine on target proteins by histone acetyltransferases (HATs).10 This post-translational modification can be removed by histone deacetylases (HDACs). The removal of this mark is crucial for maintaining low acetylation levels in heterochromatin and therefore has been associated historically with reduced gene repression. Thus, sparse nucleosome positioning, high histone acetylation levels and chromatin accessibility are structural landmarks of transcriptionally active regions.8,9,11,12 More recent global analyses of histone modifications, chromatin structure and chromatin protein composition have uncovered a highly complex landscape in which the transcriptional machinery performs its functions. While these studies have allowed chromatin to be subdivided into 5 to 9 different functional states (Fig. 1A),11,13 it is not yet known which of these chromatin features are causative vs. consequential of specific transcriptional responses.14
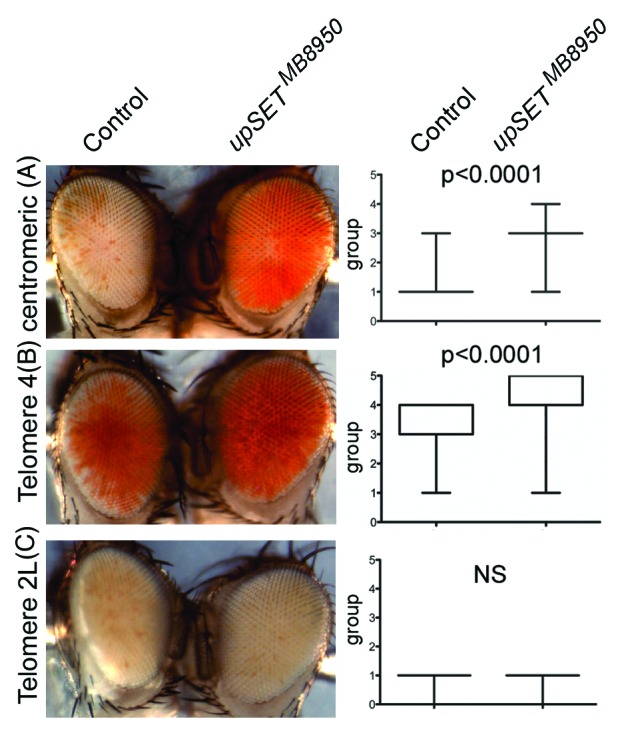
Figure 1. Transcriptional gene regulation. (A) Chromatin states based on chromatin profiling of 53 nuclear proteins. Black, Blue, and Green chromatin describe regions with low transcriptional activity and high abundance of repressive proteins, including Polycomb (Pc), Enhancer of zeste [E(z)], Lamin (Lam), Heterochromatin Protein 1 (HP1), Su(var)3–9, among others.13 Red and Yellow chromatin describe regions associated with transcription, which include histone acetyltransferases (CAF1), Histone deacetylases (RPD3), chromatin remodelers (BRM) and active chromatin marks (H3K4m3, H3K9Ac, H4K16Ac). (B) Chromatin features associated with transcribed regions that normally include high levels of RNA polymerase II and histone acetylation. In particular, active promoter regions exhibit higher sensitivity to DNaseI, higher amounts of H3K4m3 and lower nucleosome density than upstream or coding regions. Chromatin profiling of HATs and HDACs suggest that these antagonistic complexes co-exist in the Red and Yellow chromatin states in insects, although the functional role(s) associated with this co-localization is unclear.
Chromatin profiling of different co-activators and co-repressors has revealed a surprising overlap in their binding to chromatin. For example, despite their role in gene silencing, HDACs can also be found on transcribed regions overlapping with HATs (Fig. 1B).13,15 Indeed, during the last decade a more complicated functional picture for the role of HDACs in euchromatic regions has emerged. Cytological characterization of the Sin3A/Rpd3 complex on Drosophila polytene chromosomes has revealed that HDACs target the less-condensed interbands, but not heterochromatin, suggesting that this complex modulates histone acetylation in euchromatic regions.16 Genetic screening in yeast has shown that the Sin3/Rpd3 HDAC complex is required to achieve full repression and full activation of targets genes, including PHO, STE6 and TY2.17,18 The yeast Rpd3C deacetylase is found in two different complexes, Rpd3(L) and Rpd3(S), which target repressed promoters and transcribed coding sequences respectively.19 Elongation-associated histone acetylation is erased with the recruitment of Rpd3(S) over transcribed regions by either the RNA polymerase II-associated Set2 histone methyltransferase or the RNA polymerase itself.20-22 Histone deacetylation is thus required to reduce the chances of activating cryptic promoters.19 In metazoans, genome-wide analyses of HDAC binding sites have indicated that this machinery associates with repressed, as well as active, genes,13,15,23 suggesting that HDACs could be playing a role in re-setting the acetylation state. The mammalian Sin3B complex containing the Rpd3 ortholog HDAC1 was found to target chromatin during transcriptional elongation via the H3K36m3-specific chromodomain-containing protein Mrg15 and the PHD domain Pf1 protein.24 Interestingly, lack of these proteins leads to increased transcription and acetylation of four different genes examined. Together, these data suggest that histone acetylation is a tightly regulated process near transcribed regions. While HDAC recruitment to heterochromatic regions is thought to take place by direct interaction with transcription factors or chromatin-binding cofactors for repression,25,26 the mechanism(s) of its recruitment in euchromatic regions are unclear. Here, we discuss our recent work describing a conserved mechanism for the recruitment of HDAC complexes to active genes to modulate the chromatin landscape around promoter regions in metazoans.
UpSET Stabilizes the HDAC Rpd3 on Transcribed Regions
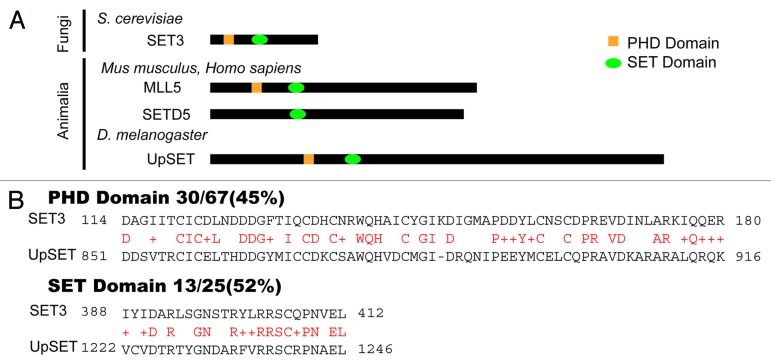
Drosophila is a model system often used to study chromatin biology from a developmental point of view, and in recent years genome-wide studies in insects have uncovered key mechanisms that govern gene expression in metazoans.11,12 We recently described UpSET (CG9007), a PHD and histone methyltransferase SET domain-containing protein, as the Drosophila ortholog of the mammalian Mixed Lineage Leukemia protein 5 (MLL5) and yeast Set3 (Fig. 2A).27,28 While all of these proteins contain methyltransferase domains, none of them exhibit histone methyltransferase activity, except for a small isoform of MLL5.29 We found that despite its targeting to active and inducible genes, UpSET interacts with the histone deacetylase Rpd3/Sin3A-containing complex such that it limits chromatin accessibility and histone acetylation to promoter regions.27 Interestingly, the human Mll5 gene is located in a region commonly deleted in several leukemias and may function as a tumor suppressor.30-35 Genome-wide RNAi screens have indirectly suggested that MLL5 is part the co-repressor NCoR complex, but no biochemical evidence supporting this assertion has yet been reported.36 Yeast Set3, initially characterized as a repressor of meiosis, forms a complex, Set3C, which includes the Hos2 and NAD-dependent Hst1 HDACs.28 Based on homology and complex composition, Set3 has also been suggested to be the ortholog of the metazoan NCoR/SMRT co-repressor.28
Figure 2. UpSET orthologs and conservation with yeast Set3. (A) The yeast Set3 protein is the founding member of the Set3 family, which also includes Drosophila UpSET and the MLL5 and SETD5 proteins in mammals. These proteins share a conserved SET domain and PHD domain, with the exception of SETD5 (which lacks the PHD domain). (B) PHD and SET domain conservation between Drosophila UpSET and yeast Set3. Amino acid comparison of the PHD and SET domains showing higher similarity over the PHD domain than over the SET domain. Red characters indicate the conserved residues.
UpSET provides a tractable genetic model in which to examine how metazoans modulate the interaction of HDACs over transcribed regions. Our results showed that UpSET preferentially binds transcribed regions around their transcriptional start sites (TSSs). These promoter regions are part of the distinct Red and Yellow transcriptionally active euchromatin states as defined previously by chromatin profiling 53 different proteins.13 In particular, chromatin profiling for UpSET, Rpd3, and Sin3A in Kc cells show high overlap around active genes. Strikingly, knock down of UpSET in these cells destabilizes the histone deacetylase Rpd3, which correlates with an increase in histone acetylation, in particular H3K9Ac and H4K16Ac, primarily around promoter regions. Although, UpSET and Rpd3/Sin3A co-immunoprecipitate, it is unclear which, if any, of these proteins physically interact. The specific reduction in Rpd3, but not Sin3A, levels around TSSs upon UpSET knockdown suggests that UpSET helps to orientate the HDAC Rpd3 toward the TSS.27 The Rpd3/Sin3A complex must be stabilized by multiple protein-protein interactions in transcribed regions, as Sin3A remains bound in the absence of UpSET. As in the mammalian Sin3B complex, candidates for such interactions include Mrg15 or Pf1, which associate with histone modifications established during elongation.24 Intriguingly, UpSET/Rpd3/Sin3A overlap occurs in approximately 50% of UpSET binding sites, suggesting that UpSET must be involved in recruiting other HDACs. In addition to Rpd3, Drosophila has at least four other HDACs (HDAC6, Hdac4, HdacX and HDAC4), as well as two NAD-dependent deacetylases (Sir2 and Sirt2).37 Alternatively, UpSET could interact with different promoter-associated factors or co-factors. Support for this view comes from a recent proteomic characterization of the dosage compensation complex (DCC), with which UpSET co-purifies, and the involvement of the DCC in H4K16 acetylation.38 Lack of UpSET increases H4K16Ac, as well as chromatin accessibility, around promoter regions. Interestingly, this histone modification has been shown to destabilize nucleosomes and to correlate with regions of chromatin decondensation.39 An exciting possibility is that UpSET performs specific functions based on this protein-protein interaction. For example, UpSET could help to stabilize components of the DCC around TSSs or modulate such DCC functions as H4K16 acetylation. Although proteomic analysis could help to clarify these questions, the unwieldy size (~330 KDa) and low stability of UpSET makes this type of approach challenging.
Modulating Chromatin Accessibility Around Active Genes
Chromatin accessibility is one of the key structural features of transcribed regions1 and is commonly evaluated by treating isolated nuclei with nucleases (e.g., DNaseI) or DNA modifying enzymes (e.g., M.SssI).40,41 Such analyses have led to the view that transcriptional regulatory elements exhibit higher chromatin accessibility than nearby or transcriptional silent regions as a consequence of the dynamic interplay among DNA-specific transcription factors, co-factors and nucleosomes.11,42 In Drosophila, DNaseI hypersensitive sites (DHS) are mainly enriched around TSSs and 5′UTRs, and moderately enriched over coding regions (Fig. 1B).43 Genome-wide approaches have revealed that promoter regions possess low levels or are depleted of nucleosomes. Promoter-associated DHSs tend to be flanked by well-positioned nucleosomes containing the histone variants H2A.Z and H3.3.44 H2A.Z-containing chromatin displays a more open chromatin configuration and it has been proposed that the replacement of canonical histones by these variants could establish regulatory element boundaries.45 In addition to histone acetylation changes, we found that lack of UpSET changes chromatin accessibility of UpSET target genes. Based on chromatin profiling studies, UpSET is enriched mainly over the TSS-associated nucleosomes on transcribed genes, where UpSET may be partially responsible for establishing TSS-associated DHS boundaries via its ability to modulate chromatin accessibility. We suggest two possibilities: (1) UpSET regulates H2A.Z-containing positioning or incorporation, and (2) UpSET/HDAC association is required for TSS boundary function. Support for these two ideas come from studies of the yeast Set3 and Rpd3 proteins. In yeast, Htz1 (H2A.Z) chromatin deposition is dependent on the ATP-remodeling complex SWR1.46,47 Genetic analysis of Set3, Htz1 (H2A.Z), and SWR1 mutants suggests a direct participation of Set3 in H2A.Z homeostasis.48 Yeast Rpd3 has also been shown to reduce nucleosome remodeling and suppress transcription.19 Yeast Rpd3 complexes inhibit nucleosome eviction caused by the ATP-dependent chromatin structure complex RSC. This activity is dependent on the presence of the complete Rpd3 core complex (Sin3, Ume1 and Rpd3), as the individual proteins do not stabilize nucleosomes.49 Remarkably, the H150A Rpd3 mutant, which lacks HDAC activity, maintains the ability to stabilize nucleosomes and prevents their eviction by RSC, suggesting that deacetylase activity is not required for nucleosome stability. If metazoan HDACs are also involved in nucleosome stabilization, the reduction in HDACs caused by the lack of UpSET would explain the increased chromatin accessibility, aberrant mis-localization or spreading of active mark-containing nucleosomes, and the aberrant activation of silent promoters (intrinsic and cryptic) located proximal to UpSET binding (Fig. 3).
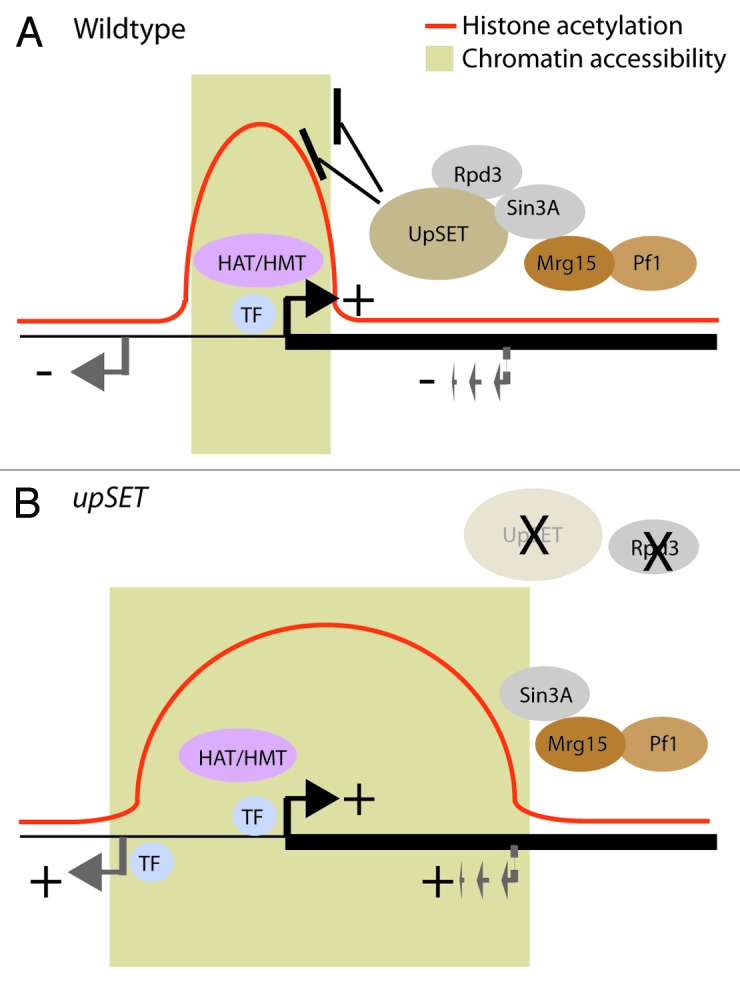
Figure 3. UpSET modulates histone acetylation and chromatin accessibility. (A) Upon promoter activation by transcription factors (TF), co-factors including histone acetyltransferases (HAT), histone methyltransferases (HMT) establish specific marks (e.g., H3K4m3), which recruit UpSET/HDACs around transcribed genes. The elongating polymerase machinery can also promote the recruitment of additional chromatin proteins (e.g., Mrg15 or Pf1). Upon recruitment, UpSET/HDAC complexes modulate the chromatin accessibility and histone acetylation generated during transcriptional initiation, thereby reducing the probability of unmasking off-target gene (gray arrow) or cryptic transcriptional start sites (dashed gray arrow). (B) Upon UpSET removal, levels of the HDAC Rpd3 are reduced, whereas Sin3A remains bound to chromatin via Mrg15 and PHD containing proteins like Pf1. These changes correlate with higher acetylation levels and higher chromatin accessibility around the transcribed gene. Increase of these open chromatin features leads to the higher probability of expressing off-target genes and repetitive elements.
Our results to date suggest that UpSET negatively modulates chromatin accessibility and predict that mutant phenotypes associated with chromatin-based silencing proteins should be enhanced, whereas those associated with position effect variegation (PEV) models should be suppressed. This turns out to be the case. Using two different upSET mutant alleles, we found that the extra sex comb phenotype caused by Polycomb mutation is enhanced upon UpSET depletion.27 Polycomb proteins are involved in silencing homeotic genes via chromatin compaction during development, and lack of these proteins cause the aberrant expression of homeotic genes in tissues where they are normally silent, causing for example, an extra sex comb phenotype.50,51 The enhancement of the Polycomb extra sex comb phenotype when UpSET activity is also reduced is likely due to increased chromatin accessibility at homeotic genes. Since UpSET does not interact with Polycomb group proteins, and its genome associations do not overlap with Polycomb Response Elements, its enhancement of this phenotype must be a direct effect of UpSET on the homeotic genes themselves.
Several Drosophila position effect variegation (PEV) models can be used to evaluate a particular protein’s ability to modulate chromatin in different contexts, such as in heterochromatin or near telomeric regions.52-55 In Drosophila, PEV of telomeric and centromeric transposon insertions are regulated by different sets of proteins, with the exception of Telomere 4 that is regulated by the same proteins that regulate centromeric (heterochromatic) silencing.56 Based on UpSET’s ability to modulate chromatin accessibility, we expected that UpSET suppressed PEV. Consistent with this, we found that the upSETMB8950 allele suppresses heterochromatic PEV associated with centromeres and Telomere 4, but not telomeric PEV associated with chromosome 2 or 3 telomeres (Fig. 4). Thus, the lack of UpSET/HDAC influences chromatin organization of the white gene by reducing the silencing effects of heterochromatin and lends support to UpSET’s role in controlling chromatin accessibility.27 Nevertheless, the lack of suppression of telomeric silencing may also suggest a physical restriction for UpSET in this context. In summary, UpSET-mediated HDAC recruitment is a mechanism used to modulate open chromatin features around transcribed regions and is likely achieved by the chromatin stabilizing activities of these recruited HDACs.
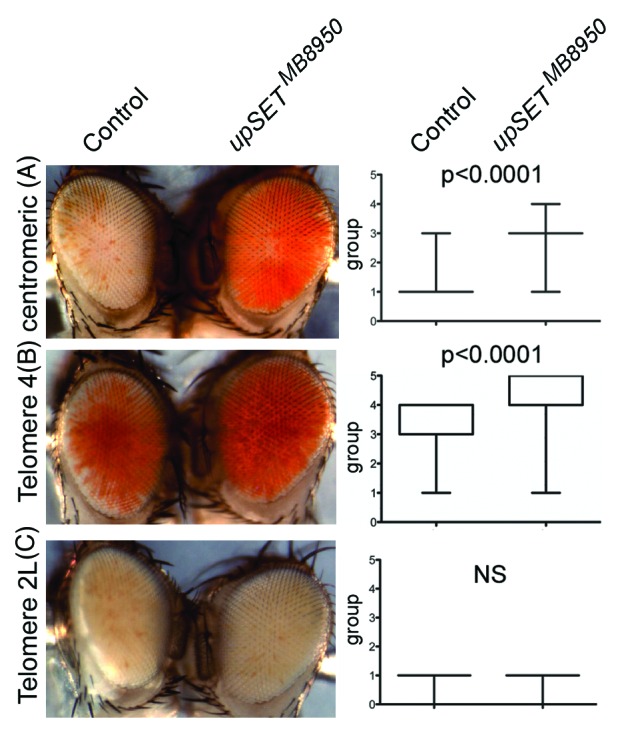
Figure 4.upSET mutants suppress heterochromatic, but not telomeric, Position Effect Variegation (PEV). (A–B) upSETMB8950 mediates suppression of heterochromatic PEV associated with w+ insertion near the X chromosome centromere (A) or the telomere on chromosome 4 (B). (C) upSETMB8950 does not affect telomeric PEV associated with w+ insertion near the telomeres on chromosomes 2 or 3 (chromosome 2L is shown). upSETMB8950 females were crossed to males from heterochromatin (centromeric or telomere 4) and telomeric (telomeres 2L or 3R) PEV models. Progeny was scored on day 3 after eclosion by placing the flies into one of 6 groups according to the percentage of the eye expressing the white gene (group 0 = 0%, group 5 = 100%). 200 flies (100 females and 100 males) were scored per genotype, except for centromeric PEV where 200 females were scored (as the white gene is on the X chromosome). Eyes shown are representative of the average phenotype for each genotype and each pair is an age-matched, sibling comparison. Box plots (right) show the distribution of phenotypes in each case. P-values were obtained by applying an unpaired t-test. This experiment was repeated twice with similar results.
How Is UpSET Targeted to TSS Regions?
Although we cannot eliminate the possibility that UpSET binding is facilitated by transcription factors or co-factors, the UpSET SET or PHD domains likely mediate UpSET binding directly to chromatin. Despite the lack of catalytic activity in vitro, the UpSET SET domain is well conserved in metazoans. UpSET could be recognized by HDACs as a mark to interact with the transcription-associated machinery, resulting in the recruitment of HDACs to active genes. Such a function may explain UpSET’s conservation among metazoans. Although SET domains have also been found to be required for protein-protein interaction,57,58 we favor the idea that UpSET targets chromatin directly via its PHD domain. For example, in yeast, Set3 requires its PHD domain to specifically bind methylated H3K4, as removal of the PHD domain or the H3K4-specific histone methyltransferase Set1 impairs Set3 recruitment to chromatin.20 Based on its conservation (Fig. 2B), the UpSET PHD domain may recognize methylation on H3K4, a mark that is usually found around promoter regions. This dependency on H3K4 methylation would suggest that UpSET is downstream of promoter H3K4 methylation encoded by Set1, Trithorax or Trithorax-related proteins. Consistent with this, we found that lack of UpSET increases histone acetylation levels and transcriptional noise as evidenced by the activation of off-target genes or repeat elements near UpSET target genes.27 Moreover, deletion of the yeast SET3 gene also triggers the activation of cryptic TSSs.59 Taken together, these results suggest that the establishment of promoter-associated histone methylation is important for the recruitment of UpSET/HDACs that modulate the features of transcribed chromatin. Such a regulatory mechanism would ensure that only the proper promoters become activated (Fig. 3).
Biological Manifestations of UpSET Proteins
Given that the described functions for UpSET and its orthologs imply key roles in gene regulation, it is surprising that organisms lacking these proteins are viable. upSET homozygous mutant flies are female sterile.27 In Mll5−/− knockout mice, hematopoietic stem cell fitness and differentiation are only mildly affected and males are sterile.60 In yeast, SET3 deletion results in normal growth, but sporulation genes are de-repressed during meiosis affecting ascus formation.28 Interestingly, the most common phenotype associated with the deletion of UpSET-like regulators occurs during gametogenesis, suggesting that gonad tissues may be less able to tolerate transcriptional noise. Our results support the idea that the developmental pathways controlling Drosophila oogenesis require UpSET to modulate the spatial and temporal regulation of key developmental regulators including Notch. In metazoans, developmentally regulated genes are usually controlled at the initial stages of transcription. Initially described in the context of viral promoters, RNA polymerase was found to be enriched at the 5′ end of cellular genes, including Drosophila hsp70 and human c-myc, regardless their transcriptional status.61,62 This paused polymerase was later shown to be a “rate-limiting step” during transcription.63 RNA polymerase pausing has been proposed to potentiate transcription and allow different transcription, elongation and pausing factors to integrate developmental signals to fine-tune gene expression.64-67 Although UpSET binds active genes independently of their pausing features, we have not eliminated the possibility that UpSET is required for RNA polymerase pausing itself. Different studies using RNA occupancy and global run-on experiments support the idea that pausing is a common feature in Drosophila and mammalian genes.62-64,66 An exciting possibility is that UpSET/HDAC complexes are involved in pausing of RNA polymerase or in downstream events in the transcriptional cycle including additional elongation-associated histone modifications (i.e., H3K36 methylation). In UpSET mutant ovaries, we observed increased levels of developmental regulators (e.g., Notch), which could be the consequence of changes in initiation, pausing and/or elongation. Future global run-on analyses should help to establish whether UpSET plays a role in pausing and whether the observed phenotypes in upSET mutant ovaries are a consequence of these roles. Unlike metazoans, yeast transcriptional pausing occurs during RNA polymerase elongation rather than in the promoter proximal region. Nevertheless, UpSET orthologs Set3 and Rpd3L in yeast have been suggested to play specific roles during transcriptional initiation: The Rpd3L HDAC has been proposed to regulate the transcriptional burst frequency, whereas the Set3C complex is thought to modulate the transcriptional burst size.68,69 It will be interesting to determine whether Rpd3L or Set3C functions are the evolutionary basis of RNA pausing in metazoans.
Perspectives
UpSET is a novel protein with conserved functions in chromatin homeostasis, and Drosophila is proving to be an ideal model organism in which to evaluate the functions of the Set3 family of proteins in metazoans. UpSET recruitment results in a modulation of open chromatin features, at least partially dependent on HDAC stabilization around transcribed genes. These functions are required to suppress activation of off-target genes and/or repeat elements. Although several questions remain, it is likely that UpSET and its interacting partners tweak gene expression. The genomic distribution of UpSET implies that this protein is globally required during active transcription. However, it is unclear whether UpSET plays similar roles on genes contained in both the RED and Yellow chromatin classes or whether it modulates association of different HDACs or other chromatin remodelers associated with these chromatin states in a developmental fashion. As upSET mutants are female sterile and viable, it remains to be determined if UpSET plays any role(s) in early embryogenesis or whether these roles are masked by the high maternal contribution of the protein (Rincon-Arano et al., unpublished observation).
Additionally, it will be interesting to address the molecular functions of the mammalian orthologs MLL5 and SETD5 to establish their functional conservation with UpSET. Murine MLL5 partially rescued Drosophila upSET-associated phenotypes. It is not yet clear if the lack of full rescue is a result of low sequence or functional conservation. Nevertheless, from a clinical point of view, it will be interesting to address whether mutations of UpSET orthologs are responsible for more aggressive leukemias as a consequence of the failure to appropriately modulate transcriptional noise resulting from the reactivation of silent genes or repetitive elements in euchromatic regions.
In summary, UpSET belongs to the Set3 family of proteins and is required to fine-tune transcription via modulating chromatin accessibility and histone acetylation. Future studies using the genetically and molecularly amenable Drosophila model are expected to provide a better understanding of the contribution of this unusual SET domain-containing protein to developmentally regulated transcription.
Acknowledgments
We thank Steve Henikoff, Lori Wallrath, the Harvard Exelixis Collection and the Bloomington/Kyoto Stock Centers for flies and other reagents used in this study. This work was supported by NIH grants GM097083 (to S.M.P.) and DK44746 and HL65440 (to M.G.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–56. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- 2.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–60. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 3.Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979;16:797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- 4.Stalder J, Groudine M, Dodgson JB, Engel JD, Weintraub H. Hb switching in chickens. Cell. 1980;19:973–80. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- 5.Banerji J, Olson L, Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33:729–40. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- 6.Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985;186:773–90. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 7.Knezetic JA, Luse DS. The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 8.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–62. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–4. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol Cell Biol. 2007;8:284–95. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 11.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Nègre N, Eaton ML, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–24. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–96. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pile LA, Wassarman DA. Chromosomal localization links the SIN3-RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 2000;19:6131–40. doi: 10.1093/emboj/19.22.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidal M, Strich R, Esposito RE, Gaber RF. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–16. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–27. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137:259–72. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buratowski S, Kim T. The role of cotranscriptional histone methylations. Cold Spring Harb Symp Quant Biol. 2010;75:95–102. doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, et al. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–46. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kidder BL, Palmer S. HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells. Nucleic Acids Res. 2012;40:2925–39. doi: 10.1093/nar/gkr1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jelinic P, Pellegrino J, David G. A novel mammalian complex containing Sin3B mitigates histone acetylation and RNA polymerase II progression within transcribed loci. Mol Cell Biol. 2011;31:54–62. doi: 10.1128/MCB.00840-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahringer J. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet. 2000;16:351–6. doi: 10.1016/S0168-9525(00)02066-7. [DOI] [PubMed] [Google Scholar]
- 26.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–8. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 27.Rincón-Arano H, Halow J, Delrow JJ, Parkhurst SM, Groudine M. UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell. 2012;151:1214–28. doi: 10.1016/j.cell.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–9. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 30.Emerling BM, Bonifas J, Kratz CP, Donovan S, Taylor BR, Green ED, et al. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21:4849–54. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Wong J, Klinger M, Tran MT, Shannon KM, Killeen N. MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood. 2009;113:1455–63. doi: 10.1182/blood-2008-05-159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Westergard TD, Hsieh JJ-D. MLL5 governs hematopoiesis: a step closer. Blood. 2009;113:1395–6. doi: 10.1182/blood-2008-11-185801. [DOI] [PubMed] [Google Scholar]
- 33.Madan V, Madan B, Brykczynska U, Zilbermann F, Hogeveen K, Döhner K, et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113:1444–54. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- 34.Heuser M, Yap DB, Leung M, de Algara TR, Tafech A, McKinney S, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113:1432–43. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 35.Damm F, Oberacker T, Thol F, Surdziel E, Wagner K, Chaturvedi A, et al. Prognostic importance of histone methyltransferase MLL5 expression in acute myeloid leukemia. J Clin Oncol. 2011;29:682–9. doi: 10.1200/JCO.2010.31.1118. [DOI] [PubMed] [Google Scholar]
- 36.Kittler R, Pelletier L, Heninger A-K, Slabicki M, Theis M, Miroslaw L, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–12. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 37.Gregoretti IV, Lee Y-M, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 38.Wang CI, Alekseyenko AA, LeRoy G, Elia AE, Gorchakov AA, Britton L-MP, et al. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol. 2013;20:202–9. doi: 10.1038/nsmb.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shogren-Knaak M, Ishii H, Sun J-M, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 40.Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, et al. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- 41.Bell O, Tiwari VK, Thomä NH, Schübeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–64. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 42.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas S, Li X-Y, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krogan NJ, Keogh M-C, Datta N, Sawa C, Ryan OW, Ding H, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–76. doi: 10.1016/S1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 47.Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–8. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 48.Hang M, Smith MM. Genetic analysis implicates the Set3/Hos2 histone deacetylase in the deposition and remodeling of nucleosomes containing H2A.Z. Genetics. 2011;187:1053–66. doi: 10.1534/genetics.110.125419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X-F, Kuryan B, Kitada T, Tran N, Li J-Y, Kurdistani S, et al. The Rpd3 core complex is a chromatin stabilization module. Curr Biol. 2012;22:56–63. doi: 10.1016/j.cub.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 51.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–32. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 52.Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 53.Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–90. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–77. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 55.Cryderman DE, Morris EJ, Biessmann H, Elgin SC, Wallrath LL. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 1999;18:3724–35. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cryderman DE, Morris EJ, Biessmann H, Elgin SCR, Wallrath LL. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 1999;18:3724–35. doi: 10.1093/emboj/18.13.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Cleary ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–7. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 58.Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontès M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum Mol Genet. 1998;7:679–84. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 59.Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–69. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yap DB, Walker DC, Prentice LM, McKinney S, Turashvili G, Mooslehner-Allen K, et al. Mll5 is required for normal spermatogenesis. PLoS One. 2011;6:e27127. doi: 10.1371/journal.pone.0027127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–6. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 62.Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986;6:3984–9. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–72. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 64.Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, et al. Defining the status of RNA polymerase at promoters. Cell Rep. 2012;2:1025–35. doi: 10.1016/j.celrep.2012.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–51. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brannan K, Bentley DL. Control of Transcriptional Elongation by RNA Polymerase II: A Retrospective. Genet Res Int. 2012;2012:170173. doi: 10.1155/2012/170173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–11. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weinberger L, Voichek Y, Tirosh I, Hornung G, Amit I, Barkai N. Expression noise and acetylation profiles distinguish HDAC functions. Mol Cell. 2012;47:193–202. doi: 10.1016/j.molcel.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hnisz D, Bardet AF, Nobile CJ, Petryshyn A, Glaser W, Schöck U, et al. A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 2012;8:e1003118. doi: 10.1371/journal.pgen.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]