Abstract
The generation of neuronal cell diversity is controlled by interdependent mechanisms, including cell intrinsic programs and environmental cues. During development, the astonishing variety of neurons is originated according to a precise timetable that is managed by a complex network of genes specifying individual types of neurons. Different neurons express specific sets of transcription factors, and they can be recognized by morphological characteristics and spatial localization, but, most importantly, they connect to each other and form functional units in a stereotyped fashion. This connectivity depends, mostly, on selective cell adhesion that is strictly regulated. While intrinsic factors specifying neuronal temporal identity have been extensively studied, an extrinsic temporal factor controlling neuronal temporal identity switch has not been shown. Our data demonstrate that pulses of steroid hormone act as a temporal cue to fine-tune neuronal cell differentiation. Here we also provide evidence that extrinsic JAK/STAT cytokine signaling acts as a spatial code in the process. Particularly, in Drosophila mushroom bodies, neuronal identity transition is controlled by steroid-dependent microRNAs that regulate spatially distributed cytokine-dependent signaling factors that in turn modulate cell adhesion. A new era of neuronal plasticity assessment via managing external temporal cues such as hormones and cytokines that specify individual types of neurons might open new possibilities for brain regenerative therapeutics.
Keywords: Drosophila mushroom body, JAK/STAT cytokine signaling, differential cell adhesion, microRNA let-7, steroid hormone ecdysone, temporal identity switch
How Multiplicity of Neuron Types Is Generated
The development of multiple compartments of the brain is a highly orchestrated process, where commitment of certain types of neurons to specific zones, layers and compartments is linked to the developmental stage, at which neurons are generated.1,2 During the last few years, significant progress has been made in the discovery of genes that identify and control development of different neuronal subtypes (reviewed in refs. 3–5). A subsequent series of intrinsic signaling programs are described in invertebrate and vertebrate organisms where neuronal progenitors in a time-dependent manner progressively acquire specific identity via expression of unique sets of genes that coordinate the generation of the multiple projection neuron subtypes.
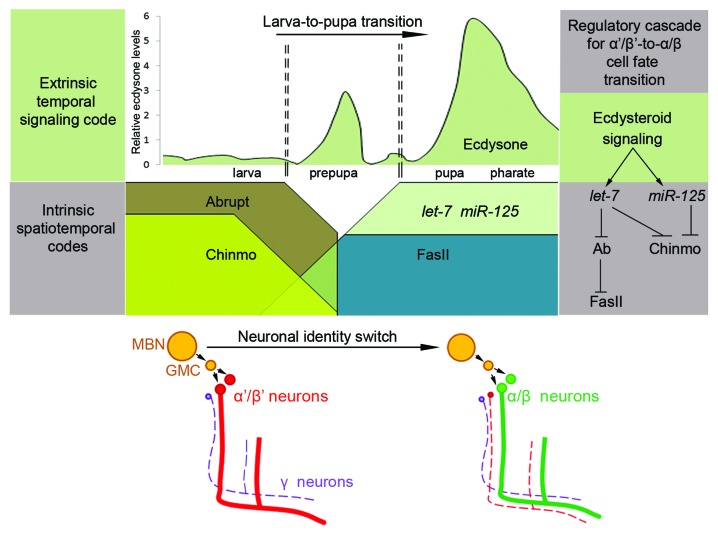
Like in vertebrates, neuronal stem cells in Drosophila produce different types of neurons depending on embryonic anterior-posterior and dorsal-ventral polarity that establish gradients of morphogens and induce expression of gap, pair-rule and Hox genes that subsequently assemble a set of differentially expressed transcription factors.6-11 Following the lineage specification, the neuronal stem cell generates a characteristic set of neuron subtypes.12-14 The exact birthdate of specialized neurons suggests an interaction between temporal cues and neuron-intrinsic cell fate factors. Despite the broad data about existence of these intrinsic programs, it is important to note that the extrinsic temporal determinants of differential morphogenesis have not been revealed in any organism. We discovered that in Drosophila steroid hormones regulate the chronological neuronal identity switch that is executed by steroid-dependent microRNAs (miRNAs) (Fig. 1).
Figure 1. Model of differential neurogenesis regulation by cooperation of developmentally controlled temporal systemic signaling and intrinsic spatiotemporal codes. Scheme represents the chronologically regulated signaling cascade controlling α′/β′ to α/β neuronal identity switch in the Drosophila MB that takes place at the larva-to-pupa developmental transition. Amount of ecdysone at different stages of development is represented as relative levels (scheme adopted from ref. 15). Developmentally regulated pulse of the steroid hormone ecdysone acts as an extrinsic temporal signaling code to activate expression of miRNAs from the let-7 complex in the differentiating MB neurons.16-18 Temporally induced miRNAs let-7 and miR-125 are intrinsic spatiotemporal codes that downregulate at least two BTB domain containing transcription factors Abrupt and Chinmo,16,19 which allows for the α′/β′ to α/β neuronal cell fate transition. Cell adhesion molecule FasII is downstream of let-7/Abrupt signaling. During larval stages Abrupt suppresses FasII expression allowing for early-born lobes to be formed, while at the pupal stage downregulation of Abrupt allows FasII expression and promote α/β neuronal differentiation.16
Steroid Hormone Regulates Chronological Neurogenesis in Drosophila
As a model to study extended neurogenesis we use Drosophila learning center or mushroom body (MB) neurons that are responsible for olfactory learning and memory.20 MB neuron subtypes are generated in the same lineages by type I neuroblasts and specified in a birth-order-dependent fashion.12 MB γ and α′/β′ neurons are produced during larval stages, while α/β neurons are born from the same neuronal precursors after transition from larval to pupal stages. This MB neuron diversification is coincident with key developmental time periods (Fig. 1). In Drosophila there are two key systemic developmental timers—steroid ecdysone and juvenile hormone15—that synchronize the genetic, morphological and behavioral changes associated with developmental transitions.21-29 Pulses of the steroid hormone ecdysone trigger major postembryonic developmental transitions, including molting and metamorphosis.15 Ecdysone interacts with a heterodimer of Ecdysone Receptor (EcR) and Ultraspiracle (Usp)—two members of nuclear receptor superfamily.30 This complex directly induces expression of primary-response targets, which in turn multiply hormonal signal by regulation of secondary-response gene transcription. These mechanisms determine stage- and tissue-specific responses to each developmentally regulated ecdysone pulse. Moreover, ecdysone signaling is patterned spatially as well as temporally; depending on the cell type and the developmental stage, the ecdysone receptor complex binds different co-activators or co-repressors that can have other binding partners, regulated by additional signaling pathways. For example, the putative transcription factor Abrupt attenuates ecdysone signaling by binding to its co-activator Taiman,31 and we showed that this interaction plays an important role in cell non-autonomous regulation of early germline progeny differentiation.29 Moreover, other signaling pathways (insulin, TGFβ, JAK/STAT) interact with ecdysone pathway components to further fine-tune the cell-type specific function.31-33 This additional level of combinatorial control allows for a highly managed regulation of gene expression by the systemic signaling. In the brain, it has been shown that ecdysone is responsible for γ neuron remodeling during metamorphosis,34 and we found that ecdysone signaling is also required for α′/β′ to α/β temporal identity switch that is accomplished via miRNAs to guarantee the specificity of this global endocrine signaling for differentiation of a certain type of neurons in the developing Drosophila brain.16
Hormones and MicroRNAs
Development of the living organism is organized into discrete temporal stages, each of which is characterized by a unique program of gene expression that controls tissue formation and differentiation. miRNAs were first found because of their role in the regulation of developmental staging of the nematode C. elegans.35,36 Multiple studies in insects also suggest an important role for miRNAs in the coordination of the developmental transitions; depletion of Dicer-1 (protein required for miRNAs biogenesis) in B. germanica37 and mutations in Drosophila miRNAs let-7 and miR-125 impair regulation of metamorphic processes.38,39 The temporal regulation of these and many other miRNAs expression is mediated by developmentally controlled hormonal signals. For example, in Drosophila, the upregulation of miR-100, miR-125, and let-7 encoded by the miRNA let-7-C locus and downregulation of miR-34,17 miR-14,40 and miR-841 require the steroid hormone ecdysone. Recent work from Chawla and Sokol18 identified and mapped three Ecdysone Response Elements within the let-7-C locus, proving that miRNAs can be first-response targets of the hormonal signaling. Importantly, not only do hormones regulate miRNA expression but also miRNAs can affect the strength of systemic signaling. For example,miR-14 has been identified to mediate a positive autoregulatory loop of EcR that amplifies ecdysone response,40 while miRNA bantam activity in ecdysone-producing cells represses hormone production and thereby promotes systemic growth.42 A number of studies in vertebrate models and cell cultures also show relationships between hormones and miRNAs. Glucocorticoids influence a variety of physiological processes in vertebrates, including adaptation to stress, metabolism, immunity and neuronal development. Kawashima et al.43 show that glucocorticosteroids regulate levels of brain-derived neurotrophic factor (BDNF) via suppression of miR-132 expression, which possibly contributes to the regulation of synaptic plasticity in the brain. On the other hand, miRs-18 and -124a can regulate levels of corticosteroid receptor and therefore modulate downstream effectors of this hormonal signaling.44 Recent work from Huang et al.45 demonstrates that the miR-21 promoter has a thyroid hormone response element that allows miRNA to be activated in response to hormonal stimuli. Thyroid hormone in vertebrates is an important regulator of development, differentiation and growth. Overactivation of miR-21 promotes hepatoma cell migration and invasion, analogous of that observed with thyroid hormone stimulation.45 In breast cancer, the estrogen receptor α (ERα) binds the miR-221/222 transcription start site and recruits co-repressors to suppress their transcriptional activity,46 while miRNAs miR-191 and miR-425 are upregulated via estrogen-mediated activation.47 Another study shows that miR-221/222 acts as a negative regulator for ERα48 supporting the idea for the existence of negative regulatory loop involving miRNAs and hormonal receptors.
Together, these data confirm that hormones and miRNAs are prone to work together in regulation of multiple processes. On one hand, cell-specific miRNAs can be used as additional factors that fine-tune the specificity of cellular responses to global hormonal signaling; on the other hand, miRNAs are also involved in feedforward and feedback loops to readjust the precision of this systemic signaling in a given cell type.
MicroRNAs in the Brain
Biogenesis of miRNAs exhibits specific temporal and spatial profiles in different types of cells and tissues and, therefore, affects a wide range of biological functions. Conditional knockout of Dicer has been extensively used to address the collective role for miRNAs in specific tissues and cell types in mice. The essential functions for the miRNA pathway have been uncovered in the brain: miRNAs regulate neuronal development and synaptic plasticity, oligodendroglia differentiation and myelin formation and are implicated in brain tumor development and in the regulation of neurodevelopmental and neurodegenerative disorders.49-58 The role of specific miRNAs in the regulation of embryonic and adult neurogenesis, particularly in the proliferation and differentiation of neural stem cells, is emerging. Recent work from Parsons et al.54 provided a genome-scale profiling of miRNA differential expression patterns in human embryonic stem cell neuronal lineages. This allowed identifying molecular miRNA signatures for human embryonic neurogenesis: the in vitro neuroectoderm-originated human neuronal cells acquire their identity by downregulation of pluripotence-associated miRNAs (such as hsa-miR-302 family). In addition, induction of high levels of expression of miRNAs required for regulation of human central nervous system development (such as hsa-miR-10 and let-7) occurs in a stage-specific manner. In a similar study Stappert et al.55 demonstrated that time-controlled modulation of specific miRNA activities not only regulates human neural stem cell self-renewal and differentiation but also contributes to the development of defined neuronal subtypes; hence miR-125b and miR-181 promote and miR-181a* inhibits generation of dopaminergic fate neurons. Boissart et al.50 found that miR-125 potentiates early neural specification of human embryonic stem cells by regulating SMAD4, a key factor for pluripotent stem cell lineage commitment. Using primary cultures derived from P1 rat cortex, neuron-enriched (miR-376a and miR-434) and glia-enriched (miR-223, miR-146a, miR-19 and miR-32) miRNAs were identified.52 MiRNAs have been also found to direct development of specific brain regions during embryogenesis. Nowakowski et al.53 showed that miR-92b is involved in the regulation of a number of intermediate progenitors populations in mice brain that give rise to the cerebral cortical neurons.
A number of studies in vertebrates reveal the role for miRNAs in the regulation of adult neurogenesis that is largely restricted to two major brain regions: subventricular zones of the ateral ventricle and of the dentate gyrus in the hippocampus. MiRNAs let-7b,59 miR-9,57 miR-106b-25 cluster,60 miR-137,61 miR-184,62 miR-124,63 and their specific targets were identified to regulate neural cell proliferation and/or neuronal differentiation during adulthood. Latest studies from Liu et al.64 uncovered the molecular mechanism by which miR-17-92 cluster regulates ischemia-induced neural progenitor cell proliferation which stimulates adult neurogenesis after injury. It has been discovered that stroke substantially upregulates mi-R17-92 cluster expression in neural progenitor cells of the adult mouse. Overexpression of miR-17-92 cluster in the cell culture and in vivo significantly increased cell proliferation, whereas inhibitions of individual members of miR-17-92 cluster, miR-18a and miR-19a suppressed cell proliferation and increased cell death. Subventricular zone neuronal fate is determined by miR-12449: in vivo inhibition of miR-124 causes a block in neurogenesis and leads to an accumulation of ectopic cells with astrocyte characteristics (neural stem cells) in the olfactory bulb, while upon miR-124 overexpression neural stem cells are not maintained in the subventricular zone of mouse brain and neurogenesis is lost.
Studies from Drosophila revealed that this evolutionary ancient miR-124 controls neural stem cells proliferation by targeting anachronism—an inhibitor of neuroblast proliferation.56 Drosophila mutant lacking miR-124 shows reduced proliferative activity of neuronal progenitor cells and decreased production of adult postmitotic neurons. We showed that ecdysteroid signaling induces expression of let-7-C in Drosophila brain, which is required for proper differentiation of the last-born MB neurons. let-7 deficiency16 or ecdysone signaling deficit65 leads to MB morphological defects that result in learning and memory disabilities.
Involvement of miRNAs in regulation of neuronal development, plasticity and maintenance provides a new additional layer of gene regulation, which has an effect on nervous system functions and contributes to therapeutic approaches toward neurological diseases. These new findings also propose miRNAs as possible candidates for innovative brain therapies. However, since the general role for miRNAs is the transcriptional repression of their targets, upcoming studies should be focused on finding functional miRNA-target pairs that are also defined at the spatiotemporal level.
BTB Transcription Factors as Temporal Codes
We established a spatiotemporal connection between the ecdysteroid-induced miRNA let-7 and its target, the BTB transcription factor Abrupt in the developing brain.16 BTB/POZ zinc finger factors are a class of nuclear DNA-binding proteins containing the BTB domain, which was first identified as a conserved element in the developmentally regulated Drosophila proteins Broad-complex, Tramtrack and Bric-a-brac.66 Afterwards, the BTB protein-protein interaction motif was found in hundreds of different proteins virtually in all organisms, ranging from yeast to humans. It is involved in the regulation of gene expression through the local control of chromatin conformation and the recruitment of degradation targets to E3 ubiquitin ligase complexes.67,68 Interestingly, the BTB domain can form dimers and mediate interactions with non-BTB domain containing proteins and can establish both stable and transient interactions. This explains the ability of BTB containing proteins to participate in multiple processes and implies that management of their proper levels is of a particular significance.68
BTB/POZ domain zinc finger factors were linked to broad range of developmental processes in vertebrates and invertebrates: chromatin remodeling, cancer development and intriguingly, regulation of cell fate specification in the nervous system.66-72 For example, the BTB/POZ zinc-finger transcription factor-encoded by gene Rp58 is required for the correct differentiation of neural progenitors into neurons, since its neural-specific deletion results in severe cerebellar hypoplasia and developmental failure of several neuronal types.71 By coherently repressing multiple proneurogenic genes in a timely manner this BTB protein supports neuronal differentiation and brain growth.72 During embryonic development of the murine cerebral cortex another mammalian BTB factor, HOF is specifically expressed in immature non-dividing cells and is downregulated in differentiated cells of the hippocampus; importantly, it is one of the factors that might be involved in early definition of hippocampal compartment within the neocortex.70
Similarly, in the Drosophila nervous system several BTB/POZ domain zinc finger transcription factors have been implicated in specifying neuronal and glial cell lineages. For example, Tramtrack proteins transcriptionally repress genes that promote transformation of neuronal support cells into neurons,73,74 while Lola, Fruitless, Abrupt, and Chinmo are intrinsically required for development of different subsets of neurons.16,19,69,75-79 Such data provide evidence that BTB/POZ zinc-finger proteins play an important role in the transcriptional program that controls differentiation of progenitors into neurons. Since the growth and organization of the brain is tightly correlated with the speed of the whole organism development, it implies that neuron differentiation should be responsive to external temporal cues. Interestingly, the neuronal temporal identity of Drosophila MB neurons is governed by two BTB transcription factors, Chinmo and Abrupt and both of them are subjects to miRNA-mediated regulation.16,19,79 We found that this regulation is chronologically induced by systemic steroid signaling that controls the major larva-to-pupa transition during Drosophila development, which also coincides with the time-point when the last-born neurons are generated.16 This demonstrated for the first time that differential neurogenesis is hierarchically regulated by extrinsic systemic signaling, which, in chronological manner, adjusts programs of intrinsic temporal determinants of neuronal cell fate and that BTB transcription factors play a role as temporal codes in the process.
Next, we aimed to understand whether intercellular environmental signaling, such as extrinsic cell-to-cell signaling would also cooperate to fine-tune the outcome of differential neurogenesis.
Concerted Action of Cytokines and Steroids in Differential Neurogenesis
Interestingly, let-7 target Abrupt that is expressed in MBs is associated with the evolutionary conserved JAK/STAT signaling pathway, which plays key roles in multiple developmental and physiological processes in the brain, ranging from the regulation of neurogenesis and stem cell fate to memory formation.80-82 In the adult brain, endogenous cytokine levels are very low under normal physiological conditions; however, various types of injuries, including trauma, seizures and ischemia induce an increase of cytokine ligand levels, which in turn promotes neuronal stem cell self-renewal.83 In the developing brain, some neuroepithelial cells become neuroblasts and generate the neuronal and glial cells, and in the Drosophila optic lobe, the timing of this transition is negatively regulated by JAK/STAT signaling. Secretion of the JAK/STAT ligand Unpaired (Upd) shapes an activity gradient in the neuroepithelium and negatively regulates the progression of the proneural wave.84 JAK/STAT signaling is further integrated with the Notch and EGFR signals to balance neuroblast self-renewal and neuron differentiation.81,84 Since the BTB transcription factor Abrupt has been shown previously to be negatively regulated by the JAK/STAT signaling pathway in ovaries,31,85 we evaluated whether JAK/STAT plays a role in Abrupt regulation during MB development.
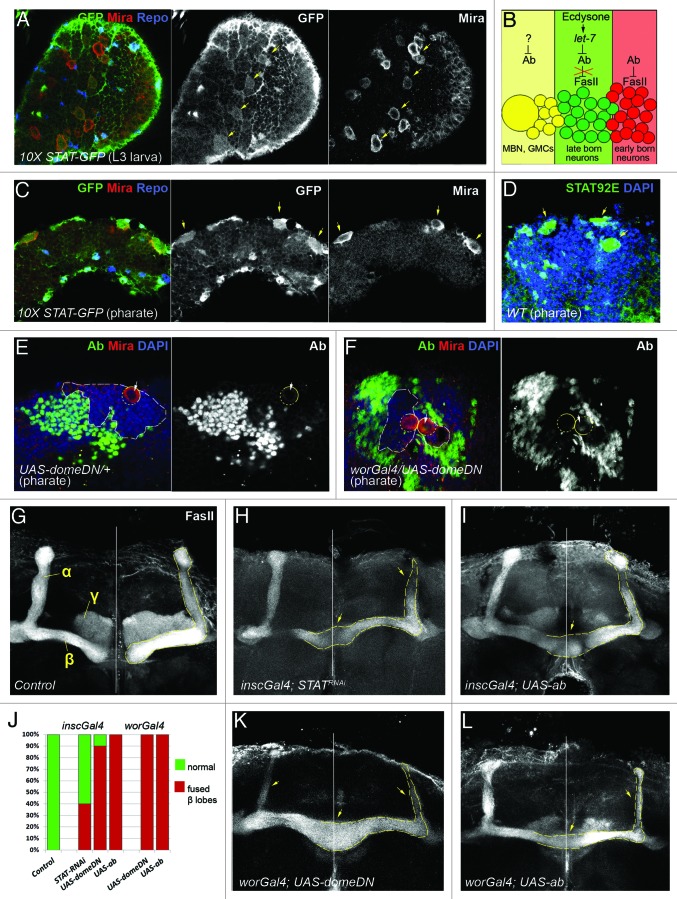
We used a 10xSTAT-GFP reporter line (Fig. 2A and C) and antibodies against STAT92E, the Drosophila homolog of mammalian STAT (signal transducer and activator of transcription) proteins (Fig. 2D) to visualize JAK/STAT signaling activity in the developing brain. At the larval stage, JAK/STAT activity was predominantly observed in neuroblasts (Miranda positive cells, arrows) and in glia (Repo positive cells) (Fig. 2A). Mushroom body neuroblasts (MBNs) are the only neuronal stem cells that continue to divide during later stages;86 interestingly, in the pupal and pharate brains, apart from glial cells, GFP signal indicating JAK/STAT activity was restricted to these mitotically active neuronal stem cells (Fig. 2C). Similar pattern of JAK/STAT signaling activity was detected with STAT92E antibodies (Fig. 2D). This expression analysis shows that the JAK/STAT signaling pathway is active in all postembryonic neuronal stem cells regardless of the developmental stage or ecdysone signaling activity.
Figure 2. JAK/STAT signaling is involved in Abrupt regulation during MB development. (A, C and D) In Drosophila brain JAK/STAT signaling activity [marked with 10xSTAT-GFP reporter in (A) and (C)] and STAT92E antibody staining (D) is detected at larval and pharate stages. JAK/STAT signaling is active in neuroblasts [marked with anti-Miranda (red) and glial cells marked with anti-Repo (blue) in (A) and (C) or determined based on morphology and nuclear DAPI staining in (D)]. Yellow arrows indicate both JAK/STAT signaling activity and neuroblast location. (B) Schematic drawing of Abrupt expression pattern and its regulation by previously described regulatory factors16 in the MB neuronal body cluster. Abrupt expression is restricted to the early born γ, α′/β′ MB (red colored) neurons where it functions as a negative regulator of FasII (cell adhesion molecule) expression. At the larva-to-pupa transition developmentally regulated ecdysteroid signaling induces expression of miRNA let-7 in the α/β (green colored) neurons. let-7 negatively regulates Abrupt which allows for FasII expression, necessary for MB neurons to undergo cell fate transition into α/β. The question mark depicted on the scheme inquires whether spatially distributed cytokine signaling acts in the concert with temporally regulated hormonal stimuli to adjust Abrupt activity in the mushroom body neuroblast (MBN) and ganglion mother cells (GMCs) (yellow). (E and F) anti-Abrupt staining (green) is elevated in the MBN upon JAK/STAT signaling downregulation achieved by overexpression of dominant negative form of dome (F) in comparison to the control (E). Circles show MBN location [marked also with anti-Miranda (red)], arrows point to anti-Abrupt staining inside the MBN, white dashed line outlines Ab-negative area in the MB cell body clusters [note smaller area in (F) in comparison to (E)]. (G-L) Both, downregulation of JAK/STAT signaling (H, J and K) and overexpression of the transcription factor Abrupt in the neuroblasts (I, J and L) causes similar morphological defects detected with anti-FasII staining in the adult brains in comparison to control [UAS-domeDN/TM6 in (G)]. White vertical line indicates position of the midline, dashed yellow line shows α/β MB lobes, yellow arrows point to slim α/β and fused β MB lobes.
Previously, we found that Abrupt is expressed in early-born γ, α′/β′ neurons and miRNA let-7 in the late-born α/β neurons and this temporally induced let-7 expression is necessary to downregulate Ab, which is critical for proper specification of the last-born neurons.16 Abrupt is a very potent cell fate regulator, since its misexpression is sufficient to even induce homeotic transformation.87 Therefore, we hypothesized the possibility that spatially distributed cytokine signaling would repress Abrupt expression in the MB neural stem cells (Fig. 2B).
To test this we analyzed different JAK/STAT pathway mutants (see Materials and Methods) and found that downregulation of JAK/STAT signaling via expression of dominant negative form of dome specifically in the neuroblasts resulted in changed Abrupt expression pattern in the MB cell body clusters and in the appearance of ectopic Abrupt protein in some of the neuroblasts (Fig. 2E and F). Next, we wanted to test if this misexpression would affect the neuronal stem cell progeny differentiation. MB neuroblasts are continuously dividing to give rise to MB neurons (Kenyon cells) that based on their birthdate and cell adhesion molecule expression, are clustered into three types of MB lobes (γ, α′/β′ and α/β) with distinct axonal projection patterns. We used FasII antibodies as a molecular marker for γ and α/β MB axons to evaluate whether downregulation of JAK/STAT signaling or overactivation of the transcription factor Abrupt in the MBNs affect overall MB morphology. We observed that downregulation of JAK/STAT activity via overexpression of a dominant negative form of dome or STAT RNAi using pan-neuronal and neuroblast-specific driver lines (inscGal4 and worGal4, respectively) indeed caused morphological changes in the adult mushroom bodies; MBs with slim α/β lobes and fused β-lobes (Fig. 2G–H and 2J–K; Table 1) were observed. Importantly, similar MB morphological defects were identified upon overexpression of Abrupt in the MB neuroblasts (Fig. 2I–J and 2L; Table 1). This evidence supports the hypothesis that spatially distributed JAK/STAT signaling represses the transcription factor Abrupt in neuronal stem cells and this downregulation is critical for proper neurogenesis. Since previously we found that ecdysone signaling also targets this BTB transcription factor via let-7 miRNA, we conclude that two extrinsic signaling pathways, global hormonal and local cytokine, collaborate to regulate extended neurogenesis during Drosophila MB development.
Table 1. Downregulation of JAK/STAT signaling and upregulation of Abrupt expression in the MBNs affects MB development.
| Driver | UAS-transgene | α/β MB lobe morphology* |
|---|---|---|
| inscGal4 x | UAS-ab | Escapers have fused β lobes (100%) Slim α/β lobes (50.0%) n = 4 |
| UAS-abRNAi | No visible morphological changes n = 18 |
|
| UAS-STATRNAi | Fused β lobes (22.2%) Slim α/β lobes (22.2%) n = 18 |
|
| UAS-domeDN | Fused β lobes (90.9%) Slim α/β lobes (50.0%) n = 22 |
|
| worGal4 x | UAS-ab | Fused β lobes (100%) Slim α/β lobes (100%) n = 16 |
| UAS-domeDN | Fused β lobes (100%) Slim α/β lobes (36.4%) n = 22 |
|
| Control | UAS-domeDN/TM6 | No visible morphological changes n = 20 |
α/β MB lobe morphology was evaluated from the maximum projections of confocal MB images based on FasII antibody staining. Fused β lobes were counted per brain; n, number of analyzed MB lobes per genotype.
Interestingly, another BTB-zinc finger protein Chinmo that has been found to control stem cell self-renewal and direct neuroblast temporal identity also depends on JAK/STAT activity and can be targeted by miRNA let-7 and miR-125.19,79,88 This implies that regulation of expression of JAK/STAT dependent BTB factors Abrupt and Chinmo should be under strict developmental control to guarantee faithful cell fate determination. Our current and previous data provide evidence that in the developing brain, the temporally induced by ecdysone miRNA let-7 negatively regulates Ab, which is additionally targeted by the local JAK/STAT cytokine signaling pathway to ensure proper MB development. The interaction between global developmental and local tissue-specific signaling results in formation of a robust spatio-temporal pattern to fine-tune the fidelity of neuronal cell differentiation, which is essential for proper brain morphogenesis (Fig. 3).
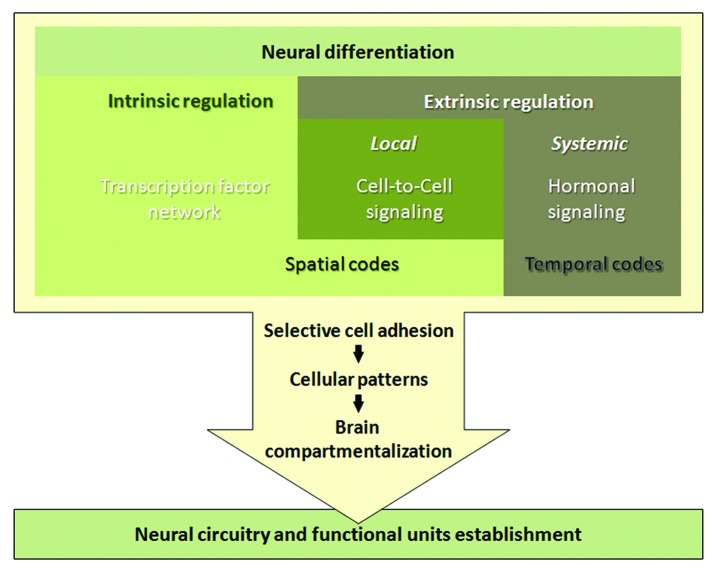
Figure 3. Model of spatiotemporal regulation of differential neurogenesis. Differentiation of the neural stem cell progenitor into a specific neuron subtype depends on concerted action of intrinsic and extrinsic programs. Intrinsic regulation is achieved via combination of multiple transcription factors that are hierarchically specified during organismal development starting from establishing the anterior-posterior and dorsal-ventral polarity that creates gradients of morphogens and induces expression of gap, pair-rule and Hox genes, and subsequently assembling a set of differentially expressed transcription factors, combination of which produces the unique code for a certain neuronal subtype. This code is additionally adjusted by extrinsic cell-to-cell signaling, for example Notch for binary cell fate decision or JAK/STAT cytokine signaling for neuronal cell type specification. This unique code constantly changes in response to internal and external conditions that coordinate the development of the whole organism. Hormones are great temporal code candidates, as they direct all major developmental steps. The combination of spatial and temporal codes in neuronal precursors allows certain types of neurons to be born at exact place and time, which is critical for brain morphogenesis. For normal brain function, these neurons must cluster and synapse in a stereotyped fashion, which predominantly depends on selective cell adhesion. As a result of establishment of brain compartments and differential neuronal connections, functional neural circuits are created that process all kinds of information and control behavior, learning, memory and plasticity of each individual.
Cell Adhesion as a Final Outcome of Differential Neurogenesis
The complexity of the brain is generated by multiple types of neurons that connect to each other in a specialized manner, which often depends on selective cell adhesion.89 Neurons expressing similar cell adhesion proteins not only cluster together to organize brain compartments that have distinct functions, selective cell adhesion is also used for establishment of synaptic connections that allow neurons to communicate and transfer information. Significant alterations in the brain structure and functions are generated even by moderate changes in the quantities of adhesion molecules on the neuronal cell surfaces. Therefore, differential cell adhesion is the final aftermath of differential neurogenesis, suggesting that timing and levels of cell adhesion protein expression must be precisely regulated (Fig. 3).
Among the most important cell adhesion molecules (CAMs) involved in the development of the nervous system, synaptic plasticity and cognition and memory are neural cell adhesion molecules (NCAMs) that belong to the immunoglobulin superfamily. Previous data show that levels of human NCAM2 that is primarily expressed in the brain to stimulate neurite outgrowth and facilitate dendritic and axonal compartmentalization are essential for normal brain development.90 For example, the increased expression of NCAM2 as a result of trisomy 21 may cause dosage-related detrimental effects in Down syndrome; also, in genome-wide association studies, NCAM2 was suggested as a candidate gene for the development of autism and Alzheimer’s disease,91-93 and multiple NCAM1 proteins are differentially altered in bipolar disorder and schizophrenia.94 Furthermore, NCAMs play a critical role in plasticity of the nervous system and in mechanisms controlling learning and memory and their expression levels are known to be highly susceptible to modulation by stress.95 Moreover, NCAM is involved in some of the bidirectional effects of stress on memory processes, where its increased synaptic expression is facilitating stress actions while its decreased expression is impairing effects of stress on memory consolidation.96 All these data imply that regulation of NCAM expression is a prerequisite for proper brain development and function. However, the question remains: What genetic machinery regulates precise expression of adhesion molecules in the brain?
Ample sets of regulatory elements are required for spatiotemporally restricted expression pattern of a given gene; however, it is not well-defined which set of transcriptional factors regulates differential expression of appropriate cell adhesion proteins that modulate the degrees to which various neurons adhere to each other to make synapses. In the Drosophila MBs, the ortholog of NCAMs, Fasciclin 2 (Fas2) displays specific temporal patterns of expression that plays a significant role in the spatial segregation of MB neurons. Low levels of Fas2 are detected in the γ and high levels in the α/β, but not α′/β′ lobes.97-99 Fas2 provides specific adhesive codes among MB neurons preventing them from intermingling and assuring formation of distinct MB lobes. We showed that the transcription factor Abrupt suppresses Fas2 expression in the earlier-born neurons, while steroid-induced miRNA let-7 via downregulation of Abrupt allows this critical adhesion molecule to be highly expressed in the late-born α/β neurons. Thus, the precise Fas2 expression is essential for proper MB morphology and function16 (Fig. 1).
Together, these data show that NCAMs are multifunctional proteins involved in neurogenesis and neurodevelopment and their expression levels are critical for dendritic and axonal compartmentalization and synaptic plasticity. This makes differential cell adhesion as a fundamental mechanism of neuronal cell differentiation that controls the finest aspects of neuronal specification (Fig. 3). Once a specific neuron is born, it must recognize and join other neurons of the correct type to assemble into a specific brain compartment that normally is determined and maintained by the system of preferential cell affinities. Even more, neurons send out axons and dendrites that via differential cell adhesion make synapses with other neurons. However, neurons do not simply reside inertly stuck together; instead, the new synapses are established and actively maintained by selective adhesion created and gradually adjusted by neurons; thus, contributing to the nervous system plasticity. We found that misexpression of Fas2 in the early-born α′/β′ MB neurons makes their axons to project into the places, where the later-born α/β neurons would send their axons.16 Since distinct MB neurons have different functions in Drosophila behavior regulation, it would be interesting to analyze, whether this alteration in the cell adhesive characteristic would change fly cognition.
Importantly, we also show that miRNAs are mediators between extrinsic temporal cues and intrinsic spatiotemporal codes that determine the precision of neuronal adhesiveness during brain development. It would be important in the future to explore the role of these factors in the adult brain plasticity. Interestingly, it has been proposed that the increased stickiness of human neurons might explain the accelerated evolution of the human brain beyond the brains of primates.100 Another factor that distinguishes humans from other primates is that developmental profiles of miRNAs, as well as their target genes, show the fastest rates of human-specific evolutionary change, which allows for the faster evolutionary rate in divergence of developmental patterns.101 One of the key features of the miRNA function is that miRNAs normally do not turn their target genes on and off, but just modulate their expression. This allows building novel networks between newly originated genes and miRNAs softly, not necessarily causing the lethality. Analysis of recently originated brain genes in Drosophila showed that numerous newly evolved genes are expressed in the brain and all of the MB-positive new genes are expressed in the α/β, but not in more ancestral γ and α′/β′ lobes.102 Since miRNAs and newly evolved genes are frequently co-expressed in the brain, the hypothesis can be put forward that the establishment of novel sets of spatiotemporal codes for differential neurogenesis that are gently fine-tuned by miRNAs is a common mechanism that might contribute to the phenotypic evolution of behavior and individual plasticity of the nervous system. Management of genetic programs that temporally specify individual subtypes of neurons could help to evaluate the true limits of progenitor plasticity within the developing and adult brain and initiate a new phase of plasticity assessment.
Materials and Methods
Fly strains and genetics
We used worGal4103 and inscGal4 (BDSC) driver lines crossed to a dominant negative form of Dome104 (UAS-domeΔCyt or UAS-domeDN) and STAT RNAi transgenic line (UAS-STAT92ERNAi, VDRC) to downregulate JAK/STAT signaling; and UAS-Abrupt (BDSC) to overexpress Abrupt in the neuroblasts. Oregon R animals were used as a wild-type control. To visualize active JAK/STAT signaling 10xSTAT-GFP reporter105 was used. All crosses were maintained at 25 °C on standard medium.
Immunohistochemistry
Brains were dissected in PBS and fixed in 4% formaldehyde (Polysciences, Inc.), adult and pupal for 30 min, larval for 15 min. Staining was performed as described.106 The following antibodies were used: mouse anti-Fas II 1:20 (marker for γ and α/β lobes) and mouse anti-Repo 1:20 (glia marker) (DSHB), rabbit anti-Abrupt 1:500,69 rabbit anti-STAT92E 1:500,88 guinea pig anti-Miranda (gift from A. Wodarz), Alexa 488, 568, or 633 goat anti-mouse, anti-rabbit, anti-guinea pig (1:500, Molecular Probes). Images were obtained with a confocal laser-scanning microscope Zeiss LSM700 and processed with ZEN 2010 and Adobe Photoshop software.
Acknowledgments
We thank all members of the Shcherbata lab, Vinodh Ilangovan, Roman Shcherbatyy for comments on the manuscript and the Max Planck Society for funding.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Passante L, Gaspard N, Degraeve M, Frisén J, Kullander K, De Maertelaer V, et al. Temporal regulation of ephrin/Eph signalling is required for the spatial patterning of the mammalian striatum. Development. 2008;135:3281–90. doi: 10.1242/dev.024778. [DOI] [PubMed] [Google Scholar]
- 3.Lin S, Lee T. Generating neuronal diversity in the Drosophila central nervous system. Dev Dyn. 2012;241:57–68. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- 4.Franco SJ, Müller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshino M. Neuronal subtype specification in the cerebellum and dorsal hindbrain. Dev Growth Differ. 2012;54:317–26. doi: 10.1111/j.1440-169X.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- 6.Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- 7.Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol. 2003;13:8–15. doi: 10.1016/S0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 8.Technau GM, Berger C, Urbach R. Generation of cell diversity and segmental pattern in the embryonic central nervous system of Drosophila. Dev Dyn. 2006;235:861–9. doi: 10.1002/dvdy.20566. [DOI] [PubMed] [Google Scholar]
- 9.Urbach R, Volland D, Seibert J, Technau GM. Segment-specific requirements for dorsoventral patterning genes during early brain development in Drosophila. Development. 2006;133:4315–30. doi: 10.1242/dev.02605. [DOI] [PubMed] [Google Scholar]
- 10.von Ohlen T, Doe CQ. Convergence of dorsal, dpp, and egfr signaling pathways subdivides the drosophila neuroectoderm into three dorsal-ventral columns. Dev Biol. 2000;224:362–72. doi: 10.1006/dbio.2000.9789. [DOI] [PubMed] [Google Scholar]
- 11.Hirth F, Hartmann B, Reichert H. Homeotic gene action in embryonic brain development of Drosophila. Development. 1998;125:1579–89. doi: 10.1242/dev.125.9.1579. [DOI] [PubMed] [Google Scholar]
- 12.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–76. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 13.Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–21. doi: 10.1016/S0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- 14.Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–89. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- 15.Riddiford LM. Hormones and Drosophila development. In: Bate M, Arias AM, eds. The Development of Drosophila Melanogaster. Plainview, NY: Cold Spring Harbor Laboratory Press, 1993. [Google Scholar]
- 16.Kucherenko MM, Barth J, Fiala A, Shcherbata HR. Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 2012;31:4511–23. doi: 10.1038/emboj.2012.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol. 2003;259:9–18. doi: 10.1016/S0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 18.Chawla G, Sokol NS. Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development. 2012;139:1788–97. doi: 10.1242/dev.077743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu YC, Chen CH, Mercer A, Sokol NS. Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell. 2012;23:202–9. doi: 10.1016/j.devcel.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–75. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 21.Riddiford LM. Hormone receptors and the regulation of insect metamorphosis. Receptor. 1993;3:203–9. [PubMed] [Google Scholar]
- 22.Kozlova T, Thummel CS. Essential roles for ecdysone signaling during Drosophila mid-embryonic development. Science. 2003;301:1911–4. doi: 10.1126/science.1087419. [DOI] [PubMed] [Google Scholar]
- 23.McBrayer Z, Ono H, Shimell M, Parvy JP, Beckstead RB, Warren JT, et al. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev Cell. 2007;13:857–71. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shirras AD, Bownes M. Separate DNA sequences are required for normal female and ecdysone-induced male expression of Drosophila melanogaster yolk protein 1. Mol Gen Genet. 1987;210:153–5. doi: 10.1007/BF00337772. [DOI] [PubMed] [Google Scholar]
- 25.Gaziova I, Bonnette PC, Henrich VC, Jindra M. Cell-autonomous roles of the ecdysoneless gene in Drosophila development and oogenesis. Development. 2004;131:2715–25. doi: 10.1242/dev.01143. [DOI] [PubMed] [Google Scholar]
- 26.Buszczak M, Freeman MR, Carlson JR, Bender M, Cooley L, Segraves WA. Ecdysone response genes govern egg chamber development during mid-oogenesis in Drosophila. Development. 1999;126:4581–9. doi: 10.1242/dev.126.20.4581. [DOI] [PubMed] [Google Scholar]
- 27.Terashima J, Bownes M. A microarray analysis of genes involved in relating egg production to nutritional intake in Drosophila melanogaster. Cell Death Differ. 2005;12:429–40. doi: 10.1038/sj.cdd.4401587. [DOI] [PubMed] [Google Scholar]
- 28.Schubiger M, Carré C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–48. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- 29.König A, Yatsenko AS, Weiss M, Shcherbata HR. Ecdysteroids affect Drosophila ovarian stem cell niche formation and early germline differentiation. EMBO J. 2011;30:1549–62. doi: 10.1038/emboj.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2000;60:1–73. doi: 10.1016/S0083-6729(00)60016-X. [DOI] [PubMed] [Google Scholar]
- 31.Jang AC, Chang YC, Bai J, Montell D. Border-cell migration requires integration of spatial and temporal signals by the BTB protein Abrupt. Nat Cell Biol. 2009;11:569–79. doi: 10.1038/ncb1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis VA, Zorzano A, Teleman AA. dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr Biol. 2010;20:1799–808. doi: 10.1016/j.cub.2010.08.055. [DOI] [PubMed] [Google Scholar]
- 33.Zheng X, Wang J, Haerry TE, Wu AY, Martin J, O’Connor MB, et al. TGF-beta signaling activates steroid hormone receptor expression during neuronal remodeling in the Drosophila brain. Cell. 2003;112:303–15. doi: 10.1016/S0092-8674(03)00072-2. [DOI] [PubMed] [Google Scholar]
- 34.Lee T, Marticke S, Sung C, Robinow S, Luo L. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron. 2000;28:807–18. doi: 10.1016/S0896-6273(00)00155-0. [DOI] [PubMed] [Google Scholar]
- 35.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 36.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 37.Gomez-Orte E, Belles X. MicroRNA-dependent metamorphosis in hemimetabolan insects. Proc Natl Acad Sci U S A. 2009;106:21678–82. doi: 10.1073/pnas.0907391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caygill EE, Johnston LA. Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol. 2008;18:943–50. doi: 10.1016/j.cub.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sokol NS, Xu P, Jan YN, Ambros V. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 2008;22:1591–6. doi: 10.1101/gad.1671708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–82. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin H, Kim VN, Hyun S. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 2012;26:1427–32. doi: 10.1101/gad.192872.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boulan L, Martín D, Milán M. bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol. 2013;23:473–8. doi: 10.1016/j.cub.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 43.Kawashima H, Numakawa T, Kumamaru E, Adachi N, Mizuno H, Ninomiya M, et al. Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience. 2010;165:1301–11. doi: 10.1016/j.neuroscience.2009.11.057. [DOI] [PubMed] [Google Scholar]
- 44.Vreugdenhil E, Verissimo CS, Mariman R, Kamphorst JT, Barbosa JS, Zweers T, et al. MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology. 2009;150:2220–8. doi: 10.1210/en.2008-1335. [DOI] [PubMed] [Google Scholar]
- 45.Huang YH, Lin YH, Chi HC, Liao CH, Liao CJ, Wu SM, et al. Thyroid hormone regulation of miR-21 enhances migration and invasion of hepatoma. Cancer Res. 2013;73:2505–17. doi: 10.1158/0008-5472.CAN-12-2218. [DOI] [PubMed] [Google Scholar]
- 46.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–21. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311. doi: 10.1371/journal.pgen.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Åkerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, et al. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879–89. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boissart C, Nissan X, Giraud-Triboult K, Peschanski M, Benchoua A. miR-125 potentiates early neural specification of human embryonic stem cells. Development. 2012;139:1247–57. doi: 10.1242/dev.073627. [DOI] [PubMed] [Google Scholar]
- 51.Feng W, Feng Y. MicroRNAs in neural cell development and brain diseases. Sci China Life Sci. 2011;54:1103–12. doi: 10.1007/s11427-011-4249-8. [DOI] [PubMed] [Google Scholar]
- 52.Jovičić A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33:5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci U S A. 2013;110:7056–61. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parsons XH, Parsons JF, Moore DA. Genome-Scale Mapping of MicroRNA Signatures in Human Embryonic Stem Cell Neurogenesis. Mol Med Ther. 2012;1 doi: 10.4172/2324-8769.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stappert L, Borghese L, Roese-Koerner B, Weinhold S, Koch P, Terstegge S, et al. MicroRNA-based promotion of human neuronal differentiation and subtype specification. PLoS One. 2013;8:e59011. doi: 10.1371/journal.pone.0059011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng R, Cohen SM. Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development. 2012;139:1427–34. doi: 10.1242/dev.075143. [DOI] [PubMed] [Google Scholar]
- 57.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–71. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marrone AK, Edeleva EV, Kucherenko MM, Hsiao NH, Shcherbata HR. Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol. 2012;13:26. doi: 10.1186/1471-2121-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107:1876–81. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3:108–24. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, et al. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–44. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu XS, Chopp M, Wang XL, Zhang L, Hozeska-Solgot A, Tang T, et al. MicroRNA-17/92 cluster mediates the proliferation and survival of neural progenitor cells after stroke. J Biol Chem. 2013;288:12478–88. doi: 10.1074/jbc.M112.449025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishimoto H, Sakai T, Kitamoto T. Ecdysone signaling regulates the formation of long-term courtship memory in adult Drosophila melanogaster. Proc Natl Acad Sci U S A. 2009;106:6381–6. doi: 10.1073/pnas.0810213106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zollman S, Godt D, Privé GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci U S A. 1994;91:10717–21. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Torrado R, Yamada D, Defossez PA. Born to bind: the BTB protein-protein interaction domain. Bioessays. 2006;28:1194–202. doi: 10.1002/bies.20500. [DOI] [PubMed] [Google Scholar]
- 68.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Privé GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu S, Fambrough D, Atashi JR, Goodman CS, Crews ST. The Drosophila abrupt gene encodes a BTB-zinc finger regulatory protein that controls the specificity of neuromuscular connections. Genes Dev. 1995;9:2936–48. doi: 10.1101/gad.9.23.2936. [DOI] [PubMed] [Google Scholar]
- 70.Mitchelmore C, Kjaerulff KM, Pedersen HC, Nielsen JV, Rasmussen TE, Fisker MF, et al. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277:7598–609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- 71.Baubet V, Xiang C, Molczan A, Roccograndi L, Melamed S, Dahmane N. Rp58 is essential for the growth and patterning of the cerebellum and for glutamatergic and GABAergic neuron development. Development. 2012;139:1903–9. doi: 10.1242/dev.075606. [DOI] [PubMed] [Google Scholar]
- 72.Xiang C, Baubet V, Pal S, Holderbaum L, Tatard V, Jiang P, et al. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 2012;19:692–702. doi: 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo M, Bier E, Jan LY, Jan YN. tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron. 1995;14:913–25. doi: 10.1016/0896-6273(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 74.Salzberg A, D’Evelyn D, Schulze KL, Lee JK, Strumpf D, Tsai L, et al. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–87. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 75.Giniger E, Tietje K, Jan LY, Jan YN. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development. 1994;120:1385–98. doi: 10.1242/dev.120.6.1385. [DOI] [PubMed] [Google Scholar]
- 76.Sugimura K, Satoh D, Estes P, Crews S, Uemura T. Development of morphological diversity of dendrites in Drosophila by the BTB-zinc finger protein abrupt. Neuron. 2004;43:809–22. doi: 10.1016/j.neuron.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 77.Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci U S A. 1996;93:9687–92. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryner LC, Goodwin SF, Castrillon DH, Anand A, Villella A, Baker BS, et al. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–89. doi: 10.1016/S0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 79.Zhu S, Lin S, Kao CF, Awasaki T, Chiang AS, Lee T. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 2006;127:409–22. doi: 10.1016/j.cell.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 80.Bauer S, Kerr BJ, Patterson PH. The neuropoietic cytokine family in development, plasticity, disease and injury. Nat Rev Neurosci. 2007;8:221–32. doi: 10.1038/nrn2054. [DOI] [PubMed] [Google Scholar]
- 81.Ngo KT, Wang J, Junker M, Kriz S, Vo G, Asem B, et al. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol. 2010;346:284–95. doi: 10.1016/j.ydbio.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Copf T, Goguel V, Lampin-Saint-Amaux A, Scaplehorn N, Preat T. Cytokine signaling through the JAK/STAT pathway is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A. 2011;108:8059–64. doi: 10.1073/pnas.1012919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bauer S, Patterson PH. Leukemia inhibitory factor promotes neural stem cell self-renewal in the adult brain. J Neurosci. 2006;26:12089–99. doi: 10.1523/JNEUROSCI.3047-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yasugi T, Umetsu D, Murakami S, Sato M, Tabata T. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135:1471–80. doi: 10.1242/dev.019117. [DOI] [PubMed] [Google Scholar]
- 85.Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev Cell. 2008;14:726–38. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 86.Siegrist SE, Haque NS, Chen CH, Hay BA, Hariharan IK. Inactivation of both Foxo and reaper promotes long-term adult neurogenesis in Drosophila. Curr Biol. 2010;20:643–8. doi: 10.1016/j.cub.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grieder NC, Charlafti I, Kloter U, Jäckle H, Schäfer U, Gehring WJ. Misexpression screen in Drosophila melanogaster aiming to reveal novel factors involved in formation of body parts. Genetics. 2007;175:1707–18. doi: 10.1534/genetics.106.064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, et al. chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell self-renewal in Drosophila. Dev Cell. 2010;18:556–68. doi: 10.1016/j.devcel.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jontes JD, Phillips GR. Selective stabilization and synaptic specificity: a new cell-biological model. Trends Neurosci. 2006;29:186–91. doi: 10.1016/j.tins.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 90.Winther M, Berezin V, Walmod PS. NCAM2/OCAM/RNCAM: cell adhesion molecule with a role in neuronal compartmentalization. Int J Biochem Cell Biol. 2012;44:441–6. doi: 10.1016/j.biocel.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 91.Han MR, Schellenberg GD, Wang LS, Alzheimer’s Disease Neuroimaging Initiative Genome-wide association reveals genetic effects on human Aβ42 and τ protein levels in cerebrospinal fluids: a case control study. BMC Neurol. 2010;10:90. doi: 10.1186/1471-2377-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hussman JP, Chung RH, Griswold AJ, Jaworski JM, Salyakina D, Ma D, et al. A noise-reduction GWAS analysis implicates altered regulation of neurite outgrowth and guidance in autism. Mol Autism. 2011;2:1. doi: 10.1186/2040-2392-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107:9270–4. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Atz ME, Rollins B, Vawter MP. NCAM1 association study of bipolar disorder and schizophrenia: polymorphisms and alternatively spliced isoforms lead to similarities and differences. Psychiatr Genet. 2007;17:55–67. doi: 10.1097/YPG.0b013e328012d850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bisaz R, Conboy L, Sandi C. Learning under stress: a role for the neural cell adhesion molecule NCAM. Neurobiol Learn Mem. 2009;91:333–42. doi: 10.1016/j.nlm.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 96.Bisaz R, Boadas-Vaello P, Genoux D, Sandi C. Age-related cognitive impairments in mice with a conditional ablation of the neural cell adhesion molecule. Learn Mem. 2013;20:183–93. doi: 10.1101/lm.030064.112. [DOI] [PubMed] [Google Scholar]
- 97.Fushima K, Tsujimura H. Precise control of fasciclin II expression is required for adult mushroom body development in Drosophila. Dev Growth Differ. 2007;49:215–27. doi: 10.1111/j.1440-169X.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- 98.Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129:409–19. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- 99.Whitlock KE. Development of Drosophila wing sensory neurons in mutants with missing or modified cell surface molecules. Development. 1993;117:1251–60. doi: 10.1242/dev.117.4.1251. [DOI] [PubMed] [Google Scholar]
- 100.Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 101.Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen S, Spletter M, Ni X, White KP, Luo L, Long M. Frequent recent origination of brain genes shaped the evolution of foraging behavior in Drosophila. Cell Rep. 2012;1:118–32. doi: 10.1016/j.celrep.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Albertson R, Chabu C, Sheehan A, Doe CQ. Scribble protein domain mapping reveals a multistep localization mechanism and domains necessary for establishing cortical polarity. J Cell Sci. 2004;117:6061–70. doi: 10.1242/jcs.01525. [DOI] [PubMed] [Google Scholar]
- 104.Brown S, Hu N, Hombría JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–5. doi: 10.1016/S0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 105.Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–31. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 106.Shcherbata HR, Yatsenko AS, Patterson L, Sood VD, Nudel U, Yaffe D, et al. Dissecting muscle and neuronal disorders in a Drosophila model of muscular dystrophy. EMBO J. 2007;26:481–93. doi: 10.1038/sj.emboj.7601503. [DOI] [PMC free article] [PubMed] [Google Scholar]