Abstract
It is well recognized that a number of proteins present within adhesion complexes perform discrete signaling functions outside these adhesion complexes, including transcriptional control. In this respect, β-catenin is a well-known example of an adhesion protein present both in cadherin complexes and in the nucleus where it regulates the TCF transcription factor. Here we discuss nuclear functions of adhesion complex proteins with a special focus on the CCM-1/KRIT-1 protein, which may turn out to be yet another adhesion complex protein with a second life.
Keywords: adhesion signaling, CCM1/Krit-1, transcription, nuclear translocation, small GTPases, Rac Rho
Adherens Junctions and Tight Junctions Regulation
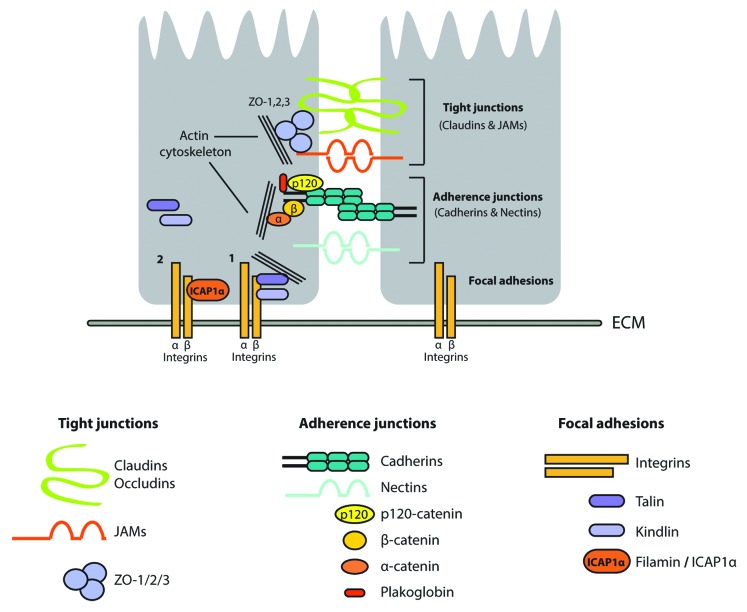
During embryonic development, endothelial cells form the network of blood vessels essential for transport of nutrients, fluids, circulating cells, gasses, and hormones to almost all tissues in our body. A tight monolayer of endothelial cells is lining the inner site of all vessel types and regulates the exchange of solutes and fluids between blood and tissue and controls entry of leukocytes in the surrounding tissue. The ability of endothelial cells to properly regulate cell–cell adhesions between themselves and neighboring cells is essential for regulation of all these functions. Endothelial cells have two specialized types of junctions to regulated cell–cell contacts, called adherens junctions (AJ) and tight junctions (TJ). In general, adherens junctions confer cell–cell contacts and tight junctions regulate the paracellular passage of ions and solutes.1,2 Proper formation of these junctions is important for tissue integrity, leukocyte extravasation, vascular permeability, and angiogenesis. In both types of junctions, adhesion is mediated through transmembrane proteins, such as cadherins and nectins in AJs and claudins, occludins, and JAMs (junction adhesion molecules) in TJs. AJs are formed at the early stages of intercellular contacts and are followed by the formation of TJs. AJs are suggested to influence the formation of TJs, as in some cases, in absence of AJs, TJs are not formed. Intracellular signaling mediated by the transmembrane proteins in AJs and TJs is mediated by a diverse set of signaling proteins. For example, in TJs, intracellular signaling can be mediated by ZO-1, ZO-2, and ZO-3 (zonula occludens), and in AJs this is mediated by the Catenins, in particular p120-catenin, β-catenin, α-catenin, and plakoglobin (γ-catenin). β-catenin interacts directly with the cytoplasmic tail of the cadherins. α-catenin can interact with β-catenin and the actin cytoskeleton, although this interaction seems mutually exclusive (reviewed in ref. 3) (depicted in Fig. 1). Cell–cell adhesion in endothelial cells is mediated by Vascular Endothelial (VE)-cadherin. The interaction between VE-cadherin and p120-catenin/β-catenin is tightly regulated by (de-) phosphorylation and binding of p120-catenin to VE-cadherin inhibits the internalization of VE-cadherin.4 Tyrosine phosphorylation of VE-cadherin reduces the interaction with p120-catenin and might therefore induce its internalization, resulting in disruption of AJs. Next to internalization, VE-cadherin is also regulated by cleavage5 and through up or downregulation of its expression.6,7
Figure 1. A simplified representation of the important mediators of cell–cell and cell–matrix adhesion. The adherens junctions consist of the Claudin and JAM families of transmembrane proteins, which are connected to the actin cytoskeleton via the ZO-family of proteins. Tight junctions consist of the catenin and nectin families of transmembrane proteins, which are connected to the cytoskeleton via the β-catenin interaction to α-catenin. Integrin-mediated cell–cell or cell–matrix interactions at focal adhesion sites is established via interaction of talins and kindlins to actin bundles (1). This can be inhibited by binding proteins such as filamin and ICAP1 to the β-integrin tail (2). For further details, see text.
Integrin Signaling
The interaction of cells to the extracellular matrix (ECM) and the link with the ECM to the actin cytoskeleton at focal adhesion sites is mediated by the transmembrane glycoprotein called integrin (Fig. 1). Integrins consist of dimers containing an α- and β-chain. There are 18 α- and eight β-integrins, and the combination of those determines the interaction with specific ECM proteins and the subsequent downstream signaling event.8 Activation of integrins occurs through both outside-in and inside-out signaling. Outside-in activation is mediated by extracellular stimulation, resulting in a conformational change that allows interaction with several cytoplasmic proteins. Inside-out activation is mediated by the interaction of intercellular activators such as talin and kindlin (Fig. 1 [1]) and also results in an open conformation. Inactive integrins adopt a closed conformation, which inhibits recruitment of extracellular ligands and intracellular proteins (reviewed in ref. 9). Several proteins are reported to compete with intercellular activators for binding to integrins, and thereby, inhibit integrin activation. For example, ICAP1 can compete with talin and kindlin for binding to β1-integrins.10 Filamin is another inhibitory protein of integrins, it interacts with the NXXY motif in β integrin tails, and thereby, inhibits talin binding (Fig. 1 [2]; reviewed in ref. 9). Crosstalk between adherens junctions and integrin signaling is postulated to be important for proper development and tissue architecture; however, the molecules and molecular mechanisms involved are still ill defined. Mainly, engagement of integrins with ECM proteins is reported to affect cadherin-containing adherens junctions, whereas cadherins that regulate integrin function is much less explored. Most of the crosstalk between cadherens and integrins is mediated by small GTPases, non-receptor kinases, cell surface receptors, and alterations of the actin network (reviewed in ref. 11).
Regulation of Junctions by Small GTPases
Adherens junctions, tight junctions, and focal adhesions are highly regulated by small GTPases belonging to the RAS superfamily of small G proteins. These small GTPases act as molecular switches by cycling between an active GTP-bound and inactive GDP-bound form. They are tightly regulated by GTPase-activating proteins (GAPs), which stimulate hydrolysis of GTP (inactivation) and guanine nucleotide exchange factors (GEFs), which stimulate GTP loading (activation).12 The small GTPase RAP1 is a member of the RAS super family important in the promotion of cell–cell adhesion through regulation of the formation and maturation of cell–cell contacts via stimulation of the adhesive function of VE-cadherin.13 In return, VE-cadherin is necessary for the recruitment of MAGI-1, a scaffold for the RAP1 guanine nucleotide-activating factor (GEF) PDZ-GEF. In addition, RAP1 activates the clustering of integrins to mediate cell adhesion to the extracellular matrix and promotes cell spreading. RAP1 is also suggested to mediate crosstalk between adherens junctions and integrin signaling, in which RAP1 is activated upon E-cadherin internalization and trafficking along the endocytic pathway. This endocytosis-dependent activation of RAP1 is required for the formation of integrin-based focal adhesions.14,15
RALA, another small GTPase belonging to the RAS super family, is important in tight junction regulation, via a GTP-dependent interaction with ZONAB (ZO-1-associated nucleic acid-binding protein). ZONAB is a Y-box transcription factor that regulates expression of genes in a cell density-dependent manner.16 Upon increase in cell density, the amount of the RALA-ZONAB complex increases, resulting in release of transcriptional repression of the ErbB-2 promotor by ZONAB.17
RHO GTPases also belong to the RAS superfamily of small GTPases and promote the formation of stress fibers and increase endothelial permeability.18 RHO induces stress fibers via activation of myosin light chain (MLC), which interacts with actin and slides along actin filaments causing contractility. MLC is regulated by myosin light chain kinases (MLCKs) and RHO kinases. RHO can induce RHO kinase-mediated phosphorylation of MLC,19 and alternatively, RHO kinase can phosphorylate, and thereby, inactivate myosin light chain phosphatase, which dephosphorylates MLC.20,21 The effect of RHO activity on endothelial permeability is less clear and is suggested to involve a fine balance between RHO and RAC, another member of the RHO family of small GTPases. Improvement of endothelial barrier function can be achieved by low RHO, high RAC activity, whereas decreased barrier function is accomplished by high RHO, low RAC activity. Although low RHO activity is beneficial for the endothelial barrier,22 long-term inactivation of RHO can also result in increased permeability.23 Similarly, RAC activity is required for endothelial barrier function, whereas long-term activation of RAC results in stress fiber formation and junction breakdown18
Cerebral Cavernous Malformations
Defects in formation of the various types of junctions described above will cause major problems in various processes and are implicated in many diseases. For example, Cerebral Cavernous Malformations (CCM), a disease characterized by a cluster of dilated blood vessels in which each individual vessel is lined with a layer of endothelium.24,25 The cerebro-vascular lesions are thought to be the result of defective endothelial cell junctions.26,27 Patients with Cerebral Cavernous Malformations (CCM) have vascular malformations predominantly in the brain and sometimes in the skin28 and retina.29 This can cause a variety of problems like severe neurological symptoms such as focal defects (20–45%), migraine-like headaches (6–52%), seizures (23–50%), and/or brain hemorrhages (9–56%); however, about 40% of the cases are asymptomatic. The prevalence of CCM has been estimated to be 0.1–0.5%, based on cerebral magnetic resonance imaging (MRI) and autopsy studies of large cohorts of patients.30 Both sporadic (80%) and familial (20%) forms of CCM have been identified. Due to studies investigating patients with sporadic and familial CCM, it is found that familial CCM patients develop larger numbers of lesions and suffer more frequently from symptoms like seizure and hemorrhage. From these data, a two-hit hypothesis has been suggested for the pathogenesis of CCM.31,32
The first gene identified related to CCM patients is called KRIT1 (Krev-interaction trapped 1) or CCM1.33,34 Later on, two other genes were found to be associated with CCM, CCM2/OSM (osmosensing protein 1)/Malcavernin35,36 and CCM3/PDCD10 (programmed cell death 10).37,38 Over 150 different germline mutations are identified in either one of these genes, predominantly resulting in loss of function. To date, it has been established that the three CCM proteins can form a complex.39-41 How and whether disruption of this complex of CCM1, CCM2, and CCM3 is involved in the pathogenesis of CCM is still highly unknown.
CCM in Model Organisms
All three CCM genes are well conserved among both vertebrates and non-vertebrates,42 and subsequently, many attempts have been made to mimic the CCM phenotype. Mice that lack Ccm1 or Ccm2 die in mid-gestation with vascular defects.43-45 Ccm1 is ubiquitously expressed until E10.5, at which point the expression becomes restricted to neural and epithelial tissues.46,47 Endothelial-specific ablation of Ccm2 results in lethality at mid-gestation due to impaired embryonic angiogenesis and endothelial-specific deletion of CCM1 produces hemorrhagic vascular lesions in the cerebellum and retina that resemble CCMs.44,48,49 However, neuronal and smooth muscle cell-specific deletion of Ccm2 does not affect vascular development.44 Also for Ccm3, both constitutive and tissue-specific deletion gave similar phenotypes.50 Mice with heterozygous knockout of Ccm1 or Ccm2 do not develop CCM-like vascular lesions in the brain with any useful frequency, which makes these mice unsuitable to study CCM pathogenesis. Because of the suggestions of a two-hit hypothesis for the disease phenotype of CCM patients, other mice studies used mice lacking either p5351 or Msh252 in addition to heterozygosity of CCM1. These mice have high-mutation frequencies and were therefore chosen to function for second hit generation. Indeed, these mice develop CCM-like lesions, indicating that these mice have a second mutation resulting in CCM-like lesions. The combined data of the existing CCM mice models all indicate an important role for CCM in endothelial barrier function and vasculogenesis, but until now they do not provide sufficient insight into the molecular function of the CCM proteins in CCM pathogenesis.
Similar to mice, all three CCM proteins are expressed in zebrafish. Depletion of zebrafish CCM1 (Santa), CCM2 (valentine), and CCM3 (ccm3a and ccm3b) results in a dilated heart phenotype combined with vascular defects.53-56 Interestingly, this phenotype is similar to that of Heart of glass (heg) mutations, suggesting that they are functioning in the same molecular pathway.150 HEG1 is a transmembrane protein of unknown function that is expressed specifically in the endothelium and endocardium. Also in mice, CCM2 and HEG1 were found to interact genetically. Heg1−/−; Ccm2lacz/+ mice, like Ccm2−/− mice, have severe cardiovascular defects and die early in development.57 It is also shown that in human umbilical vein, ECs (HUVECs) CCM1 needs HEG1 to localize to endothelial cell–cell junctions.58 Recently, a novel gene with sequence identity to ccm2, ccm2l, was described in zebrafish and mice. Whereas the Ccm2L-knockout mice are viable with no gross cardiovascular defects,59 in zebrafish, injection of ccm2l morpholino results in cardiac dilation.60 In both mice and zebrafish, knockdown of ccm2l in addition to mutations in the HEG–CCM pathway enhances heart defects. Therefore, investigation of ccm2l might provide further insight in the CCM disease phenotype.
Molecular details of CCM1
Of the three CCM proteins, KRIT1/CCM1, is the first protein identified related to CCM33,34 and is most extensively studied compared with CCM2 and CCM3. Therefore, we will predominantly focus on the molecular details described for CCM1.
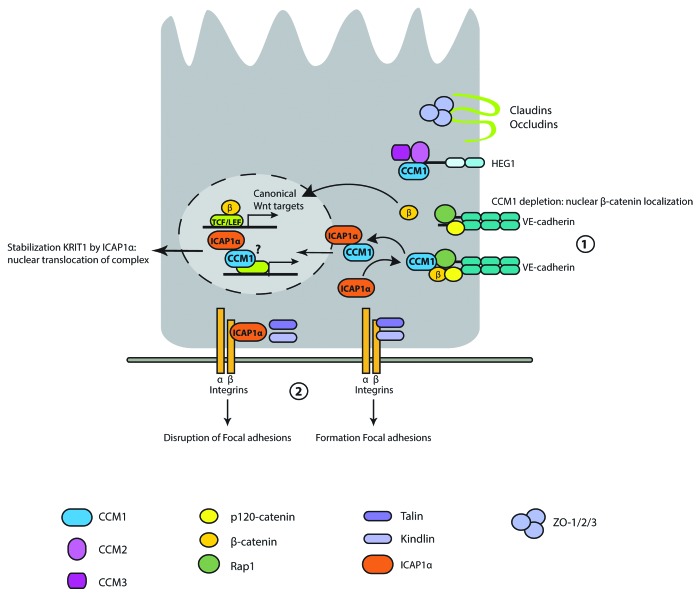
CCM1 was first identified in a yeast two-hybrid screen for interaction partners of the small GTPase RAP1.61 As RAP1 plays an important role in cell–cell adhesion,62 cell–matrix adhesion,63 and cell polarity,64 the interaction with CCM1 led to the suggestion of a role of CCM1 in maintaining junction integrity together with RAP1.65-67 In the past couple of years, several groups indeed show a role for CCM1 in the junctions. CCM1 is found in complex with β-catenin and RAP1 at the junctions and loss of CCM1 results in release of β-catenin from VE-cadherin in AJs68 (Fig. 2 [1]).
Figure 2. Molecular details of CCM1 biological function. Loss of CCM1 results in release of β-catenin from VE-cadherin, and subsequent activation of TCF/LEF-dependent transcription (1). Interaction of ICAP1 to β1-integrins disturbs focal adhesions by preventing binding of talin and kindlin. CCM1 inhibits binding of ICAP1 to β1-integrins and ICAP1 stabilizes CCM1 followed by nuclear translocation of the complex (2). CCM1 is located to the plasma membrane through interaction with the HEG1 transmembrane receptor.
CCM1 in integrin signaling
Next to RAP1, an integrin binding protein was identified to interact with CCM1, called ICAP1 (integrin cytoplasmic domain-associated protein 1).69-71 ICAP1 binds to the cytoplasmic domain of integrin β1, and thereby, prevents binding of talin72-74 and kindlin.10,75 Binding of talin and kindlin to integrin β1 is essential for proper integrin-mediated cell adhesion and formation of focal adhesions;76 hence, inhibition of this binding disrupts proper cell adhesion. Binding of ICAP1 to β1-integrins is mutually exclusive with its binding to CCM1.69,70 These data would fit a model where CCM1 prevents binding of ICAP1 to β1-integrins and preserves proper talin- and kindlin-mediated integrin signaling77 (Fig. 2 [2]). ICAP1 mediates stability of CCM1 by binding the first NPxY motif of CCM1 (CCM1 contains three NPxY motifs10), resulting in an open and more stable conformation.78,79 Furthermore, CCM1 is shown to interact with microtubules,78,80 and from this, a model is suggested where CCM1 is brought toward the plasma membrane via microtubules,78,80 where subsequently RAP1 and ICAP1 can capture CCM1. Indeed, Liu et al. show that RAP1 binding to CCM1 releases CCM1 from microtubules, enabling the translocation to cell–cell junctions.81
Inhibition of RHO signaling
As mentioned above, improvement of endothelial barrier function can be achieved by low RHO, high RAC activity. Recently, several groups have reported an inhibitory role of CCM1 toward RHO signaling, which would improve the endothelial barrier function.82,83 Activation of RHO results in ROCK-mediated phosphorylation of several substrates involved in regulation of actin cytoskeletal dynamics, like myosin light chain and LIM kinase. Furthermore, ROCK is described to phosphorylate Occludin and Claudin-5 in brain endothelium, and thereby, enhances leakiness.84 Inhibition of RHO by CCM1 is beneficial to keep endothelial cells in a quiescent state and maintain the endothelial monolayer. In addition, Nd1-L, an actin binding protein that negatively regulates RHO activity, is reported to induce cytoplasmic localization of CCM1, providing an extra layer of CCM1-mediated RHO regulation.85 Also, for both CCM2 and CCM3, an inhibitory role toward RHO has been described.82,83,86 How inhibition of RHO/ROCK by the CCM proteins is achieved is not known and it will be of interest to further investigate whether CCM proteins might influence the balance between RHO and RAC signaling.
Inhibition of angiogenesis
Loss of VE-cadherin signaling results in weakened cell contacts, but it also results in initiation of angiogenesis.87 Loss of CCM1 results in release of β-catenin from VE-cadherin, subsequent nuclear translocation and transcriptional activation, ultimately resulting in cell cycle re-entry and potential activation of angiogenesis. The Notch signaling pathway plays an important role in regulation of angiogenesis.88 In mammals, there are four Notch receptors (Notch1–4) and five ligands (DLL1, DLL3–4, and Jagged 1–2). DLL4-Notch can inhibit endothelial sprouting by inhibition of excessive tip-cell formation and is shown to inhibit sprouting in culture cells, animal embryos, and during tumor angiogenesis.88-91 Interestingly, loss of CCM1, CCM3, or ICAP1 impairs DLL4-Notch signaling, resulting in excessive angiogenesis.92-94 Furthermore, induction of DLL4-NOTCH signaling by CCM1 results in increased PKB signaling and inhibition of ERK. Also, protein lysates from human CCM1 lesions show increased phospho-ERK levels,92 indicating that CCM1 suppresses ERK activation. Altogether, this suggests that CCM proteins activate DLL4-Notch signaling, and thereby, inhibit excessive angiogenesis, but also here molecular details are lacking on how CCM proteins activate DDL4-Notch signaling.
CCM in cell polarity
Next to maintaining the endothelial monolayer, adherens junctions are important for cell polarization and lumen formation. In various cell types and organisms, cell polarity is established by a protein complex consisting of: the partitioning defective (PAR) proteins PAR-3 and PAR-6 and atypical protein kinase C (aPKC).95,96 PAR-3 assembles PAR-6, aPKC, and the RAC1 guanine nucleotide exchange factor TIAM1.97,98 PAR-6 can interact with proteins from other cell polarity complexes like Crumbs and Pals1 from the CRB3-Pals1-PATJ (Pals1-associated tight junction protein) complex and Lgl (Lethal giant larvae) from the Scribble-Disc large (Dlg)-Lgl complex.99-102 In vertebrate epithelial cells, the PAR complex is localized to the tight junctions and disruption of this complex result in defects in tight junctions and polarity. VE-cadherin is co-distributed with members of the Par polarity complex, like Par-3 and Par-6.103 Integrin β1-matrix interactions at the basal EC surface regulate PAR-3 expression and junctional localization. Loss of CCM1 results in loss of apicobasal polarity and disturbance of proper vascular lumen formation, indicating an important role for CCM1 in polarity.104 However, how CCM1 regulates polarity is unknown. Because integrin β1-matrix interactions regulate PAR-3 expression, it is possible that ICAP1a, together with CCM1, plays a role polarity. Alternatively, serine threonine kinase (STK) 24, STK25, and mammalian sterile 2like 4 (MST4), were identified as interaction partners of CCM3 in a yeast-two-hybrid screen.39,105-107 Combined with the connection of MST4 with LKB1 function in cell polarity,108 this indicates a potential role for CCM3 in cell polarity.
Endothelial to mesenchymal transition
Loss of apicobasal polarity and cell–cell contacts is also associated with the induction of endothelial to mesenchymal transition (EndMT). EndMT is characterized by the acquisition of mesenchymal- and stem cell-like characteristics by the endothelium.109,110 By use of an endothelial-specific tamoxifen-inducible Ccm1 loss of function mice (iCCM1), it was demonstrated that the endothelial cells lining the vascular lesions associated with CCM, showed highly disorganized VE-cadherin expression and upregulated N-cadherin expression.111 Furthermore, CCM1 downregulation in lung and brain microvascular endothelial cells showed increased proliferation and enhanced invasive/sprouting capacitiy, which is mediated by Notch inhibition and subsequent BMP6 (bone morphogenetic protein 6) upregulation. Upregulation of BMP6 activates transforming growth factor-β (TGF-β) and BMP signaling pathways and results in increased EndMT.111 As Wnt/β-catenin signaling plays an important role in EndMT in myocardial cells112 and loss of CCM1 enhances β-catenin-dependent activation of the Wnt signaling pathway,68 this might be another pathway that contributes the EndMT phenotype.
Dual Role of Proteins in Adhesion Complexes and Transcription
Signals from junctions are transmitted toward the cell interior via two different mechanisms: by regulation of intercellular signaling cascades or via shuttling of proteins between adhesions sites at the plasma membrane and the nucleus. Proteins involved in the latter type of signaling are called NACos; proteins that can localize to the nucleus and adhesion complexes.113 All of the above described junction complexes contain proteins that can fulfill such a dual function. For example, the transcription factor ZONAB, which is found in tight junctions in high density, confluent cells where it is retained by the tight junction protein ZO-1 and binds to the small GTPase RALA.17,114 In proliferating cells, ZONAB accumulates in the nucleus where it interacts with the cell cycle regulator CDK4 and controls expression of cell cycle regulators like cyclin D1 and PCNA.16,115
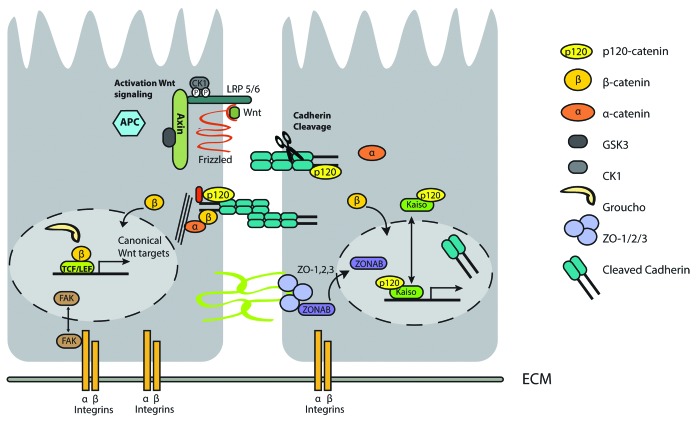
In adherens junctions, the protein β-catenin is an extensively studied example with dual localization. β-catenin stability is predominantly regulated by Wnt signaling. In absence of Wnt signaling, β-catenin is targeted for degradation by a multi-protein destruction complex consisting of the scaffold proteins Axin and Adenoma Polyposis Coli (APC), the serine/threonine kinases Casein Kinase 1 (CK1) and Glycogen Synthase-3β (GSK-3β), and the protein phosphatase 2A (PP2A). Phosphorylation of β-catenin by CK1 and GSK-3β target β-catenin for β-TRCP-mediated ubiquitination and degradation by the proteasome.116 Activation of Wnt signals result in inactivation of GSK3β activity and stabilization of β-catenin, which subsequently mediates transcription via the TCF (T cell factor)/LEF (lymphocyte enhancer binding factor 1) family of transcription factors. In absence of Wnt signaling, TCF/LEF transcription factors bind groucho and act as transcriptional repressors. Whereas in presence of Wnt signals, β-catenin displaces groucho and binds other co-factors to form a transcriptionally active complex with TCF/LEF117 (Fig. 3, left panel).
Figure 3. Dual role of proteins in adhesion complexes and transcription regulation. In absence of wnt signaling, β-catenin is degraded by the APC-destruction complex, whereas in presence of wnt signaling, degradation of β-catenin by APC is prevented and β-catenin activates TCF/LEF-mediated transcription (Left panel). Disruption of E-cadherin signaling by, for example, ADAM10-mediated cleavage of E-cadherin, results in nuclear translocation of β-catenin, and subsequent activation of the Wnt signaling pathway. However, following complete disintegration of the E-cadherin complex, the cytoplasmic domain of E-cadherin, derived after proteolytic cleavage and in addition p120ctn, may also translocate to the nucleus (Right panel). Combined or in isolation this nuclear shuttling of E-cadherin complex proteins may have different biological outcome. Multiple signals, like stress signals can result in nuclear localization of FAK, a key component of integrin signaling (left panel).
In absence of a Wnt stimulus, the majority of β-catenin is located at the plasma membrane where it binds to the cytoplasmic domain of type 1 cadherins. Association of E-cadherin to β-catenin prevents proteosomal degradation of both proteins. β-catenin shields a PEST sequence motif on E-cadherin, which when available is recognized by an ubiquitin ligase that marks E-cadherin for degradation,118 whereas E-cadherin prevents binding of APC and Axin to β-catenin, and thereby, prevents degradation of β-catenin.
Interestingly, E-cadherin itself can also accumulate in the nucleus (Fig. 3, right panel). Proteolytic cleavage of E-cadherin results in the disassembly of the cadherin, α-catenin, β-catenin complex, and may thus result in the release of the cytoplasmic domain of E-cadherin into the cytosol. Here the cytoplasmic domain may compete with APC for binding of β-catenin, and consequently, modulate Wnt signaling by determining β-catenin availability. Alternatively, cytoplasmic E-cadherin fragments have been shown to translocate to the nucleus by an, at present unknown, mechanism. Nuclear E-cadherin is detected in several tumor types and serves as prognostic marker, but molecular details with respect to its function within the nucleus are lacking.119
Next to TCF/LEF-mediated transcription, β-catenin also mediates transcription via other transcription factors, like the Forkhead box O (FOXO) family. During reactive oxygen species (ROS) signaling, β-catenin switches form TCF/LEF toward FOXO-dependent transcription.120,121 Furthermore, in absence of VE-cadherin signaling, β-catenin relocates to the nucleus, and together with FOXO1 mediates inhibition of Claudin-5 transcription.122 In this manner, FOXO acts as a mediator between adherens and tight junctions.
The catenin p120 (p120ctn) is also an armadillo repeat-containing member of the cadherin–catenin cell–cell adhesion complex.123 The structural homology between p120ctn and β-catenin has led to the discovery that similar to β-catenin, p120ctn binds to the E-cadherin complex, can translocate to the nucleus, and binds to a specific transcription factor, named Kaiso (Fig. 3, right panel).124 Kaiso belongs to the BTB/POZ (Broad complex, Tramtrak, Bric a brac/POx virus and Zinc finger) family of transcription factors. The POZ domain of these transcription factors acts as a protein–protein interaction domain and mediates homo-dimerization as well as binding of Kaiso to other transcriptional regulators such as CTCF125 and the NCoR repressor complex.126 Kaiso also contains highly acidic regions and these regions are associated with activation of transcription. Thus, Kaiso can act both as an activator and repressor of transcription.
Interestingly, nuclear localization of p120ctn was initially difficult to detect, but treatment of cells with leptomycin B, a specific inhibitor of CRM1-mediated nuclear export, results in robust detection of nuclear p120ctn.127 This indicates that nuclear import is nullified by nuclear export, and that therefore nuclear localization requires specific nuclear retention. Binding to Kaiso may serve in this way as a nuclear anchor for p120ctn. In addition, Kaiso itself also displays nucleo-cytoplasmic shuttling and cytoplasmic localization of Kaiso appears to correlate with cytoplasmic localization of p120ctn.128,129 This correlation may suggest that p120ctn binding to Kaiso serves to facilitate export of Kaiso out of the nucleus. In agreement, Kaiso does harbor a classical basic Nuclear Localization Sequence (NLS) and binds importin-α.130 This suggests that Kaiso independently of p120ctn can re-enter the nucleus.
The protein tyrosine kinase (PTK), focal adhesion kinase (FAK), is a key component in the signaling downstream of integrins. The presence of both a NLS131 and nuclear export signal (NES),132 suggest a role for FAK in the nucleus. Indeed, multiple signals result in nuclear FAK localization (Fig. 3, left panel).133 For example, stress signals (H2O2) induce nuclear localization of FAK where it promotes muscle cell differentiation.134 Nuclear localization of FAK does not appear to require its kinase activity, but rather depends on its FERM (band 4.1,ezrin, radixin, moesin homology) domain. In agreement, other FERM domain proteins e.g., moesin, can also be found in the nucleus (reviewed in ref. 133). Mutational analysis has suggested that the FERM domain harbors a NLS sequence,131 whereas a NES sequence is identified within the kinase domain.132 The FERM domain of nuclear FAK is suggested to facilitate the interaction between p53 and its E3 ligase mdm2.131 This will result in a reduction in p53 levels, and thus, inhibition of p53 transcriptional activity. Other nuclear interaction partners of FAK may include transcription complexes containing the methyl CpG-binding protein MBD2 e.g., NuRD complex.135
Taken together, the examples described in brief above indicate that a nuclear role for cell–cell adhesion or, for that matter cell–matrix adhesion complex members such as FAK, is likely to be more common than initially anticipated. Exploring such a nuclear role for other adhesion complex members besides the ones described above is therefore warranted.
Spatial Regulation of CCM1
Interestingly, CCM1 harbors a putative NLS and NES,136,137 which suggests CCM1 might have a function in the nucleus as well, next to its role in junction maintenance. Mutation of the NLS sequence reduces the nuclear localization of CCM1 to ~10%.41 Furthermore, a role for ICAP1 in mediating nuclear localization of CCM1 has been described. As described above, the β1-integrin binding protein ICAP1 affects CCM1 stabilization through binding to the first NPxY motive in CCM1, which results in an open, more stable conformation of CCM1.78 ICAP1α binds to the cytoplasmic domain of integrin β1, and thereby, prevents binding of talin72-74 and kindlin.10,75 This subsequently inhibits proper formation of focal adhesions. ICAP1α acts as a negative regulator of integrin function by competing with kindlin for binding to the β1-intergrin tail. This suggests that integrin activation and concomitant release of ICAP1α results in CCM1 nuclear translocation. Next to ICAP1-induced nuclear localization of CCM1, CCM1, and Nd1-L are described to mediate cytoplasmic localization of CCM1. ICAP1-mediated nuclear translocation of CCM1 is dominant over the cytoplasmic localization induced by CCM2.138 What function is mediated by nuclear CCM1 is however at present completely unknown.
What is the function of nuclear CCM1?
Nuclear localization of CCM1 can be induced by co-transfection of ICAP1; however, the mechanism behind this translocation is still unknown. As described above, loss of CCM1 or VE-cadherin results in nuclear translocation of β-catenin and subsequent increase of β-catenin-dependent transcription regulation. Whether loss of VE-cadherin or β-catenin also results in CCM1 nuclear localization will be important to determine to get more insight into the signals that mediate nuclear CCM1.
In C. elegans, it is shown that KRI-1 (C. elegans CCM1) is important for nuclear localization of DAF16 (C. elegans FOXO), which results in subsequent lifespan extension during lack of germ-line signals.139 Whether CCM1 is also important in regulation of mammalian FOXO localization has not been described yet. However, recently, a function for CCM1 in regulation of ROS homeostasis via FOXO is shown.140 CCM1-knockout MEFs (mouse embryonic fibroblasts) show increased ROS levels and decreased levels of FOXO1 and SOD2 (super oxide dismutase 2). The authors suggest that CCM1 exerts its effect on FOXO through regulation of FOXO1 stability. However, they also show increased PKB-mediated FOXO phosphorylation upon add-back of CCM1. Phosphorylation of FOXO by PKB results in ubiquitination and degradation of FOXOs,141,142 which contradicts the conclusions of the authors. Also, others have shown increased PKB phosphorylation upon CCM1 overexpression in HUVEC cells,92 whereas this is not shown for CCM3.94 Based on these data, it is unclear what effect CCM1 could have on FOXO function and if this involves the nuclear localization of either FOXO or CCM1.
Interestingly, FOXO1 and FOXO3a also have an important function in the regulation of angiogenesis. Foxo1−/− mice die from severe vascular defects143,144 and inducible Foxo 1-, 3-, and 4-knockout mice show upregulation of Sprouty and PBX1, among others, as FOXO-regulated mediators of endothelial cell morphogenesis and vascular homeostasis.145 Furthermore, in endothelial cells, it is shown by microarray analysis that FOXO1 induces many genes associated with vascular destabilization and apoptosis.146 For example, Angiopoietin-2 (Ang-2) is an important FOXO1 target.146 Ang-2 is an antagonist of the receptor tyrosine kinase Tie2 and its activating ligand Ang-1. Tie2 and Ang-1 are required for vascular development.147-149 Ang-1 promotes survival of endothelial cells by activation of PKB and subsequent inhibition of FOXO1.146 Hence, regulation of Ang-2 by FOXO inhibits its inhibition through Ang-1-mediated PKB activation.
Concluding Remarks
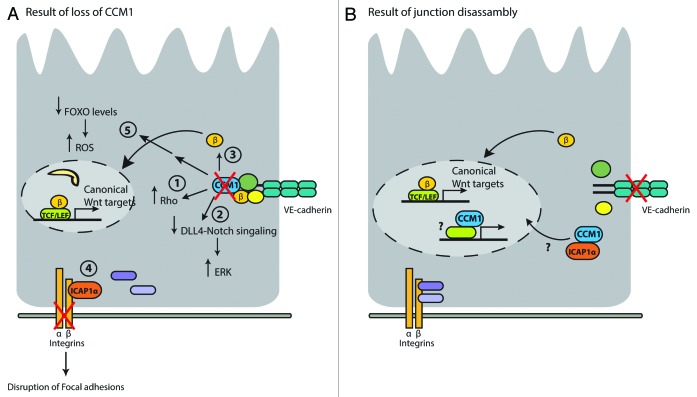
Until now and discussed above, previous work on CCM1 is predominantly focused on the role of CCM1 in maintenance of the endothelial monolayer. The experiments performed in both tissue culture and model organisms have provided valuable information on the potential function of the CCM1 protein. Based on these experiments, the following conclusions can be drawn (summarized in Fig. 4A):
Figure 4. (A) Loss of CCM1 (depicted with a red cross in the figure) results in activation of Rho signaling (1), inhibition of DLL4-Notch signaling (2), nuclear localization of β-catenin (3), disruption of focal adhesions (indicated by red cross through integrins) as a result of ICAP binding (4), and decreased FOXO protein levels, resulting in increased ROS levels (5). (B) Suggested model in a situation where cadherin signaling is disrupted (depicted by a red cross through cadherin). See text for more details.
(1) Loss of CCM1 results in increased RHO activity and leakiness of the endothelial barrier.
(2) Next to induction of RHO activity, loss of CCM1 results in decreased activity of the DLL4-Notch pathway, resulting in increased proliferation. This is also indicated by increased activity of ERK in CCM patient material.
(3) Loss of CCM1 disturbs the adherens junctions due to release of β-catenin from VE-cadherin. Release of β-catenin from VE-cadherin stimulates its function as transcription co-factor, and thereby, enhances activation of the Wnt pathway and induction of proliferation.
(4) Loss of CCM1 results in increased binding of ICAP1 to the β1-integrin tail and subsequent disruption of focal adhesions.
(5) Loss of CCM1 results in increased ROS due to decreased FOXO levels and subsequent decreased SOD-mediated ROS scavenging.
These data provide us with important information on the potential role of CCM in maintaining the endothelial barrier; however, they provide little molecular details on a potential nuclear function of CCM1. Based on the knowledge gained from other junctional proteins and on the observed nuclear localization of CCM1, we suggest a role of CCM1 in transcription regulation. We propose a similar role as described for β-catenin, which upon release of cadherins, translocates to the nucleus to mediate transcription. Furthermore, ICAP1 shows a stabilizing function toward CCM1 and induces its nuclear localization. Therefore, examining the circumstances in which ICAP1 interacts with CCM1 and mediates KRIT1 nuclear localization will provide more clues on the nuclear function of CCM1 (suggested model in Fig. 4B). In addition, experiments addressing directly a role of CCM1 in nuclear functions, such as transcription regulation, may shed light onto the possible nuclear function of CCM1. By analogy to b-catenin and p120-catenin binding of CCM1 to transcriptional regulators appears the most likely nuclear role for CCM1, but by no means precludes other roles such as e.g., a role in nuclear architecture or mRNA nuclear export. As for now this second life of CCM1 is still largely unknown.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Bazzoni G, Dejana E. Endothelial cell-to-cell junctions: molecular organization and role in vascular homeostasis. Physiol Rev. 2004;84:869–901. doi: 10.1152/physrev.00035.2003. [DOI] [PubMed] [Google Scholar]
- 2.Steed E, Balda MS, Matter K. Dynamics and functions of tight junctions. Trends Cell Biol. 2010;20:142–9. doi: 10.1016/j.tcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Dejana E, Tournier-Lasserve E, Weinstein BM. The control of vascular integrity by endothelial cell junctions: molecular basis and pathological implications. Dev Cell. 2009;16:209–21. doi: 10.1016/j.devcel.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Nanes BA, Chiasson-MacKenzie C, Lowery AM, Ishiyama N, Faundez V, Ikura M, Vincent PA, Kowalczyk AP. p120-catenin binding masks an endocytic signal conserved in classical cadherins. J Cell Biol. 2012;199:365–80. doi: 10.1083/jcb.201205029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dejana E, Orsenigo F, Lampugnani MG. The role of adherens junctions and VE-cadherin in the control of vascular permeability. J Cell Sci. 2008;121:2115–22. doi: 10.1242/jcs.017897. [DOI] [PubMed] [Google Scholar]
- 6.Birdsey GM, Dryden NH, Amsellem V, Gebhardt F, Sahnan K, Haskard DO, Dejana E, Mason JC, Randi AM. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111:3498–506. doi: 10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deleuze V, Chalhoub E, El-Hajj R, Dohet C, Le Clech M, Couraud PO, Huber P, Mathieu D. TAL-1/SCL and its partners E47 and LMO2 up-regulate VE-cadherin expression in endothelial cells. Mol Cell Biol. 2007;27:2687–97. doi: 10.1128/MCB.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–87. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 9.Bouvard D, Pouwels J, De Franceschi N, Ivaska J. Integrin inactivators: balancing cellular functions in vitro and in vivo. Nat Rev Mol Cell Biol. 2013;14:430–42. doi: 10.1038/nrm3599. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, Draheim KM, Zhang R, Calderwood DA, Boggon TJ. Mechanism for KRIT1 release of ICAP1-mediated suppression of integrin activation. Mol Cell. 2013;49:719–29. doi: 10.1016/j.molcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J Cell Sci. 2011;124:1183–93. doi: 10.1242/jcs.064618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–38. doi: 10.1016/S0092-8674(00)00115-X. [DOI] [PubMed] [Google Scholar]
- 13.Kooistra MR, Dubé N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 14.Retta SF, Balzac F, Avolio M. Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol. 2006;85:283–93. doi: 10.1016/j.ejcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 15.Balzac F, Avolio M, Degani S, Kaverina I, Torti M, Silengo L, Small JV, Retta SF. E-cadherin endocytosis regulates the activity of Rap1: a traffic light GTPase at the crossroads between cadherin and integrin function. J Cell Sci. 2005;118:4765–83. doi: 10.1242/jcs.02584. [DOI] [PubMed] [Google Scholar]
- 16.Balda MS, Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000;19:2024–33. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel P, Aronheim A, Kavanagh E, Balda MS, Matter K, Bunney TD, Marshall CJ. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2005;24:54–62. doi: 10.1038/sj.emboj.7600497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wojciak-Stothard B, Ridley AJ. Rho GTPases and the regulation of endothelial permeability. Vascul Pharmacol. 2002;39:187–99. doi: 10.1016/S1537-1891(03)00008-9. [DOI] [PubMed] [Google Scholar]
- 19.van Nieuw Amerongen GP, van Hinsbergh VW. Cytoskeletal effects of rho-like small guanine nucleotide-binding proteins in the vascular system. Arterioscler Thromb Vasc Biol. 2001;21:300–11. doi: 10.1161/01.ATV.21.3.300. [DOI] [PubMed] [Google Scholar]
- 20.Essler M, Hermann K, Amano M, Kaibuchi K, Heesemann J, Weber PC, Aepfelbacher M. Pasteurella multocida toxin increases endothelial permeability via Rho kinase and myosin light chain phosphatase. J Immunol. 1998;161:5640–6. [PubMed] [Google Scholar]
- 21.Kawano Y, Fukata Y, Oshiro N, Amano M, Nakamura T, Ito M, Matsumura F, Inagaki M, Kaibuchi K. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J Cell Biol. 1999;147:1023–38. doi: 10.1083/jcb.147.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbajal JM, Schaeffer RC., Jr. RhoA inactivation enhances endothelial barrier function. Am J Physiol. 1999;277:C955–64. doi: 10.1152/ajpcell.1999.277.5.C955. [DOI] [PubMed] [Google Scholar]
- 23.Hordijk PL, Anthony E, Mul FP, Rientsma R, Oomen LC, Roos D. Vascular-endothelial-cadherin modulates endothelial monolayer permeability. J Cell Sci. 1999;112:1915–23. doi: 10.1242/jcs.112.12.1915. [DOI] [PubMed] [Google Scholar]
- 24.Shenkar R, Venkatasubramanian PN, Wyrwicz AM, Zhao JC, Shi C, Akers A, Marchuk DA, Awad IA. Advanced magnetic resonance imaging of cerebral cavernous malformations: part II. Imaging of lesions in murine models. Neurosurgery. 2008;63:790–7, discussion 797-8. doi: 10.1227/01.NEU.0000315862.24920.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shenkar R, Venkatasubramanian PN, Zhao JC, Batjer HH, Wyrwicz AM, Awad IA. Advanced magnetic resonance imaging of cerebral cavernous malformations: part I. High-field imaging of excised human lesions. Neurosurgery. 2008;63:782–9, discussion 789. doi: 10.1227/01.NEU.0000325490.80694.A2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clatterbuck RE, Eberhart CG, Crain BJ, Rigamonti D. Ultrastructural and immunocytochemical evidence that an incompetent blood-brain barrier is related to the pathophysiology of cavernous malformations. J Neurol Neurosurg Psychiatr, 2001:188-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong JH, Awad IA, Kim JH. Ultrastructural pathological features of cerebrovascular malformations: a preliminary report. Neurosurgery. 2000;46:1454–9. doi: 10.1097/00006123-200006000-00027. [DOI] [PubMed] [Google Scholar]
- 28.Toll A, Parera E, Giménez-Arnau A, Pou A, Lloreta J, Limaye N, et al. Cutaneous Venous Malformations in Familial Cerebral Cavernomatosis Caused by KRIT1 Gene Mutations. Dermatology (Basel) 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toldo I, Drigo P, Mammi I, Marini V, Carollo C. Vertebral and spinal cavernous angiomas associated with familial cerebral cavernous malformation. Surg Neurol. 2009;71:167–71. doi: 10.1016/j.surneu.2007.07.067. [DOI] [PubMed] [Google Scholar]
- 30.Otten P, Pizzolato GP, Rilliet B, Berney J. [131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies] Neurochirurgie. 1989;35:82–3, 128-31. [PubMed] [Google Scholar]
- 31.Pagenstecher A, Stahl S, Sure U, Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum Mol Genet. 2009;18:911–8. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18:919–30. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahoo T, Johnson EW, Thomas JW, Kuehl PM, Jones TL, Dokken CG, Touchman JW, Gallione CJ, Lee-Lin SQ, Kosofsky B, et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum Mol Genet. 1999;8:2325–33. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- 34.Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF, Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23:189–93. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 35.Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73:1459–64. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, et al. Société Française de Neurochirurgie Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet. 2004;74:326–37. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, et al. Société Française de Neurochirurgie Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guclu B, Ozturk AK, Pricola KL, Bilguvar K, Shin D, O’Roak BJ, Gunel M. Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery. 2005;57:1008–13. doi: 10.1227/01.NEU.0000180811.56157.E1. [DOI] [PubMed] [Google Scholar]
- 39.Voss K, Stahl S, Schleider E, Ullrich S, Nickel J, Mueller TD, Felbor U. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8:249–56. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- 40.Hilder TL, Malone MH, Bencharit S, Colicelli J, Haystead TA, Johnson GL, Wu CC. Proteomic identification of the cerebral cavernous malformation signaling complex. J Proteome Res. 2007;6:4343–55. doi: 10.1021/pr0704276. [DOI] [PubMed] [Google Scholar]
- 41.Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson GL, Marchuk DA. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14:2521–31. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- 42.Chan AC, Li DY, Berg MJ, Whitehead KJ. Recent insights into cerebral cavernous malformations: animal models of CCM and the human phenotype. FEBS J. 2010;277:1076–83. doi: 10.1111/j.1742-4658.2009.07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131:1437–48. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- 44.Whitehead KJ, Chan AC, Navankasattusas S, Koh W, London NR, Ling J, Mayo AH, Drakos SG, Jones CA, Zhu W, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15:177–84. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plummer NW, Squire TL, Srinivasan S, Huang E, Zawistowski JS, Matsunami H, Hale LP, Marchuk DA. Neuronal expression of the Ccm2 gene in a new mouse model of cerebral cavernous malformations. Mamm Genome. 2006;17:119–28. doi: 10.1007/s00335-005-0098-8. [DOI] [PubMed] [Google Scholar]
- 46.Denier C, Gasc J-M, Chapon F, Domenga V, Lescoat C, Joutel A, Tournier-Lasserve E. Krit1/cerebral cavernous malformation 1 mRNA is preferentially expressed in neurons and epithelial cells in embryo and adult. Mech Dev. 2002;117:363–7. doi: 10.1016/S0925-4773(02)00209-5. [DOI] [PubMed] [Google Scholar]
- 47.Kehrer-Sawatzki H, Wilda M, Braun VM, Richter H-P, Hameister H. Mutation and expression analysis of the KRIT1 gene associated with cerebral cavernous malformations (CCM1) Acta Neuropathol. 2002;104:231–40. doi: 10.1007/s00401-002-0552-6. [DOI] [PubMed] [Google Scholar]
- 48.Boulday G, Blécon A, Petit N, Chareyre F, Garcia LA, Niwa-Kawakita M, Giovannini M, Tournier-Lasserve E. Tissue-specific conditional CCM2 knockout mice establish the essential role of endothelial CCM2 in angiogenesis: implications for human cerebral cavernous malformations. Dis Model Mech. 2009;2:168–77. doi: 10.1242/dmm.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boulday G, Rudini N, Maddaluno L, Blécon A, Arnould M, Gaudric A, Chapon F, Adams RH, Dejana E, Tournier-Lasserve E. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J Exp Med. 2011;208:1835–47. doi: 10.1084/jem.20110571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He Y, Zhang H, Yu L, Gunel M, Boggon TJ, Chen H, Min W. Stabilization of VEGFR2 signaling by cerebral cavernous malformation 3 is critical for vascular development. Sci Signal. 2010;3:ra26. doi: 10.1126/scisignal.2000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plummer NW, Gallione CJ, Srinivasan S, Zawistowski JS, Louis DN, Marchuk DA. Loss of p53 sensitizes mice with a mutation in Ccm1 (KRIT1) to development of cerebral vascular malformations. Am J Pathol. 2004;165:1509–18. doi: 10.1016/S0002-9440(10)63409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonald DA, Shenkar R, Shi C, Stockton RA, Akers AL, Kucherlapati MH, Kucherlapati R, Brainer J, Ginsberg MH, Awad IA, et al. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum Mol Genet. 2011;20:211–22. doi: 10.1093/hmg/ddq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mably JD, Chuang LP, Serluca FC, Mohideen M-APK, Chen J-N, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–46. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- 54.Yoruk B, Gillers BS, Chi NC, Scott IC. Ccm3 functions in a manner distinct from Ccm1 and Ccm2 in a zebrafish model of CCM vascular disease. Dev Biol. 2012;362:121–31. doi: 10.1016/j.ydbio.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Zheng X, Xu C, Di Lorenzo A, Kleaveland B, Zou Z, Seiler C, Chen M, Cheng L, Xiao J, He J, et al. CCM3 signaling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120:2795–804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hogan BM, Bussmann J, Wolburg H, Schulte-Merker S. ccm1 cell autonomously regulates endothelial cellular morphogenesis and vascular tubulogenesis in zebrafish. Hum Mol Genet. 2008;17:2424–32. doi: 10.1093/hmg/ddn142. [DOI] [PubMed] [Google Scholar]
- 57.Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, Sweeney SM, Chen M, Guo L, Lu MM, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15:169–76. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gingras AR, Liu JJ, Ginsberg MH. Structural basis of the junctional anchorage of the cerebral cavernous malformations complex. J Cell Biol. 2012;199:39–48. doi: 10.1083/jcb.201205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng X, Xu C, Smith AO, Stratman AN, Zou Z, Kleaveland B, Yuan L, Didiku C, Sen A, Liu X, et al. Dynamic regulation of the cerebral cavernous malformation pathway controls vascular stability and growth. Dev Cell. 2012;23:342–55. doi: 10.1016/j.devcel.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosen JN, Sogah VM, Ye LY, Mably JD. ccm2-like is required for cardiovascular development as a novel component of the Heg-CCM pathway. Dev Biol. 2013;376:74–85. doi: 10.1016/j.ydbio.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21-22. Oncogene. 1997;15:1043–9. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- 62.Pannekoek WJ, Kooistra MR, Zwartkruis FJ, Bos JL. Cell-cell junction formation: the role of Rap1 and Rap1 guanine nucleotide exchange factors. Biochim Biophys Acta. 2009;1788:790–6. doi: 10.1016/j.bbamem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 63.Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J Biol Chem. 2009;284:10995–9. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwamborn JC, Püschel AW. The sequential activity of the GTPases Rap1B and Cdc42 determines neuronal polarity. Nat Neurosci. 2004;7:923–9. doi: 10.1038/nn1295. [DOI] [PubMed] [Google Scholar]
- 65.Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell cell junctions. J Cell Biol. 2007;179:247–54. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X, Zhang R, Draheim KM, Liu W, Calderwood DA, Boggon TJ. Structural basis for small G protein effector interaction of Ras-related protein 1 (Rap1) and adaptor protein Krev interaction trapped 1 (KRIT1) J Biol Chem. 2012;287:22317–27. doi: 10.1074/jbc.M112.361295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gingras AR, Puzon-McLaughlin W, Ginsberg MH. The structure of the ternary complex of Krev interaction trapped 1 (KRIT1) bound to both the Rap1 GTPase and the heart of glass (HEG1) cytoplasmic tail. J Biol Chem. 2013;288:23639–49. doi: 10.1074/jbc.M113.462911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glading AJ, Ginsberg MH. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis Model Mech. 2010;3:73–83. doi: 10.1242/dmm.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang J, Clatterbuck RE, Rigamonti D, Chang DD, Dietz HC. Interaction between krit1 and icap1alpha infers perturbation of integrin beta1-mediated angiogenesis in the pathogenesis of cerebral cavernous malformation. Hum Mol Genet. 2001;10:2953–60. doi: 10.1093/hmg/10.25.2953. [DOI] [PubMed] [Google Scholar]
- 70.Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum Mol Genet. 2002;11:389–96. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- 71.Liu W, Boggon TJ. Cocrystal structure of the ICAP1 PTB domain in complex with a KRIT1 peptide. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69:494–8. doi: 10.1107/S1744309113010762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouvard D, Vignoud L, Dupé-Manet S, Abed N, Fournier HN, Vincent-Monegat C, Retta SF, Fassler R, Block MR. Disruption of focal adhesions by integrin cytoplasmic domain-associated protein-1 alpha. J Biol Chem. 2003;278:6567–74. doi: 10.1074/jbc.M211258200. [DOI] [PubMed] [Google Scholar]
- 73.Millon-Frémillon A, Bouvard D, Grichine A, Manet-Dupé S, Block MR, Albiges-Rizo C. Cell adaptive response to extracellular matrix density is controlled by ICAP-1-dependent beta1-integrin affinity. J Cell Biol. 2008;180:427–41. doi: 10.1083/jcb.200707142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouvard D, Aszodi A, Kostka G, Block MR, Albigès-Rizo C, Fässler R. Defective osteoblast function in ICAP-1-deficient mice. Development. 2007;134:2615–25. doi: 10.1242/dev.000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brunner M, Millon-Frémillon A, Chevalier G, Nakchbandi IA, Mosher D, Block MR, Albigès-Rizo C, Bouvard D. Osteoblast mineralization requires beta1 integrin/ICAP-1-dependent fibronectin deposition. J Cell Biol. 2011;194:307–22. doi: 10.1083/jcb.201007108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moser M, Legate KR, Zent R, Fässler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–9. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Basu S, Rigamonti D, Dietz HC, Clatterbuck RE. Krit1 modulates beta 1-integrin-mediated endothelial cell proliferation. Neurosurgery. 2008;63:571–8, discussion 578. doi: 10.1227/01.NEU.0000325255.30268.B0. [DOI] [PubMed] [Google Scholar]
- 78.Béraud-Dufour S, Gautier R, Albiges-Rizo C, Chardin P, Faurobert E. Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. FEBS J. 2007;274:5518–32. doi: 10.1111/j.1742-4658.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Faurobert E, Rome C, Lisowska J, Manet-Dupé S, Boulday G, Malbouyres M, Balland M, Bouin AP, Kéramidas M, Bouvard D, et al. CCM1-ICAP-1 complex controls β1 integrin-dependent endothelial contractility and fibronectin remodeling. J Cell Biol. 2013;202:545–61. doi: 10.1083/jcb.201303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gunel M, Laurans MSH, Shin D, DiLuna ML, Voorhees J, Choate K, Nelson-Williams C, Lifton RP. KRIT1, a gene mutated in cerebral cavernous malformation, encodes a microtubule-associated protein. Proc Natl Acad Sci U S A. 2002;99:10677–82. doi: 10.1073/pnas.122354499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu JJ, Stockton RA, Gingras AR, Ablooglu AJ, Han J, Bobkov AA, Ginsberg MH. A mechanism of Rap1-induced stabilization of endothelial cell--cell junctions. Mol Biol Cell. 2011;22:2509–19. doi: 10.1091/mbc.E11-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borikova AL, Dibble CF, Sciaky N, Welch CM, Abell AN, Bencharit S, Johnson GL. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J Biol Chem. 2010;285:11760–4. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207:881–96. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yamamoto M, Ramirez SH, Sato S, Kiyota T, Cerny RL, Kaibuchi K, Persidsky Y, Ikezu T. Phosphorylation of claudin-5 and occludin by rho kinase in brain endothelial cells. Am J Pathol. 2008;172:521–33. doi: 10.2353/ajpath.2008.070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guazzi P, Goitre L, Ferro E, Cutano V, Martino C, Trabalzini L, Retta SF. Identification of the Kelch family protein Nd1-L as a novel molecular interactor of KRIT1. PLoS One. 2012;7:e44705. doi: 10.1371/journal.pone.0044705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richardson BT, Dibble CF, Borikova AL, Johnson GL. Cerebral cavernous malformation is a vascular disease associated with activated RhoA signaling. Biol Chem. 2013;394:35–42. doi: 10.1515/hsz-2012-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vestweber D, Winderlich M, Cagna G, Nottebaum AF. Cell adhesion dynamics at endothelial junctions: VE-cadherin as a major player. Trends Cell Biol. 2009;19:8–15. doi: 10.1016/j.tcb.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 88.Limbourg FP, Takeshita K, Radtke F, Bronson RT, Chin MT, Liao JK. Essential role of endothelial Notch1 in angiogenesis. Circulation. 2005;111:1826–32. doi: 10.1161/01.CIR.0000160870.93058.DD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hellström M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–80. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 90.Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–7. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 91.Ehebauer M, Hayward P, Arias AM. Notch, a universal arbiter of cell fate decisions. Science. 2006;314:1414–5. doi: 10.1126/science.1134042. [DOI] [PubMed] [Google Scholar]
- 92.Wüstehube J, Bartol A, Liebler SS, Brütsch R, Zhu Y, Felbor U, Sure U, Augustin HG, Fischer A. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc Natl Acad Sci U S A. 2010;107:12640–5. doi: 10.1073/pnas.1000132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brütsch R, Liebler SS, Wüstehube J, Bartol A, Herberich SE, Adam MG, Telzerow A, Augustin HG, Fischer A. Integrin cytoplasmic domain-associated protein-1 attenuates sprouting angiogenesis. Circ Res. 2010;107:592–601. doi: 10.1161/CIRCRESAHA.110.217257. [DOI] [PubMed] [Google Scholar]
- 94.You C, Sandalcioglu IE, Dammann P, Felbor U, Sure U, Zhu Y. Loss of CCM3 impairs DLL4-Notch signalling: implication in endothelial angiogenesis and in inherited cerebral cavernous malformations. J Cell Mol Med. 2013;17:407–18. doi: 10.1111/jcmm.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Macara IG. Parsing the polarity code. Nat Rev Mol Cell Biol. 2004;5:220–31. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- 96.Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. J Cell Sci. 2006;119:979–87. doi: 10.1242/jcs.02898. [DOI] [PubMed] [Google Scholar]
- 97.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–9. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 98.Mertens AE, Rygiel TP, Olivo C, van der Kammen R, Collard JG. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J Cell Biol. 2005;170:1029–37. doi: 10.1083/jcb.200502129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nat Cell Biol. 2003;5:137–42. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]
- 100.Plant PJ, Fawcett JP, Lin DC, Holdorf AD, Binns K, Kulkarni S, Pawson T. A polarity complex of mPar-6 and atypical PKC binds, phosphorylates and regulates mammalian Lgl. Nat Cell Biol. 2003;5:301–8. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 101.Yamanaka T, Horikoshi Y, Sugiyama Y, Ishiyama C, Suzuki A, Hirose T, Iwamatsu A, Shinohara A, Ohno S. Mammalian Lgl forms a protein complex with PAR-6 and aPKC independently of PAR-3 to regulate epithelial cell polarity. Curr Biol. 2003;13:734–43. doi: 10.1016/S0960-9822(03)00244-6. [DOI] [PubMed] [Google Scholar]
- 102.Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Médina E, Arsanto JP, Le Bivic A. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell. 2004;15:1324–33. doi: 10.1091/mbc.E03-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iden S, Rehder D, August B, Suzuki A, Wolburg-Buchholz K, Wolburg H, Ohno S, Behrens J, Vestweber D, Ebnet K. A distinct PAR complex associates physically with VE-cadherin in vertebrate endothelial cells. EMBO Rep. 2006;7:1239–46. doi: 10.1038/sj.embor.7400819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lampugnani MG, Orsenigo F, Rudini N, Maddaluno L, Boulday G, Chapon F, Dejana E. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J Cell Sci. 2010;123:1073–80. doi: 10.1242/jcs.059329. [DOI] [PubMed] [Google Scholar]
- 105.Ma X, Zhao H, Shan J, Long F, Chen Y, Chen Y, Zhang Y, Han X, Ma D. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18:1965–78. doi: 10.1091/mbc.E06-07-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Goudreault M, D’Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–71. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Voss K, Stahl S, Hogan BM, Reinders J, Schleider E, Schulte-Merker S, Felbor U. Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Hum Mutat. 2009;30:1003–11. doi: 10.1002/humu.20996. [DOI] [PubMed] [Google Scholar]
- 108.ten Klooster JP, Jansen M, Yuan J, Oorschot V, Begthel H, Di Giacomo V, Colland F, de Koning J, Maurice MM, Hornbeck P, et al. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev Cell. 2009;16:551–62. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 109.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–61. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 110.Medici D, Shore EM, Lounev VY, Kaplan FS, Kalluri R, Olsen BR. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat Med. 2010;16:1400–6. doi: 10.1038/nm.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, et al. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–6. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- 112.Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E. Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J Cell Biol. 2004;166:359–67. doi: 10.1083/jcb.200403050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Balda MS, Matter K. Epithelial cell adhesion and the regulation of gene expression. Trends Cell Biol. 2003;13:310–8. doi: 10.1016/S0962-8924(03)00105-3. [DOI] [PubMed] [Google Scholar]
- 114.Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003;160:423–32. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26:2387–98. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Polakis P. Casein kinase 1: a Wnt’er of disconnect. Curr Biol. 2002;12:R499–501. doi: 10.1016/S0960-9822(02)00969-7. [DOI] [PubMed] [Google Scholar]
- 117.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 118.Huber AH, Stewart DB, Laurents DV, Nelson WJ, Weis WI. The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin. A possible mechanism for regulating cadherin turnover. J Biol Chem. 2001;276:12301–9. doi: 10.1074/jbc.M010377200. [DOI] [PubMed] [Google Scholar]
- 119.Chetty R, Serra S, Salahshor S. Nuclear expression of E-cadherin. Am J Surg Pathol. 2008;32:1269–70. doi: 10.1097/PAS.0b013e31816a0cdd. [DOI] [PubMed] [Google Scholar]
- 120.Hoogeboom D, Essers MAG, Polderman PE, Voets E, Smits LMM, Burgering BMT. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J Biol Chem. 2008;283:9224–30. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- 121.Almeida M, Han L, Martin-Millan M, O’Brien CA, Manolagas SC. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting beta-catenin from T cell factor- to forkhead box O-mediated transcription. J Biol Chem. 2007;282:27298–305. doi: 10.1074/jbc.M702811200. [DOI] [PubMed] [Google Scholar]
- 122.Taddei A, Giampietro C, Conti A, Orsenigo F, Breviario F, Pirazzoli V, Potente M, Daly C, Dimmeler S, Dejana E. Endothelial adherens junctions control tight junctions by VE-cadherin-mediated upregulation of claudin-5. Nat Cell Biol. 2008;10:923–34. doi: 10.1038/ncb1752. [DOI] [PubMed] [Google Scholar]
- 123.Reynolds AB, Herbert L, Cleveland JL, Berg ST, Gaut JR. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 1992;7:2439–45. [PubMed] [Google Scholar]
- 124.Daniel JM, Reynolds AB. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–23. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Defossez PA, Kelly KF, Filion GJ, Pérez-Torrado R, Magdinier F, Menoni H, Nordgaard CL, Daniel JM, Gilson E. The human enhancer blocker CTC-binding factor interacts with the transcription factor Kaiso. J Biol Chem. 2005;280:43017–23. doi: 10.1074/jbc.M510802200. [DOI] [PubMed] [Google Scholar]
- 126.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–34. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 127.Roczniak-Ferguson A, Reynolds AB. Regulation of p120-catenin nucleocytoplasmic shuttling activity. J Cell Sci. 2003;116:4201–12. doi: 10.1242/jcs.00724. [DOI] [PubMed] [Google Scholar]
- 128.Soubry A, van Hengel J, Parthoens E, Colpaert C, Van Marck E, Waltregny D, Reynolds AB, van Roy F. Expression and nuclear location of the transcriptional repressor Kaiso is regulated by the tumor microenvironment. Cancer Res. 2005;65:2224–33. doi: 10.1158/0008-5472.CAN-04-2020. [DOI] [PubMed] [Google Scholar]
- 129.Daniel JM. Dancing in and out of the nucleus: p120(ctn) and the transcription factor Kaiso. Biochim Biophys Acta. 2007;1773:59–68. doi: 10.1016/j.bbamcr.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 130.Kelly KF, Otchere AA, Graham M, Daniel JM. Nuclear import of the BTB/POZ transcriptional regulator Kaiso. J Cell Sci. 2004;117:6143–52. doi: 10.1242/jcs.01541. [DOI] [PubMed] [Google Scholar]
- 131.Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ossovskaya V, Lim ST, Ota N, Schlaepfer DD, Ilic D. FAK nuclear export signal sequences. FEBS Lett. 2008;582:2402–6. doi: 10.1016/j.febslet.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lim ST. Nuclear FAK: a new mode of gene regulation from cellular adhesions. Mol Cells. 2013;36:1–6. doi: 10.1007/s10059-013-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Luo SW, Zhang C, Zhang B, Kim CH, Qiu YZ, Du QS, Mei L, Xiong WC. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. EMBO J. 2009;28:2568–82. doi: 10.1038/emboj.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mei L, Xiong WC. FAK interaction with MBD2: A link from cell adhesion to nuclear chromatin remodeling? Cell Adh Migr. 2010;4:77–80. doi: 10.4161/cam.4.1.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang J, Clatterbuck RE, Rigamonti D, Dietz HC. Cloning of the murine Krit1 cDNA reveals novel mammalian 5′ coding exons. Genomics. 2000;70:392–5. doi: 10.1006/geno.2000.6410. [DOI] [PubMed] [Google Scholar]
- 137.Zhang J, Rigamonti D, Dietz HC, Clatterbuck RE. Interaction between krit1 and malcavernin: implications for the pathogenesis of cerebral cavernous malformations. Neurosurgery. 2007;60:353–9, discussion 359. doi: 10.1227/01.NEU.0000249268.11074.83. [DOI] [PubMed] [Google Scholar]
- 138.Francalanci F, Avolio M, De Luca E, Longo D, Menchise V, Guazzi P, Sgrò F, Marino M, Goitre L, Balzac F, et al. Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp Cell Res. 2009;315:285–303. doi: 10.1016/j.yexcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 139.Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–68. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 140.Goitre L, Balzac F, Degani S, Degan P, Marchi S, Pinton P, Retta SF. KRIT1 regulates the homeostasis of intracellular reactive oxygen species. PLoS One. 2010;5:e11786. doi: 10.1371/journal.pone.0011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JMA, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–54. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fu W, Ma Q, Chen L, Li P, Zhang M, Ramamoorthy S, Nawaz Z, Shimojima T, Wang H, Yang Y, et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J Biol Chem. 2009;284:13987–4000. doi: 10.1074/jbc.M901758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, Yoshida-Araki K, Hisatsune H, Nishikawa S, Nakayama K, Nakayama K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J Biol Chem. 2004;279:34741–9. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 144.Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–80. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Daly C, Wong V, Burova E, Wei Y, Zabski S, Griffiths J, Lai KM, Lin HC, Ioffe E, Yancopoulos GD, et al. Angiopoietin-1 modulates endothelial cell function and gene expression via the transcription factor FKHR (FOXO1) Genes Dev. 2004;18:1060–71. doi: 10.1101/gad.1189704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sato TN, Tozawa Y, Deutsch U, Wolburg-Buchholz K, Fujiwara Y, Gendron-Maguire M, Gridley T, Wolburg H, Risau W, Qin Y. Distinct roles of the receptor tyrosine kinases Tie-1 and Tie-2 in blood vessel formation. Nature. 1995;376:70–4. doi: 10.1038/376070a0. [DOI] [PubMed] [Google Scholar]
- 148.Davis S, Aldrich TH, Jones PF, Acheson A, Compton DL, Jain V, Ryan TE, Bruno J, Radziejewski C, Maisonpierre PC, et al. Isolation of angiopoietin-1, a ligand for the TIE2 receptor, by secretion-trap expression cloning. Cell. 1996;87:1161–9. doi: 10.1016/S0092-8674(00)81812-7. [DOI] [PubMed] [Google Scholar]
- 149.Suri C, Jones PF, Patan S, Bartunkova S, Maisonpierre PC, Davis S, Sato TN, Yancopoulos GD. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–80. doi: 10.1016/S0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 150.Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, Sweeney SM, Chen M, Guo L, Lu MM, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15:169–76. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]