Abstract
Circadian rhythms have a profound influence on most bodily functions: from metabolism to complex behaviors. They ensure that all these biological processes are optimized with the time-of-day. They are generated by endogenous molecular oscillators that have a period that closely, but not exactly, matches day length. These molecular clocks are synchronized by environmental cycles such as light intensity and temperature. Drosophila melanogaster has been a model organism of choice to understand genetically, molecularly and at the level of neural circuits how circadian rhythms are generated, how they are synchronized by environmental cues, and how they drive behavioral cycles such as locomotor rhythms. This review will cover a wide range of techniques that have been instrumental to our understanding of Drosophila circadian rhythms, and that are essential for current and future research.
1. Introduction
Circadian rhythms are biological events that occur with a period length of about 24 hours. The name is derived from the Latin words “circa” and “diem”, which means “about a day”. They are driven by molecular clocks and are found in most organisms, from cyanobacteria to humans. These molecular pacemakers allow organisms to accurately predict rhythmic changes in their environment and thus increase their fitness. Anticipation of dawn, for example, helps a nocturnal animal to avoid predators active during the day and provides a safe window for activities such as feeding, sleep and reproduction. In mammals, the circadian clock in the brain orchestrates behavioral, hormonal and other physiological rhythms throughout the body [1]. In Drosophila, it gates eclosion and courtship, determines the period of rest and activity, the timing of feeding and influence temperature preference [2, 3]. Besides controlling various behaviors, the Drosophila circadian clock also coordinate many rhythms in peripheral organs, such as olfactory and gustatory sensitivity rhythms [4, 5], and the mitotic response of gut stem cells to damage [6]. Clocks help organisms in unexpected ways, too. For example, navigation using the sun as a compass requires a functional circadian clock in insects and birds [7–9]. The position of the sun changes throughout the day and circadian clocks provide the essential timing information to compensate for this change and adjust flight direction accordingly. The broad impact of circadian clocks makes them of particular importance in the general field of biology, and uncovering the mechanisms involved in their generation, regulation and output pathways is essential.
Interestingly, although the individual molecular components of the circadian clock are not always homologous, its features, organization and the molecular mechanism that generates rhythmicity are very similar across kingdoms [10, 11]. In all organisms, circadian clocks are endogenous and can sustain their rhythmicity in the absence of environmental cues. This rhythmicity is also independent of ambient temperature. However, various time cues (also called Zeitgebers, which means time-givers in German) such as light and temperature cycles, and in many cases nutrient availability, can synchronize (entrain) the clock. At the molecular level, circadian rhythms are generated by a negative transcriptional feedback loop, which involves transcription factors that drive their own repressors. These repressors are modified throughout the day by various means (such as phosphorylation) and eventually degraded, thereby starting a new cycle.
Our knowledge of the basis of circadian rhythm generation and its entrainment by environmental cycles has been profoundly influenced by research using Drosophila. The roots of this influence can be traced back to Colin Pittendrigh, one of the founding fathers of chronobiology, who used various Drosophila species to study fundamental aspects of circadian clocks, such as entrainment and temperature compensation [12–15]. Further critical influence came from the work of Seymour Benzer and Ronald Konopka and their initial forward mutagenesis screen using Drosophila eclosion, in which they identified the first circadian gene: period [16]. Their work and that of many others following these seminal studies, as well as the powerful techniques developed by other Drosophila scientists, made fruit flies especially suited to investigate circadian rhythms.
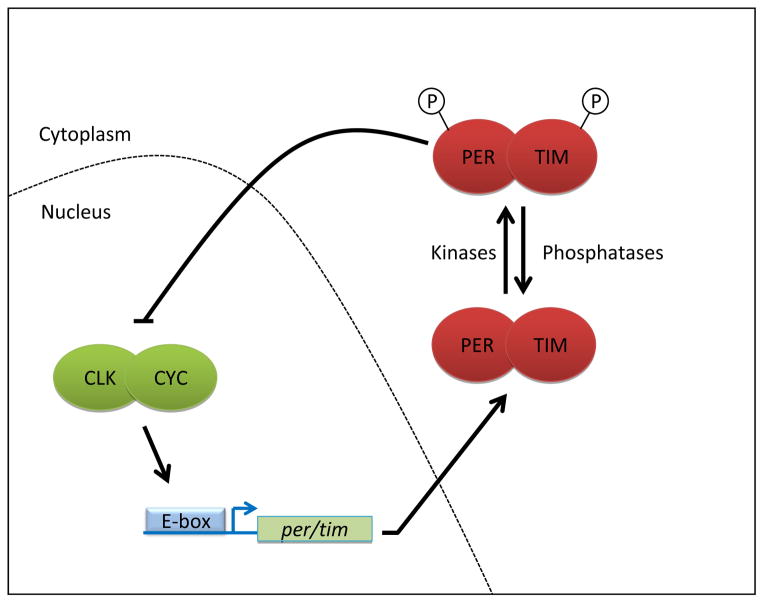
We have now a deep understanding of the Drosophila circadian pacemaker (Figure 1, for review, see for example [2, 17]). The circadian transcription factors CLOCK (CLK) and CYCLE (CYC) form a heterodimeric complex and promote period (per) and timeless (tim) transcription [18–20]. PER and TIM accumulate during the night and form a heterodimer as well [21, 22]. The PER/TIM complex enters the nucleus and promotes the phosphorylation of CLK/CYC, which inhibits its activity, and reduces its affinity for DNA [23, 24]. However, PER and TIM are also gradually modified by phosphorylation during the day [21, 25]. This eventually results in their degradation and releases CLK/CYC from repression to start a new cycle. This molecular clock receives one of the strongest environmental inputs, light, through the activation of CRYPTOCHROME (CRY) [26–28]. Upon photon absorption, this blue-light photoreceptor undergoes a conformational change that allows it to bind TIM [29–31]. This promotes TIM ubiquitination and proteasomal degradation, thereby resetting the clock [21, 32–37]. The neurons that control circadian locomotor behavior can also receive light information through an as yet poorly characterized neuronal pathway that originates from visual photoreceptors [38].
Figure 1. The transcriptional feedback loop of the Drosophila circadian clock.
CLK/CYC drive expression of their own repressors PER and TIM. PER/TIM go through various modifications during the day, until they are eventually turned over to release CLK/CYC from repression, starting the next cycle.
The pacemaker mechanisms we just described are remarkably well conserved in mammals and humans [1]. Actually, conservation extends to the neural circuits controlling circadian behavior. Indeed, homologous neuropeptides and receptors are involved in the control of rhythmic behavior [39]. Drosophila is thus a fantastic model organism to understand the basic molecular and neural underpinnings of circadian rhythms. Here, we will attempt to review the many approaches that have been developed to understand these pathways and the molecular mechanisms of rhythm generation. We hope our review will provide a solid background into the history of these techniques as well as their strengths and caveats.
2. Circadian behaviors
2.1. Eclosion
Eclosion is a key event in the development of Drosophila and is defined as the emergence of the adult fly from the pupal case. This event occurs primarily in the morning and has been very successfully used to study circadian rhythms by Colin Pittendrigh. He demonstrated that the mechanism that drives eclosion is a true biological clock, because it satisfied all three fundamental properties of a circadian clock: being endogenous, entrainable and temperature compensated [11–15]. Pittendrigh’s eclosion monitors have been improved, computerized and have become available from private manufacturers. A modern version produced by Trikinetics (Waltham, MA) consists of a reusable pupae disk above a funnel and an electronic solenoid attached to a computer that taps this disk. The flies that emerge are counted by an infrared sensor as they fall. This version allows unattended monitoring for days, occupies significantly less space and generates digital data.
Eclosion was the read-out used by Konopka and Benzer in their ground-breaking study that transformed the field of chronobiology and moved it into the genetics era [16]. In a later study, M.W. Young and his colleagues identified timeless as a clock gene also using eclosion [40]. However, eclosion is a population rhythm that occurs only once in the lifetime of Drosophila. This is a significant limitation, particularly if high throughput is required. Thus, single fly assays have progressively replaced eclosion. Adult locomotor behavior and luciferase based methods are now much more commonly used read-outs for the study of circadian rhythms (see sections 2.2 and 3.5). This said, the study of eclosion and its circadian control has important implications, such as population control of insects in the wild.
2.2. Locomotion
2.2.1. Monitoring rhythmic locomotor behavior
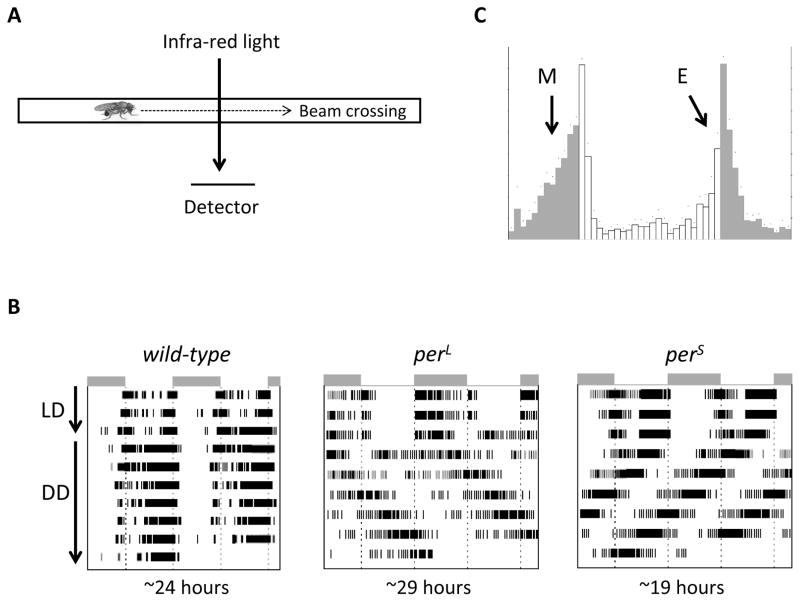
Contributions by Pittendrigh on the eclosion rhythm and its dependence on light have spurred a wide range of studies that followed. In one such study, the wavelength of light that is required to shift the circadian clock was characterized and found to be in the blue region of visible light [41]. An unintended consequence of this study was cleverly utilized by Dr. Yoshiki Hotta, a post-doctoral researcher in Seymour Benzer’s laboratory. He developed a device to monitor the locomotor activity of a single fly based on the fact that infra-red light has no effect on the circadian clock. In his contraption, flies were housed in individual tiny glass tubes (with food) which was crossed by an infra-red light beam hitting photovoltaic cells that emitted an electronic signal each time the fly crossed the beam (figure 2A). This method overcame the two major limitations of eclosion: being a single life event and a population rhythm. Konopka and Benzer used adult locomotor behavior to demonstrate that in addition to eclosion, per also affects this behavior, further supporting its role as a core clock gene [16]. This method, like eclosion monitoring, has been modernized and is ubiquitously used today to monitor locomotor activity rhythms of single flies [42]. The most commonly used monitors are those produced by Trikinetics. While usually small tubes are used, modifications of the system exists for longer recordings of locomotor behavior such as “comfortable” cells that are larger, with access to water in addition to food [43]. Also, groups of flies can be monitored using large vials and monitors (Trikinetics).
Figure 2. Locomotor activity monitoring.
(A) Fly locomotion is detected when a fly breaks an infra-red beam crossing the small glass tube in which it is housed (B) Double-plotted actogram showing the activity of flies entrained to a 12/12-hr LD cycle and then released in constant darkness for period determination. Each day is plotted twice, first on the right and duplicated on the left half of the next line, except for the first day. Note the progressive drift of circadian behavior in constant conditions in per mutant flies, corresponding to long and short periods (C) Eduction plot of fly activity after entrainment to an LD cycle. The Morning (M) and Evening (E) anticipatory behavior driven by the circadian clock are shown with arrows.
The high-throughput nature of locomotor monitoring has considerably facilitated the study of all aspects of circadian rhythms: from input detection to output mechanisms. Its most salient success has been the discovery of many pacemaker genes through forward genetic screens. Because of the importance of locomotion as a circadian read-out, we describe in this section how to determine circadian period, study light inputs and the role of natural cycles for entrainment. However, similar principles apply to any read-out, whether behavioral or molecular.
2.2.2. Determining circadian period
One of the fundamental properties of a circadian clock is that it is endogenous to the organism and it persists in constant conditions [11]. Studying the period length of circadian rhythms provides critical insight into the workings of the clock. Therefore, determination of period length in constant darkness (and constant temperature) has become a standard assay to test whether a given perturbation has any effect on the clock. In this assay, flies are loaded into individual tubes and housed in incubators with temperature (usually 25°C) and lightning control. They are then subjected to three days (at least) of 12 h/12 h light/dark (LD) cycles followed by a release into constant darkness (DD). Mathematical determination of period length can be based on different approaches (periodogram, MESA, auto-correlation) and requires at least 5 days in DD [44, 45]. It is also recommended that the assay be run with at least 8 flies per genotype and repeated three times for satisfactory statistical analysis. The locomotor behavior of individual flies or group averages is often plotted in an “actogram” format where period length, phase and amplitude can be visually observed. In these plots, flies with a period length shorter or longer than 24 hours will seem to drift towards left or right in the actogram, respectively (figure 2b). Multiple program and toolboxes are available for mathematical analysis of the properties of circadian behavior (period, but also phase and amplitude) and for generation of actograms (e.g. Matlab (Mathwork, Natick, MA) toolbox [44], FAAS [46], ClockLab (Actimetrics, Willmette, IL); figure 2B).
2.2.3. Studying circadian light (or temperature) responses
Although endogenous, circadian clocks are synchronized by environmental cues. A shift in the Light: Dark (LD) (or temperature) cycle results in the clock gradually aligning itself to the new cycle. This is called entrainment. To verify that flies have really entrained to an environmental input, behavioral phase should be measured in constant conditions before and after entrainment, or behavioral phase should be compared between flies subjected or not to the entraining cue. Indeed, locomotor behavior can also respond to change in the environment independently of the circadian clock, a phenomenon called masking [47]. Masking disappears once flies are returned to constant conditions. The kinetics of entrainment can be used as a measure of the sensitivity of the circadian pacemaker to an input. For example, the circadian behavior of flies defective for CRY-dependent photoreception takes longer to entrain to a shifted LD cycle than control flies [26, 27, 37].
Once a stable phase relationship with the environmental cycle is reached, the activity profile during a circadian cycle under entrained conditions can be shown in “eduction” graphs (figure 2c), where the activity is plotted against time of day. Most frequently, eductions are average daily activity of a group of flies over several days. Under normal LD conditions (12 h of light and 12 h of dark), an anticipatory increase in activity is observed before dawn and dusk (Figure 2c), which are aptly named the “morning” and the “evening” peaks. The photoperiod (light phase duration) can be varied, and flies adapt to photoperiod length by adjusting the timing of the evening and morning anticipation [48]. Interestingly, flies also adapt the phase of their behavior to ambient temperature, earlier under cold temperature and later under warm conditions [49]. This adjustment is dependent on a splicing in the 3′-UTR of per.
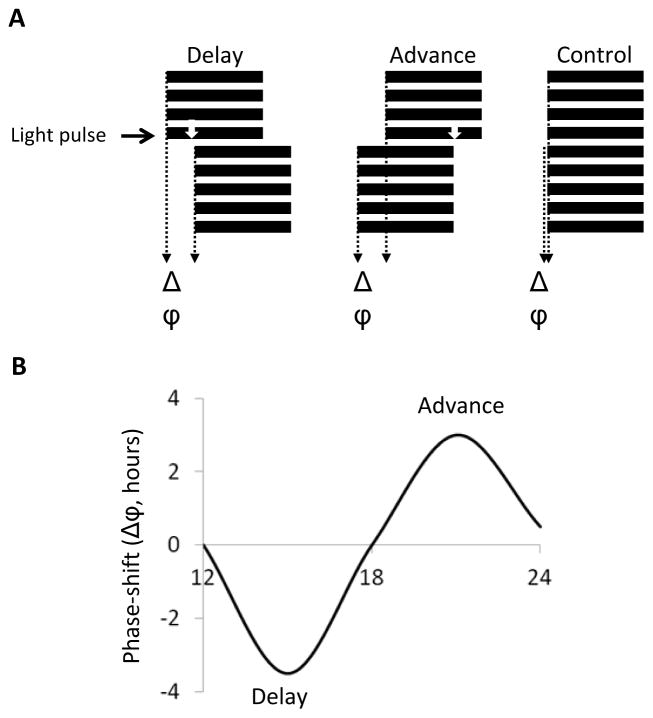
The phase of circadian clocks can also be shifted by brief pulses of stimuli. Here, flies are usually entrained to a LD cycle, and then pulsed with light or an increase in temperature during the night of the last LD cycle, or the “subjective” day that follows [50–53]. Phase (Φ) is then measured in constant conditions, and compared to that of flies that have not been pulsed (figure 3a). To obtain precise phase measurement, it is best to use at least 16 flies per time point and per genotypes, as phase is more variable than period in individual flies. Different phase markers can be used, such as peak of activity or the midpoint of the descending part of the evening peak (see for example [29, 53]). The programs mentioned above for period measurements all have functions to calculate phase.
Figure 3. Analyzing phase-shifts.
(A) Flies are entrained to an LD cycle and pulsed with a brief light pulse during the early or late night (white arrow). The phase in pulsed flies is compared to non-pulsed controls (Δϕ) (B) A Phase-response curve (PRC) can be generated by plotting phase delays (−) and phase advances (+) as a function of time of light pulse (or any other stimuli).
The direction (+ for advances, − for delays) and strength of the change in the phase of the circadian clock can be depicted in “Phase-response curves” (PRC, Figure 3b). Indeed, the amplitude and directionality (advance, delays) of the phase shift is a function of time, but also of pulse intensity and duration. High intensity light pulses as short as 1 minute given during the early or late night induce >3 h phase delays or advances [54]. Usually however, 5–10 minute light pulses are used (e.g. [27, 53]). Recent studies indicate that the duration/intensity relationship is not linear, as the circadian photoreceptor CRY somehow integrates photon detection during long pulses [55]. Flies are less responsive to temperature input. 30-minutes of a 37°C pulse are required to elicit a ca. 2.5 h phase delay [51, 52]. Longer pulses are required at temperatures that flies more frequently encounter. This translates into slower entrainment to shifted temperature cycles, compared to light cycles [56].
PRCs have been an invaluable tool in elucidating the light-input pathway to the clock in Drosophila (e.g. [26, 29, 37, 53, 57–59]. For example, light input pathways mediated by CRY or the eyes can be dissociated with the use of a 5-minute light pulse given at night. Indeed, acute photic responses are totally dependent on CRY [27], while entrainment to a LD cycle can rely on both [38]. Sensitivity of the CRY input pathway can be tested by varying light intensities, or the duration of a pulse at a given light intensity [28, 54, 55].
Another way to probe specifically CRY-dependent photoreception is to place flies in constant light (LL). Wild-type flies are arrhythmic under these conditions because of constant CRY activation and hence TIM degradation, but cry mutant flies are rhythmic [60, 61]. The LL assay has resulted in the isolation of an additional cry mutant (crym) [29], as well as identification of the jet gene, which encodes a subunit of an E3-ubiquitin ligase responsible for light-dependent TIM degradation [37].
2.2.4. Natural cycles
It is common practice in the field of circadian biology to use square shaped stimuli to entrain the clock (e.g. cycles with 12 hours of bright light and 12 hours of complete darkness). Furthermore, ambient temperature is usually kept constant, flies are individually housed and food is provided ad libitum. However, these conditions are far from what flies experience in their natural environment. Light and temperature are well known to work synergistically to entrain the Drosophila clock [62, 63]. In addition, flies do not live in isolation and are exposed to various social stimuli [64]. Understanding how environmental cues are integrated by the clock is important if we are to elucidate the mechanisms required for entrainment under natural conditions. Therefore, several new approaches have been taken recently to more closely mimic natural conditions.
Under natural conditions, temperature rises gradually during the day. A thorough examination of such temperature cycles have shown that clocks are very sensitive to this stimulus and can entrain to a temperature cycle with an amplitude of 4°C [65]. As observed with more square temperature cycles however, circadian clocks re-entrain slowly (days) if a large phase shift is applied. Light also gradually increases at dawn and dusk, and light is never null. Recent studies have thus used for example moonlight intensities during the night, and progressive photic dawn and dusk. Behavior phase and entrainment were in some cases affected, and molecular correlates identified [66, 67].
Perhaps the most surprising results were obtained when the traditional activity monitors were placed outside the laboratory environment and the flies were exposed to natural variables. Under these conditions (or when these conditions are mimicked more closely in the laboratory), the morning anticipatory activity seems much less dependent on a functional clock [68]. Interestingly, temperature seems to be a strong Zeitgeber in the wild, while under lab conditions light is dominant [68]. Flies also exhibit an afternoon peak when temperature is high, which might be actually an escape response [68, 69]. Whether this peak is actually controlled by circadian clocks is disputed [69]. Results are not surprisingly quite complex and very dependent on daily physical properties of the environment [68, 70]. Incubators in which light, temperature and humidity can be computer-controlled to better mimic natural conditions should prove in the future important to understand the intricacies of entrainment under natural conditions.
2.3. Courtship, feeding and temperature preference
Circadian clocks also control reproductive behaviors such as courtship in Drosophila. Loss of clock function results in arrhythmic courtship behavior [71, 72]. Moreover, specific circadian neurons drive rhythmic courtship [73, 74]. Such circadian outputs were detected by visual observations of courtship or approaches made by the male fly to female (close-proximity rhythm) [71]. In the case of nocturnal sex drive, locomotor behavior was used as a proxy [72]. These techniques have also been computerized and it is now possible to monitor a large number of flies for reproductive behavior using camera-based systems [75].
Drosophila is poikilothermic and thus cannot internally regulate its body temperature. Instead, they move towards a preferred temperature [76]. This behavior is clearly observed when flies are air blown into a chamber with a temperature gradient. Interestingly, the preferred temperature is a function of time-of-day and under circadian control [3]. This might be a functional homolog to the body temperature rhythm of mammals.
In many organisms, feeding behavior is driven by the circadian clock and Drosophila is no exception. Rhythmic feeding of flies depends on functional and coordinated actions of circadian clocks in the brain and digestive system [77]. This behavior can be measured by either feeding flies with a dye and analyzing the dye content of the homogenates or by measuring the consumption of a sucrose solution fed through a capillary tube (CAFÉ assay) [77, 78].
These and other recent developments in behavioral monitoring allow researchers to continuously follow even the most miniscule events in the life of a fly [79, 80]. This is an exciting time for Drosophila circadian research as novel questions can be tackled using new behavioral assays to understand the specific role and mechanisms by which the circadian clock control daily behavioral rhythms.
3. Molecular clocks
3.1. Basic approaches to measure circadian mRNA and protein levels
Over the ca. 8 years that followed the cloning of the per gene by M. Rosbash and J. C. Hall at Brandeis, and M. W. Young at Rockefeller [81–83], it was discovered that per transcription, mRNA and protein abundance cycle [84–87]. This led to the central concept of the circadian transcriptional feedback loop (section 1). The expression of several other pacemaker genes (clk, tim, pdp1, vri), as well as cry (input) and many output genes are under circadian control [2]. Finally, protein modifications such as phosphorylation are also frequently rhythmic. Thus, methods to measure transcriptional, mRNA and protein rhythms are essential for circadian rhythm research.
Rhythmic gene expression is frequently measured from whole heads, as this part of the fly contains a high concentration of tissue with circadian clocks and is easily separated from the rest of the fly by vortexing frozen flies. mRNA and proteins can then be isolated with standard extraction approaches (see for example [88, 89]). Northern blots or RNAse protection assays have been largely replaced by quantitative real-time PCR to measure mRNA levels. For proteins, traditional Western Blots are mostly used, and can detect phosphorylation cycles through changes in mobility [21, 25, 90]. Time points are usually collected every 2–4 hours. It is essential to normalize data to a constantly expressed mRNA or protein control to compensate for variation in samples.
While much of what we have learned about the molecular circadian pacemaker is based on whole head extracts, what is mostly detected is circadian proteins from the eyes [91]. The neurons that control circadian behavior contribute to a small fraction of the signal. Thus, it is also important to use immunohistochemistry for proteins and in situ hybridization for mRNAs to directly correlate circadian behavioral observation with molecular phenotypes (for methods, see for example [66, 92–94]. There are in fact differences between circadian pacemakers in peripheral tissues and circadian neurons. For example, translation control of PER expression is particularly important for the pacemaker neurons driving circadian behavior, the small ventral lateral neurons (see section 4) [95–98].
3.2 S2 cell-based assays
S2 cells are a very commonly used embryonically-derived cell line. They do not have circadian clocks though, and do not express several important clock genes. The absence of a cell line with a circadian clock is actually the only weakness of Drosophila as a model to study circadian rhythms. Nevertheless, S2 cells have been very frequently used to recapitulate certain aspects of the circadian clock. The establishment of a transcriptional assay that showed that PER and TIM repress the activity of the CLK/CYC dimer was a major development [20]. This is a classic transcriptional assay, in which fragments of per and tim promoters are placed in front of luciferase (see also section 3.5). CLK is co-transfected with the luc constructs and increases their expression. CYC is expressed endogenously in S2 cells. PER and TIM coexpression represses CLK/CYC activity. This assay has been and is still widely used to understand the mechanisms of circadian transcription. PER phosphorylation and degradation have been studied in detail in S2 cells as well. For example, DBT expression can be placed under the control of the inducible Metallothioneine promoter. Soon after DBT induction with copper, PER becomes hyperphosphorylated and is degraded in a SLIMB-dependent manner, as in vivo [46, 99]. CRY and TIM degradation can be recapitulated in these cells as well [36, 37]. Although very useful for initial studies, any conclusion needs to be validated in vivo, since S2 cells do not contain a functional circadian clock
3.3 Genomic approaches
A critical development in molecular biology has been the powerful approaches to interrogate expression profile at the genomic level. They are having an important impact on the field of chronobiology. Microarray studies have shown that up to 10 percent of the mRNAs of the genome might be driven by the clock, which in turn controls various downstream pathways, further increasing the involvement and impact of the circadian clock [100–104]. However, it should be noted that these studies surprisingly showed little overlap in the mRNAs that cycle, probably because of methodology differences. A meta-analysis of all these data indicated that about 200 genes show robust cycling in heads [105]. Recently, methods to isolate specific population of neurons and extract mRNAs from them have been tremendously improved. This has allowed profiling genome-wide expression in a few groups of circadian neurons (see section 4) and has revealed interesting differences between them [106–109]. These approaches might prove particularly potent to understand how different circadian neurons perform their respective functions.
The binding of CLK/CYC heterodimer to E-boxes on their target promoters and the rhythmic repression of this activity by PER/TIM play a critical role in the generation of circadian mRNA rhythms. In fact, CLK/CYC binding to E-boxes was shown to be rhythmic initially by Hardin and colleagues using Chromatin Immunoprecipitation techniques (ChIP) [90]. PER association to the chromatin has also been followed [23]. ChiP was used to map CLK/CYC and PER binding sites on the genome and the timing of this binding, using tiling arrays. Unexpectedly, RNA polymerase II binding indicates that only 30% of CLK targets show rhythmic transcription [110]. This type of approaches could be extended to rhythmically expressed transcription factor such as CWO, PDP1 and VRI to understand further the circadian transcriptional network.
Recently, deep sequencing (RNA-seq) has also been used to study whole genome circadian expression in Drosophila [111, 112]. This allows precise measurements of expression levels and a detailed study of expression of alternatively spliced mRNAs, and usage of alternative promoters, polyA and termination signals. It can also detect mRNA editing. Isolation and sequencing of nascent transcripts – that have not yet been post-transcriptionally processed - revealed yet another layer of regulation: post-transcriptional regulatory mechanisms amplify mRNA oscillations for many genes, including circadian pacemaker genes [112].
3.4 Proteomic approaches
Analysis of post-translational modifications such as phosphorylation and ubiquitination can be greatly facilitated by mass-spectrometry analysis of purified proteins. PER phosphorylation is by far best understood among Drosophila circadian proteins. PER phosphorylation sites were mapped by mass spectrometry through different approaches. One study used phosphorylated PER with DBT in vitro [113], while two groups isolated PER from S2 cells. The first group focused on DBT-dependent phosphorylation [114], while the other looked for phosphatase-dependent sites [115]. Identification of putative phosphorylation sites was then followed by mutagenesis of these sites and observations of effects of PER phosphorylation, activity and stability in S2 cells and in vivo. Also, whether these mutant PER could or not rescue circadian behavioral and molecular rhythms in per0 flies was tested. Several important phosphorylation sites have now been identified, and it appears that there is a sequence of interdependent phosphorylation event that regulates PER function. Of course, mass spectrometry could also be used to identify interactors of circadian proteins, but there is no such report in Drosophila yet.
3.5 Luciferase-based methods
Luciferases are a class of enzymes that produce light upon oxidation of their substrates. They are commonly used in biology as reporters of transcription or translation and the circadian clock field is no exception. Luciferase-based methods are broadly used to monitor circadian clocks in various organisms. Luciferase has several advantages over other biological reporters such as green fluorescent protein (GFP) that make them particularly suited for circadian research [116, 117]. The most important is the fact that luciferase does not require excitation by light for its readout and therefore avoids phototoxicity and any complications with entrainment or resetting of the clock by repetitive light exposure. It also as a short half-life and there is no significant background luminescence from biological tissues, so the signal to noise ratio is higher than GFP. Its substrate luciferin is non-toxic, relatively cheap (nowadays) and reasonably stable at room temperature and can be mixed in the food or tissue culture medium.
We have briefly described above the basic methods to measure circadian gene expression (section 3.1). These methods are still critical once a particular gene of interest is found. However, they are not suitable for automated measurement of circadian rhythmicity in the early phases of a study. They require samples that are taken at regular intervals, at least every 4 hours and are practically hard since samples need to be collected over several days for proper analysis of phase, period and amplitude in mRNA or protein expression. They also require sacrificing animals or the lysis of the tissue sample, and require many flies (i.e. 20 heads per time-point) or transfected cells, which increase costs and labor even more. Instead, Luciferase enzyme driven by candidate promoters or fusions with a target protein can be used. Flies or cells that are transgenic with such constructs are supplied with luciferin and luminescence is recorded with the help of a photo-multiplier tube based scintillation counter or a CCD camera. Up to 384 flies can be reliably recorded simultaneously in current generation luminescence readers for up to two weeks in a single multi-well plate and phase, period length and amplitude of their rhythms can be analyzed (Figure 4). Use of plate stackers, albeit with caution since they can fail due to the high number of readings, can further increase this throughput.
Figure 4. Luciferase recording.
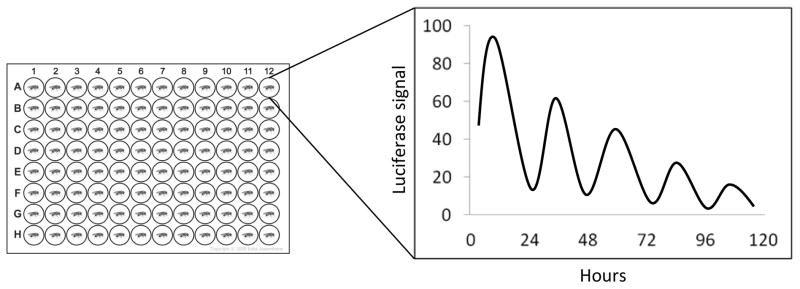
Individual flies are housed in wells of a multiplate containing food with luciferin. The light generated by the luciferase activity is recorded over several days with a photomultiplier-based device. The data is plotted to show the phase, amplitude and period length of the rhythm.
The first luciferase reporter based recording in Drosophila was performed by Brandes et al. in J.C. Hall’s laboratory [118]. A fragment of the period promoter was used to drive the luciferase protein (plo-luc). This study showed that the circadian rhythmicity of individual flies, or heads, can easily be recorded and reflects the circadian expression of endogenous period mRNA. Later, construct including different part of the per gene, including untranslated regions and coding regions, indicated the existence of multiple regulatory elements for per expression [119, 120]. The promoter region of tim was also dissected using luc reporters, and multiple cooperating regulatory sequences were identified [121]. Drosophila strains that harbor CRY [122] and PER [119] translational fusions with luciferase are also available. BG-LUC, for example is a fusion of LUCIFERASE with PER. It accurately reports the circadian expression profile of the PER protein [119]. Therefore, it has been a useful tool in determining the period length of the clock in DD as well as its entrainment.
Our understanding of entrainment has greatly benefited from screens based on luc. First, it was possible with dissected body part to show that peripheral clocks can tissue-autonomously entrain to light and temperature cycles, indicating the presence of cell-autonomous thermal and photic sensors [123, 124]. Importantly, the first mutation in cry was discovered from a screen using BG-LUC recordings in EMS-mutagenized flies [27]. This was possible because most peripheral clocks are strictly dependent on CRY for their entrainment to light, and the luc assay essentially measures peripheral clock oscillations. LUC signal was arrhythmic in cryb mutants under LD cycles. This mutant would not have been identified in a regular LD/DD screen, because circadian behavior is also entrained by the eyes [27, 38]. The first temperature entrainment gene (nocte) was also identified in a luc screen [124, 125].
Like any technique, luciferase-based methods of circadian clock measurement have limitations. Although the throughput using BG-LUC or plo-luc is high (especially when coupled with plate stackers), the rhythms do not persist for more than 3–4 days in constant darkness. This is because rhythms in the periphery are not as robust as locomotor behavior, as also observed with western blots. In addition, unlike GFP, light emitted by luciferase diffuses, which affects the quality of the spatial resolution. Luciferase is thus not suitable for recording from individual Drosophila neurons. Lastly, the activity of luciferase is temperature dependent, which means that appropriate controls need to be used if temperature cycles are used. These limitations, however, are likely to be overcome with the development of new and improved luciferases [126].
4. Probing the circadian neural network
The previous section focused on molecular methods aimed at understanding circadian rhythms. We now turn to approaches that can be used to understand the neural network controlling circadian behavioral rhythms. Note that although peripheral clocks function autonomously and can be directly entrained by light or temperature [123, 124], recent results indicate that circadian clocks in the oenocytes (the pheromone-producing cells) are modulated by brain circadian neurons [127]. Thus the role of circadian neurons extends beyond the direct control of rhythmic behaviors.
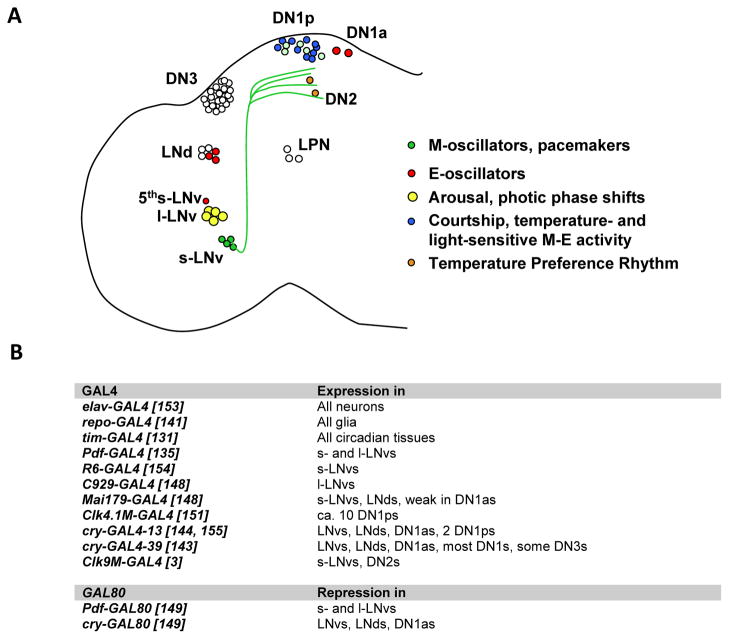
Circadian neurons were first characterized through PER and later TIM immunostaining [86, 87, 128–130]. The use of GAL4 lines (see below) combined with markers such as GFP considerably improved identification of circadian neurons and resolved their neural projections [131]. These neurons have been divided into different groups based on their anatomical localization (Figure 5A): the ventral Lateral Neurons (LNvs), the dorsal Lateral Neurons (LNds), the Dorsal Neurons (DN) 1, 2 and 3, and more recently identified Lateral Posterior Neurons (LPNs). It is important to realize that these are not functional groups. In fact, most of these groups are very heterogeneous in terms of size, pattern of neural projections and neurotransmitter content [2, 132]. For example, the 6 LNds contain at least four different types of circadian neurons [133]. Most LNvs express the critical circadian neuropeptide Pigment Dispersing Factor (PDF), which synchronizes oscillations throughout the brain, and also influence oenocyte rhythms in the fly body. However, the LNvs can be further divided into large and small LNvs (l-LNvs and s-LNvs, respectively), based on their cell body size. These two sets of neurons have very different arborizations and functions. Moreover, one of the five s-LNvs does not express PDF and is referred to as the “5th sLNv”. Determining the function of these different types of neurons thus require methods to specifically target them.
Figure 5. Targeting circadian neurons.
(A) The circadian neurons of Drosophila and their best-known functions. Projection from the s-LNvs are shown in green. (B) Important driver and repressor transgenes and their expression patterns in circadian neurons. References are indicated in brackets.
4.1. Neuron-specific GAL4 and LexA drivers
Heroic efforts based on the study of gynandromorphs and anatomical optic lobe mutants, as well as a promoterless per rescue construct expressed only in few circadian neurons, all pointed to a critical role of the Lateral Neurons in the control of circadian behavior, particularly the PDF positive LNvs [43, 130, 134]. The development of the GAL4/UAS system dramatically facilitated the analysis of the role of circadian neurons, and their anatomy. Indeed, over the last 15 years, a plethora of tissue-specific drivers targeting different subset of circadian neurons, have been developed or isolated from enhancer trap screens (figure 5b). For example, with the isolation of the Pdf gene, it was possible to generate a GAL4 driver expressed in the brain the PDF positive LNvs [135, 136]. This allowed researchers to definitely establish that these neurons are critical pacemaker neurons that drive circadian behavior in constant darkness. However, it should be noted that PDF and thus Pdf-GAL4 are also expressed in a few non-circadian neurons in the abdominal ganglion. In general, it should always be kept in mind that drivers might not just be expressed in clock neurons, in particular if promoters from circadian genes are used, since circadian clocks are found in most organs [123]. Moreover, these drivers might also be active in glial cells in the brain. This is important since glial cells impact circadian locomotor behavior [130, 137, 138]. All glia (with repo-GAL4) or subsets of glial cells can actually also be targeted by specific GAL4 drivers [139–142], and thus determining whether a given phenotype is caused by neurons or glia is easily testable. Finally, GAL4 drivers can show ectopic expression. Also different genomic insertions of the same GAL4 driver can result in different patterns of expression, because the chromatin environment is different. This is the case for example with cry-GAL4 drivers. cry-GAL4-13 shows a much more restricted brain expression than cry-GAL4-39 [143, 144].
The most commonly used circadian GAL4 drivers are Pdf-GAL4 and tim-GAL4 [131, 135, 145]. Both show robust expression. Pdf-GAL4 faithfully reproduces PDF expression and is thus quite specific to the PDF-positive LNvs. tim-GAL4 is expressed in all circadian tissues and thus all circadian neurons. A great advantage of Pdf-GAL4 – besides being expressed quite specifically in key pacemaker neurons - is that it targets tissues that are not necessary for viability. Thus, the role of essential genes such as kay in the control of circadian behavior can be tested using RNA interference [146]. Before, the role of essential genes had to be established using complicated genetic approaches, such as the use of heat-inducible transgenic rescue construct activated only during development [147]. This approach does not work if expression of the gene of interest is required during adulthood for viability.
Other commonly used GAL4 drivers target the LNds (as well as the LNvs), for example the enhancer trap Mai179-GAL4 [148] and cry-GAL4-13 [26, 149]. Dissecting circadian promoters in different pieces can prove very potent to limit expression to different circadian neurons. Fragments of the clk promoter have yielded very specific drivers: the Clk-4.1M-GAL4 driver targets only a subset of DN1s [150, 151], while the Clk-9M-GAL4 driver is restricted to the DN2s and the sLNvs [3]. Recently, a pdf-LexA driver was developed to drive expression of genes of interests independently of GAL4 [59, 152]. More circadian LexA drivers will undoubtedly be generated to express one gene in a subset of circadian neurons, and a different gene in another subset. A non-exhaustive list of important drivers is given in figure 5b.
4.2. Intersectional and exclusion approaches
While having all these GAL4 lines is extremely helpful to dissect the function of circadian neurons, they are frequently not specific enough. Tools to restrict expression further are however available. A common approach is to use the GAL80 repressor [156], which blocks GAL4 transactivation. GAL80 can be placed under the control of a circadian promoter. For example, Pdf-GAL80 is widely used [149]. Combining cry-GAL4-13 with pdf-GAL80 allows targeting only 3 LNds, the 5th s-LNv and two DN1s (the so-called E-oscillators, see below).
Lethality problem may arise when an interesting driver is expressed in essential tissues. This is the case of c929-GAL4, which within circadian neurons is only expressed in l-LNvs [148]. To target the l-LNvs specifically, Shang et al devised a very sophisticated approach [59]. They used a ubiquitously expressed tubulin-GAL80 flanked by FRT sites. In the same flies, they introduced the c929-GAL4 driver and a UAS transgene of interest (the proapoptotic gene hid for example). In addition, they also had Pdf-LexA and LexAop-FLP. Thus, FLP was expressed in all PDF-positive LNvs, and it was therefore only in these cells that GAL80 was excised. This allowed expression of HID controlled by c929-GAL4 only in the l-LNvs. The caveat with this approach is that FLP/FRT is far from being 100% efficient at excising GAL80. Thus, the phenotype obtained can vary dramatically from fly to fly, depending on the number of cells in which the GAL80 excision occurs. For example, when measuring the phase shifts occurring after short light pulses (see section 2.2.3); Shang et al. dissected brains at the end of the behavioral experiments and correlated the amplitude of the phase shifts with the number of hid-ablated cells [59].
Another intersectional approaches that could be used is split-GAL4, but to our knowledge, this has not yet been applied to study circadian rhythms. The idea is to divide GAL4 in two transgenes: the DNA binding domain with a dimerization domain in one, and the activation domain with a complementary dimerization domain in the other. GAL4 is reconstituted only in tissue expressing both transgenes [157].
4.3. Manipulation of circadian neurons
With these approaches to target specific circadian neurons, we have learned enormously about the circadian neural network. Using UAS-hid (or UAS-rpr) to specifically eliminate groups of circadian neurons can reveal the function of specific neurons if they are essential. Eliminating the PDF positive LNvs demonstrated their role as circadian pacemaker neurons [135]. It also showed that there are necessary for morning anticipatory activity. Eliminating the LNds, the 5th sLNv and 2 DN1s showed that these cells are required for the evening anticipation [149]. The opposite experiments, restoring PER expression only in specific circadian neurons in arrhythmic per0 mutant flies beautifully complemented these results [148]. These studies led to current model that the morning and Evening anticipatory behavior is controlled by two separate groups of neurons, the M- and E-oscillators. Rescuing CRY expression in cry mutant flies was used to demonstrate the cell-autonomous nature of circadian photoreception, even in the brain [26].
The battery of neuronal manipulation has recently dramatically increased. RNAi can be used in different clock neurons to inhibit specific genes. Neuronal activity can be manipulated in different ways. Neurons can be silenced with Kir2.1, a potassium channel [158]. They can be hyperexcited with NachBac, a bacterial Sodium channels [159]. Circadian neurons can be acutely activated with TrpA1 [76, 160], a temperature sensitive cationic channels. Use of temperature pulses has however to be done with careful controls, since temperature is an input to the clock. Optogenetics is tricky, given the sensitivity of the circadian clock to light, but use of red-shifted channel-rhodopsins [161] might in the future avoid activation of CRY, the dedicated circadian photoreceptor. Also note that the commonly used UAS-shibirets, which inhibits synaptic transmission, seems somewhat problematic as well, as its expression alters circadian period [162]. With careful controls, it might nevertheless be possible to use it in experiments in which synaptic transmission is acutely inhibited.
Interestingly, manipulation of neural excitability was combined recently with the whole genome approaches mentioned in section 3. LNvs isolated from larvae in which the sLNvs had been either hyperexcited with NachBac or silenced with Kir showed interesting alteration of the pattern of expression of circadianly-controlled genes [106]. These studies indicated that electrical activity of circadian neurons can drive expression of many genes under circadian control, suggesting that membrane properties contribute to or even reprogram circadian transcription. It should however be kept in mind that long-term manipulation of neural excitability might have unintended consequences in circadian neurons. Constitutive Kir2.1 expression, starting early during development, had been shown to completely disrupt oscillations of the circadian pacemaker in LNvs, and to result in behavioral arrhythmicity [158]. However, a recent study used an inducible system. GAL4 can be fused to a fragment of the progesterone receptor, and its activity becomes dependent on the presence of RU486 [163]. When using such an inducible Pdf-GAL4 driver to express Kir only in adult flies, results were quite different from those obtained with constitutive Kir expression [164]. Although flies were arrhythmic during induction, they returned to rhythmicity after removal of RU486, and the phase of behavior was identical to that observed before Kir induction. Immunohistochemistry confirmed that the circadian clock kept running during Kir expression. Thus, acute manipulation of circadian neurons is preferable when possible. Note that the use of a temperature-sensitive GAL80 (GAL80ts) can also be used to induce expression of a gene or an RNAi of interest [165]. As mentioned before it is however important to take into account the effect of temperature on circadian oscillations, and thus have appropriate controls to ensure that any observed effects are due to the transgene of interest and not to a phase-shift caused by the temperature change.
4.4. Visualizing activity in the circadian neural circuit
A recent development in neuroscience has been the ability to visualize the activity of neurons in intact circuits. This approach is particularly important in a small animal like Drosophila in which electrophysiology is challenging. For example, it is very difficult to record from the s-LNvs, the pacemaker neurons of the fly brain. In only one report was electrical activity recorded from these neurons, and this only in wild-type flies [166].
The first live-imaging of the circadian neural network used a FRET-based reporter for cAMP/cGMP: Epac1-camp [167, 168]. This is a fusion between the cAMP-responsive Guanine nucleotide exchange factor EPAC, YFP and CFP. Upon cAMP binding, loss of FRET is observed. The use of Epac1-camp revealed widespread response of circadian neurons to the neuropeptide PDF, which was bath applied in these experiments. In addition, the genetically encoded Calcium sensor GCaMP is a powerful tool to detect neural activity [169].
Recently, O. Shafer’s group has begun to interrogate the connectivity between circadian neurons using GCaMP and Epac1-camp in combination with the purinoreceptor P2X2 [170]. This receptor allows Sodium and Calcium entry in the presence of ATP, and thus excites the neurons in which it is expressed. There are no such endogenous receptors in flies. P2X2 was expressed in the sLNvs with Pdf-LexA, and GCamp3.0 was expressed throughout the circadian neural network with Clk(856)-GAL4 [171]. Brains were dissected, and ATP bath-applied to activate the LNvs. Nothing happened in the LNds, even though these cells are targeted by the LNvs. However, when the Epac1-camp sensor was used instead of GCaMP3, then functional connectivity was observed. This makes sense, since PDF is thought to be the critical synchronizing circadian neurons with the s-LNvs, and its receptor is a GPCR positively coupled to cAMP [172].
These results are the proof-of-principle that circadian neuron connectivity can be interrogated. Much will probably learned about the circadian “connectome” in the near future. It should be noted that methods to remote-control neurons and to monitor neural activity keeps improving at a very fast pace. A very recent and important development is a FRET-based Voltage sensor that allows the visualization of membrane potential and thus electrical activity, virtually replacing electrophysiology [173]. This method was tested successfully in circadian neurons and could dramatically improve our capacity of understanding neural network (circadian or not) in fruit flies.
5. Non-behavioral circadian outputs
We have given great attention to circadian behavioral output throughout this review. Locomotor activity, in particular, has been studied in great detail. However, as mentioned above, circadian clocks are found in most organs of the flies, and thus must control local physiology and metabolism. For example, rhythms in olfactory and gustatory sensitivity have been observed [4, 5]. These rhythms can be recorded electrophysiologically through extracellular recordings in different types of sensilla of the antennae and the labellum. Interestingly, in the olfactory system, different sensilla have different phase of sensitivity rhythms, suggesting that the fly tune its olfactory system to different odors at different time of the day [174]. This could help explaining preferential times for courtship or food search. Internal organs such as the gut and malpighian tubules also have circadian clocks [123, 175]. In an interesting recent study, it was demonstrated that the circadian clock controls the mitotic response of gut stem cells to chemical damage, and is actually important for the survival of the animals exposed to those damages [6].
The approaches to study the function of peripheral clocks are similar to many of those used in the brain. Tissue-specific rescue or RNA interference can be used to determine the necessity and sufficiency of peripheral clocks to defined clock-controlled physiological responses. In the case of the antennae, dominant-negative mutants of the transcription factors CLK and CYC were expressed tissue-specifically to disrupt the circadian clock in antennae [176]. The antennal clock proved necessary and sufficient for normal olfactory sensing rhythms. Interestingly however, the circadian clock in the Intestinal Stem Cells (ISCs) are necessary, but not sufficient for proper ISC mitotic responses to chemically-induced gut damages [6]. Clocks in both ISCs and Enteroblasts are required, indicating that the mitotic response requires communication between circadian oscillators in multiple cell types. The mechanisms by which clocks control ISC mitosis is not yet clear, but whole genome expression monitoring has identified multiple rhythmically expressed genes in the gut. Among those must be genes important for communication between cell types.
In summary, the study of peripheral clocks require the identification of specific rhythmic physiological or metabolic processes, and the role of local circadian clocks can then be determined with local manipulations, that of course require the necessary GAL4 (or LexA) drivers for specific targeting. We expect that whole genome expression studies combined with local genetic manipulation will reveal the functions of circadian clocks all over the body of fruit flies.
6. Conclusions
43 years after the genetic identification of the per gene, and 30 years after its cloning, Drosophila remains a powerhouse for the study of circadian rhythms, from input pathways to circadian outputs. The combination of biochemical, genetic, genomic, neural, and behavioral approaches is permitting to understand with remarkable depth how circadian rhythms are generated, and why they are important for fruit flies. This, of course, is greatly facilitated by the relative simplicity of the Drosophila genome and brain, which makes fruit flies such a fantastic animal model. We anticipate that advances in behavioral monitoring, neural imaging, genomics and proteomics will considerably accelerate the pace of discoveries in the upcoming years. It is therefore an exciting time to study circadian clock in Drosophila.
Acknowledgments
Patrick Emery is supported by NIH R01 grants GM066777, GM079182 and GM100091 from the National Institute of General Medicine Science
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weaver DR, Emery P. Circadian Timekeeping. In: Squire LR, editor. Fundamental Neuroscience. Elsevier; 2013. pp. 819–846. [Google Scholar]
- 2.Zhang Y, Emery P. Molecular and Neural Control of Insects Circadian Rhythms. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic Press; 2012. pp. 513–551. [Google Scholar]
- 3.Kaneko H, Head LM, Ling J, Tang X, Liu Y, Hardin PE, Emery P, Hamada FN. Circadian rhythm of temperature preference and its neural control in Drosophila. Curr Biol. 2012;22:1851–1857. doi: 10.1016/j.cub.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatterjee A, Tanoue S, Houl JH, Hardin PE. Regulation of gustatory physiology and appetitive behavior by the Drosophila circadian clock. Curr Biol. 2010;20:300–309. doi: 10.1016/j.cub.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnan B, Dryer SE, Hardin PE. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature. 1999;400:375–378. doi: 10.1038/22566. [DOI] [PubMed] [Google Scholar]
- 6.Karpowicz P, Zhang Y, Hogenesch JB, Emery P, Perrimon N. The circadian clock gates the intestinal stem cell regenerative state. Cell Rep. 2013;3:996–1004. doi: 10.1016/j.celrep.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reppert SM, Gegear RJ, Merlin C. Navigational mechanisms of migrating monarch butterflies. Trends Neurosci. 2010;33:399–406. doi: 10.1016/j.tins.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloch G. The social clock of the honeybee. J Biol Rhythms. 2010;25:307–317. doi: 10.1177/0748730410380149. [DOI] [PubMed] [Google Scholar]
- 9.Cassone VM, Westneat DF. The bird of time: cognition and the avian biological clock. Frontiers in molecular neuroscience. 2012;5:32. doi: 10.3389/fnmol.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 11.Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–182. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Pittendrigh CS. On temperature independence in the clock-system controlling emergence time in Drosophila. Proc Natl Acad Sci U S A. 1954;40:1018–1029. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmerman WF, Pittendrigh CS, Pavlidis T. Temperature compensation of the circadian oscillation in Drosophila pseudoobscura and its entrainment by temperature cycles. J Insect Physiol J1 - JIP. 1968;14:669–684. doi: 10.1016/0022-1910(68)90226-6. [DOI] [PubMed] [Google Scholar]
- 14.Pittendrigh CS. Temporal organization: Reflections of a Darwinian clock-watcher. Annu Rev Physiol. 1993;55:17–54. doi: 10.1146/annurev.ph.55.030193.000313. [DOI] [PubMed] [Google Scholar]
- 15.Pittendrigh CS. Circadian systems. I. The driving oscillation and its assay in Drosophila pseudoobscura. Proc Natl Acad Sci U S A. 1967;58:1762–1767. doi: 10.1073/pnas.58.4.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 19.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS protein essential for circadian transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 20.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA. Closing the Circadian Loop: CLOCK-induced Transcription of Its Own Inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 21.Zeng H, Qian Z, Myers MP, Rosbash M. A light-entrainment mechanism for the Drosophila circadian clock. Nature. 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 22.Gekakis N, Saez L, Delahaye-Brown A-M, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interactions: Defective interaction between timeless protein and long-period mutant PERL. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 23.Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 2010;24:358–367. doi: 10.1101/gad.1883910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu W, Zheng H, Price JL, Hardin PE. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol Cell Biol. 2009;29:1452–1458. doi: 10.1128/MCB.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edery I, Zwiebel LJ, Dembinska ME, Rosbash M. Temporal phosphorylation of the Drosophila period protein. 1994;91(6):2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 27.Stanewsky R, Kaneko M, Emery P, Beretta M, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 28.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 29.Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- 30.Ozturk N, Selby CP, Annayev Y, Zhong D, Sancar A. Reaction mechanism of Drosophila cryptochrome. Proc Natl Acad Sci U S A. 2011;108:516–521. doi: 10.1073/pnas.1017093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaidya AT, Top D, Manahan CC, Tokuda JM, Zhang S, Pollack L, Young MW, Crane BR. Flavin reduction activates Drosophila cryptochrome. Proc Natl Acad Sci U S A. 2013;110:20455–20460. doi: 10.1073/pnas.1313336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 33.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 34.Hunter-Ensor M, Ousley A, Sehgal A. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 35.Naidoo N, Song W, Hunter-Ensor M, Sehgal A. A role for the proteasome in the light response of the timeless clock protein. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- 36.Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A. Photic signaling by cryptochrome in the drosophila circadian system. Mol Cell Biol. 2001;21:7287–7294. doi: 10.1128/MCB.21.21.7287-7294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helfrich-Forster C, Winter C, Hofbauer A, Hall JC, Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- 39.Helfrich-Forster C. PDF has found its receptor. Neuron. 2005;48:161–163. doi: 10.1016/j.neuron.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Sehgal A, Price JL, Man B, Young MW. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 41.Frank KD, Zimmerman WF. Action spectra for phase shifts of a circadian rhythm in Drosophila. Science. 1969;163:688–689. doi: 10.1126/science.163.3868.688. [DOI] [PubMed] [Google Scholar]
- 42.Chiu JC, Low KH, Pike DH, Yildirim E, Edery I. Assaying locomotor activity to study circadian rhythms and sleep parameters in Drosophila. Journal of visualized experiments: JoVE. 2010 doi: 10.3791/2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helfrich-Forster C. Robust Circadian Rhythmicity of Drosophila Melanogaster Requires the Presence of Lateral Neurons: A Brain Behavioral Study of disconnected mutants. J Comp Physiol J1 - JCP. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- 44.Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3 doi: 10.1186/1471-2202-3-1. electronic publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamblen M, Zehring WA, Kyriacou CP, Reddy P, Yu Q, Wheeler DA, Zwiebel LJ, Konopka RJ, Rosbash M, Hall JC. Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: Overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per-mutants. J Neurogenet. 1986;3:249–291. doi: 10.3109/01677068609106855. [DOI] [PubMed] [Google Scholar]
- 46.Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- 47.Mrosovsky N. Masking: history, definitions, and measurement. 1999;16:415–429. doi: 10.3109/07420529908998717. [DOI] [PubMed] [Google Scholar]
- 48.Yoshii T, Wulbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Forster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Majercak J, Sidote D, Hardin PE, Edery I. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron. 1999;24:219–230. doi: 10.1016/s0896-6273(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 50.Levine JD, Casey CI, Kalderon DD, Jackson FR. Altered circadian pacemaker functions and cyclic AMP rhythms in the drosophila learning mutant dunce. Neuron. 1994;13:967–974. doi: 10.1016/0896-6273(94)90262-3. [DOI] [PubMed] [Google Scholar]
- 51.Edery I, Rutila JE, Rosbash M. Phase shifting of the circadian clock by induction of the Drosophila period protein. Science. 1994;263:237–240. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]
- 52.Kaushik R, Nawathean P, Busza A, Murad A, Emery P, Rosbash M. PER-TIM Interactions with the Photoreceptor Cryptochrome Mediate Circadian Temperature Responses in Drosophila. PLoS Biol. 2007;5:e146. doi: 10.1371/journal.pbio.0050146. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Suri V, Qian Z, Hall JC, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 54.Egan ES, Franklin TM, Hilderbrand-Chae MJ, McNeil GP, Roberts MA, Schroeder AJ, Zhang X, Jackson FR. An extraretinally expressed insect cryptochrome with similarity to the bluelight photoreceptors of mammals and plants. J Neurosci. 1999;19:3665–3673. doi: 10.1523/JNEUROSCI.19-10-03665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vinayak P, Coupar J, Hughes SE, Fozdar P, Kilby J, Garren E, Yoshii T, Hirsh J. Exquisite light sensitivity of Drosophila melanogaster cryptochrome. PLoS Genet. 2013;9:e1003615. doi: 10.1371/journal.pgen.1003615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Busza A, Murad A, Emery P. Interactions between circadian neurons control temperature synchronization of Drosophila behavior. J Neurosci. 2007;27:10722–10733. doi: 10.1523/JNEUROSCI.2479-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Emerson M, Su HS, Sehgal A. Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron. 1998;21:215–223. doi: 10.1016/s0896-6273(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 58.Tang CH, Hinteregger E, Shang Y, Rosbash M. Light-mediated TIM degradation within Drosophila pacemaker neurons (s-LNvs) is neither necessary nor sufficient for delay zone phase shifts. Neuron. 2010;66:378–385. doi: 10.1016/j.neuron.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang Y, Griffith LC, Rosbash M. Feature Article: Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci U S A. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 61.Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177:329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Integration of Light and Temperature in the Regulation of Circadian Gene Expression in Drosophila. PLoS Genet. 2007;3:e54. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshii T, Vanin S, Costa R, Helfrich-Forster C. Synergic entrainment of Drosophila’s circadian clock by light and temperature. J Biol Rhythms. 2009;24:452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- 64.Billeter JC, Levine JD. Who is he and what is he to you? Recognition in Drosophila melanogaster. Curr Opin Neurobiol. 2013;23:17–23. doi: 10.1016/j.conb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 65.Currie J, Goda T, Wijnen H. Selective entrainment of the Drosophila circadian clock to daily gradients in environmental temperature. BMC Biol. 2009;7:49. doi: 10.1186/1741-7007-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachleitner W, Kempinger L, Wulbeck C, Rieger D, Helfrich-Forster C. Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:3538–3543. doi: 10.1073/pnas.0606870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoshii T, Hermann C, Helfrich-Forster C. Cryptochrome-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. J Biol Rhythms. 2010;25:387–398. doi: 10.1177/0748730410381962. [DOI] [PubMed] [Google Scholar]
- 68.Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, Sandrelli F, Costa R, Kyriacou CP. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature. 2012;484:371–375. doi: 10.1038/nature10991. [DOI] [PubMed] [Google Scholar]
- 69.De J, Varma V, Saha S, Sheeba V, Sharma VK. Significance of activity peaks in fruit flies, Drosophila melanogaster, under seminatural conditions. Proc Natl Acad Sci U S A. 2013;110:8984–8989. doi: 10.1073/pnas.1220960110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Menegazzi P, Yoshii T, Helfrich-Forster C. Laboratory versus nature: the two sides of the Drosophila circadian clock. J Biol Rhythms. 2012;27:433–442. doi: 10.1177/0748730412463181. [DOI] [PubMed] [Google Scholar]
- 71.Sakai T, Ishida N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc Natl Acad Sci U S A. 2001;98:9221–9225. doi: 10.1073/pnas.151443298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–251. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujii S, Amrein H. Ventral lateral and DN1 clock neurons mediate distinct properties of male sex drive rhythm in Drosophila. Proc Natl Acad Sci U S A. 2010;107:10590–10595. doi: 10.1073/pnas.0912457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamasaka Y, Suzuki T, Hanai S, Ishida N. Evening circadian oscillator as the primary determinant of rhythmic motivation for Drosophila courtship behavior. Genes Cells. 2010;15:1240–1248. doi: 10.1111/j.1365-2443.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- 75.Pan Y, Robinett CC, Baker BS. Turning males on: activation of male courtship behavior in Drosophila melanogaster. PLoS One. 2011;6:e21144. doi: 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci U S A. 2007;104:8253–8256. doi: 10.1073/pnas.0702726104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dankert H, Wang L, Hoopfer ED, Anderson DJ, Perona P. Automated monitoring and analysis of social behavior in Drosophila. Nat Methods. 2009;6:297–303. doi: 10.1038/nmeth.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Branson K, Robie AA, Bender J, Perona P, Dickinson MH. High-throughput ethomics in large groups of Drosophila. Nat Methods. 2009;6:451–457. doi: 10.1038/nmeth.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- 82.Zehring WA, Wheeler DA, Reddy P, Konopka RJ, Kyriacou CP, Rosbash M, Hall JC. P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell. 1984;39:369–376. doi: 10.1016/0092-8674(84)90015-1. [DOI] [PubMed] [Google Scholar]
- 83.Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- 84.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 85.Hardin PE, Hall JC, Rosbash M. Circadian oscillations in period gene mRNA levels are transcriptionally regulated. Proc Natl Acad Sci U S A. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 87.Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Emery P. Protein extraction from Drosophila heads. Methods Mol Biol. 2007;362:375–377. doi: 10.1007/978-1-59745-257-1_27. [DOI] [PubMed] [Google Scholar]
- 89.Emery P. RNA extraction from Drosophila heads. Methods Mol Biol. 2007;362:305–307. doi: 10.1007/978-1-59745-257-1_20. [DOI] [PubMed] [Google Scholar]
- 90.Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006;20:723–733. doi: 10.1101/gad.1404406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zeng H, Hardin PE, Rosbash M. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoshii T, Todo T, Wulbeck C, Stanewsky R, Helfrich-Forster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 93.Shafer OT, Rosbash M, Truman JW. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci. 2002;22:5946–5954. doi: 10.1523/JNEUROSCI.22-14-05946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R. Drosophila clock can generate ectopic circadian clocks. Cell. 2003;113:755–766. doi: 10.1016/s0092-8674(03)00400-8. [DOI] [PubMed] [Google Scholar]
- 95.Lim C, Allada R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science. 2013;340:875–879. doi: 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- 96.Lim C, Lee J, Choi C, Kilman VL, Kim J, Park SM, Jang SK, Allada R, Choe J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature. 2011;470:399–403. doi: 10.1038/nature09728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bradley S, Narayanan S, Rosbash M. NAT1/DAP5/p97 and Atypical Translational Control in the Drosophila Circadian Oscillator. Genetics. 2012;192:943–957. doi: 10.1534/genetics.112.143248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Ling J, Yuan C, Dubruille R, Emery P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science. 2013;340:879–882. doi: 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- 100.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 101.Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 103.Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young M. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 105.Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mizrak D, Ruben M, Myers GN, Rhrissorrakrai K, KCG, Blau J. Electrical activity can impose time-of-day on the circadian transcriptome of pacemaker neurons. Curr Biol. 2012;22:1871–1880. doi: 10.1016/j.cub.2012.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ruben M, Drapeau MD, Mizrak D, Blau J. A mechanism for circadian control of pacemaker neuron excitability. J Biol Rhythms. 2012;27:353–364. doi: 10.1177/0748730412455918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci U S A. 2010;107:13497–13502. doi: 10.1073/pnas.1002081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagoshi E, Sugino K, Kula E, Okazaki E, Tachibana T, Nelson S, Rosbash M. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat Neurosci. 2009;13:60–68. doi: 10.1038/nn.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, Rosbash M. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hughes ME, Grant GR, Paquin C, Qian J, Nitabach MN. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 2012;22:1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rodriguez J, Tang CH, Khodor YL, Vodala S, Menet JS, Rosbash M. Nascent-Seq analysis of Drosophila cycling gene expression. Proc Natl Acad Sci U S A. 2013;110:E275–284. doi: 10.1073/pnas.1219969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kivimae S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chiu JC, Vanselow JT, Kramer A, Edery I. The phospho-occupancy of an atypical SLIMB-binding site on PERIOD that is phosphorylated by DOUBLETIME controls the pace of the clock. Genes Dev. 2008;22:1758–1772. doi: 10.1101/gad.1682708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Garbe DS, Fang Y, Zheng X, Sowcik M, Anjum R, Gygi SP, Sehgal A. Cooperative interaction between phosphorylation sites on PERIOD maintains circadian period in Drosophila. PLoS Genet. 2013;9:e1003749. doi: 10.1371/journal.pgen.1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Welsh DK, Imaizumi T, Kay SA. Real-time reporting of circadian-regulated gene expression by luciferase imaging in plants and mammalian cells. Methods Enzymol. 2005;393:269–288. doi: 10.1016/S0076-6879(05)93011-5. [DOI] [PubMed] [Google Scholar]
- 117.Welsh DK, Kay SA. Bioluminescence imaging in living organisms. Current opinion in biotechnology. 2005;16:73–78. doi: 10.1016/j.copbio.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 118.Brandes C, Plautz JD, Stanewsky R, Jamison CF, Straume M, Wood KV, Kay SA, Hall JC. Novel features of Drosophila period transcription revealed by real-time luciferase reporting. Neuron. 1996;16:687–692. doi: 10.1016/s0896-6273(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 119.Stanewsky R, Jamison CF, Plautz JD, Kay SA, Hall JC. Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 1997;16:5006–5018. doi: 10.1093/emboj/16.16.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stanewsky R, Lynch KS, Brandes C, Hall JC. Mapping of elements involved in regulating normal temporal period and timeless RNA expression patterns in Drosophila melanogaster. J Biol Rhythms. 2002;17:293–306. doi: 10.1177/074873002129002609. [DOI] [PubMed] [Google Scholar]
- 121.McDonald MJ, Rosbash M, Emery P. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol. 2001;21:1207–1217. doi: 10.1128/MCB.21.4.1207-1217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19:241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 123.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 124.Glaser FT, Stanewsky R. Temperature synchronization of the Drosophila circadian clock. Curr Biol. 2005;15:1352–1356. doi: 10.1016/j.cub.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 125.Sehadova H, Glaser FT, Gentile C, Simoni A, Giesecke A, Albert JT, Stanewsky R. Temperature entrainment of Drosophila’s circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron. 2009;64:251–266. doi: 10.1016/j.neuron.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 126.Welsh DK, Noguchi T. Cellular bioluminescence imaging. Cold Spring Harbor protocols. 2012;2012 doi: 10.1101/pdb.top070607. [DOI] [PubMed] [Google Scholar]
- 127.Krupp JJ, Billeter JC, Wong A, Choi C, Nitabach MN, Levine JD. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in drosophila. Neuron. 2013;79:54–68. doi: 10.1016/j.neuron.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Helfrich-Forster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila malanogaster. Proc Natl Acad Sci U S A. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kaneko M, Helfrich-Forster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemakers candidates and novel features of clock gene product cycling. J Neurosci. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 132.Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- 133.Johard HA, Yoishii T, Dircksen H, Cusumano P, Rouyer F, Helfrich-Forster C, Nassel DR. Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol. 2009;516:59–73. doi: 10.1002/cne.22099. [DOI] [PubMed] [Google Scholar]
- 134.Frisch B, Hardin PE, Hamblen-Coyle MJ, Rosbash M, Hall JC. A promoterless DNA fragment from the period locus rescues behavioral rhythmicity and mediates cyclical gene expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 135.Renn SCP, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf Neuropeptide Gene Mutation and Ablation of PDF Neurons Each Cause Severe Abnormalities of Behavioral Circadian Rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 136.Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ng FS, Tangredi MM, Jackson FR. Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Curr Biol. 2011;21:625–634. doi: 10.1016/j.cub.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Awasaki T, Lai SL, Ito K, Lee T. Organization and postembryonic development of glial cells in the adult central brain of Drosophila. J Neurosci. 2008;28:13742–13753. doi: 10.1523/JNEUROSCI.4844-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Doherty J, Logan MA, Tasdemir OE, Freeman MR. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci. 2009;29:4768–4781. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leiserson WM, Harkins EW, Keshishian H. Fray, a Drosophila serine/threonine kinase homologous to mammalian PASK, is required for axonal ensheathment. Neuron. 2000;28:793–806. doi: 10.1016/s0896-6273(00)00154-9. [DOI] [PubMed] [Google Scholar]
- 142.Silies M, Yuva Y, Engelen D, Aho A, Stork T, Klambt C. Glial cell migration in the eye disc. J Neurosci. 2007;27:13130–13139. doi: 10.1523/JNEUROSCI.3583-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Klarsfeld A, Malpel S, Michard-Vanhee C, Picot M, Chelot E, Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep-brain circadian photoreceptor. Neuron. 2000;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 145.Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC. Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol. 2000;43:207–233. doi: 10.1002/(sici)1097-4695(20000605)43:3<207::aid-neu1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 146.Ling J, Dubruille R, Emery P. KAYAK-alpha modulates circadian transcriptional feedback loops in Drosophila pacemaker neurons. J Neurosci. 2012;32:16959–16970. doi: 10.1523/JNEUROSCI.1888-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martinek S, Inonog S, Manoukian AS, Young MW. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell. 2001;105:769–779. doi: 10.1016/s0092-8674(01)00383-x. [DOI] [PubMed] [Google Scholar]
- 148.Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 149.Stoleru D, Peng Y, Agusto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 150.Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner RA, Hardin PE, Allada R. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 153.Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev J1 - GD. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- 154.Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, Taghert P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol. 2007;500:47–70. doi: 10.1002/cne.21146. [DOI] [PubMed] [Google Scholar]
- 155.Shafer OT, Helfrich-Forster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 157.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 159.Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sivachenko A, Li Y, Abruzzi KC, Rosbash M. The transcription factor Mef2 links the Drosophila core clock to Fas2, neuronal morphology, and circadian behavior. Neuron. 2013;79:281–292. doi: 10.1016/j.neuron.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–1508. doi: 10.1038/nn.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kilman VL, Zhang L, Meissner RA, Burg E, Allada R. Perturbing dynamin reveals potent effects on the Drosophila circadian clock. PLoS One. 2009;4:e5235. doi: 10.1371/journal.pone.0005235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci U S A. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol. 2011;21:1783–1793. doi: 10.1016/j.cub.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 166.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nikolaev VO, Bunemann M, Hein L, Hannawacker A, Lohse MJ. Novel single chain cAMP sensors for receptor-induced signal propagation. J Biol Chem. 2004;279:37215–37218. doi: 10.1074/jbc.C400302200. [DOI] [PubMed] [Google Scholar]
- 168.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol. 2012;108:684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gummadova JO, Coutts GA, Glossop NR. Analysis of the Drosophila Clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. J Biol Rhythms. 2009;24:353–367. doi: 10.1177/0748730409343890. [DOI] [PubMed] [Google Scholar]
- 172.Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 173.Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Krishnan P, Chatterjee A, Tanoue S, Hardin PE. Spike amplitude of single unit responses in antennal sensillae is controlled by the Drosophila circadian clock. Curr Biol. 2008:803–807. doi: 10.1016/j.cub.2008.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hege DM, Stanewsky R, Hall JC, Giebultowicz JM. Rhythmic expression of a PER-reporter in the Malpighian tubules of decapitated Drosophila: evidence for a brain-independent circadian clock. 1997;12:300–308. doi: 10.1177/074873049701200402. [DOI] [PubMed] [Google Scholar]
- 176.Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]