Abstract
Bacterial two-component systems (TCSs) are of vital importance in the translation of rapidly changing environmental conditions into appropriate cellular regulatory responses enabling adaptation, growth, and survival. The diverse range of environmental signals that TCSs can process, coupled with discrete modular domains within TCS proteins, offers considerable potential for the rational design of bio-sensor and/or bio-reporter strains. In this study we functionally characterize the multi-domain StyS sensor kinase associated with sensing of the aromatic pollutant styrene by Pseudomonas putida CA-3. Deletion analysis of discrete domains was performed and the ability of the truncated StyS sensor proteins to activate a cognate reporter system in an E. coli host assessed. The essential histidine kinase and PAS input domains were identified for StyS dependent activation of the reporter system. However, co-expression of an ABC-transporter protein StyE, previously linked to styrene transport in P. putida CA-3, enabled activation of the reporter system with a StyS construct containing a non-essential PAS input domain, suggesting a novel role for intracellular detection and/or activation. Site directed mutagenesis and amino acid deletions were employed to further characterize the PAS sensing domains of both input regions. The potential implications of these findings in the use of multi-domain sensor kinases in rational design strategies and the potential link between transport and intracellular sensing are discussed.
Keywords: styrene, PAS domains, histidine kinase, two-component, Pseudomonas putida, ABC-transporter
Introduction
In Pseudomonas species characterized to date, degradation of the alkenylbenzene styrene involves two distinct catabolic operons: the sty operon, encoding the enzymes necessary for an initial, stepwise conversion of styrene to phenylacetic acid (PAA), and the PACoA catabolic operon involved in the metabolism of PAA to krebs cycle intermediates.2,21,29,30 The sty operon is positively regulated at the transcriptional level by a TCS, composed of a hybrid histidine kinase sensor, StyS, and a DNA-binding transcriptional activator, StyR.20,41 The StySR system is one of only five characterized TCS systems currently known to be involved in the degradation of aromatic compounds.11,39 The others include the TodST and TutBC systems involved in toluene degradation in P. putida F1 and Thauera sp. strain T1, respectively;9,19 the BpdST system involved in degradation of biphenyls in Rhodococcus sp. strain M5;18 and the TdiSR system regulating benzylsuccinate synthase which is involved in the anaerobic catabolism of toluene in Azoarcus sp. strain T.1 Members of these TCSs, and others, have been incorporated into the rational design of biosensors by integrating the sensing apparatus with appropriate reporter genes (e.g., lux, gfp) for in situ monitoring of aromatic pollutant presence in both aquatic and soil environments.33 The application of such biosensors is not without challenges, however, as they are typically characterized under laboratory conditions, which are often not reflective of real-time settings. Furthermore, issues of sensitivity persist, as most cannot detect inducers below µM range40, and signal strength and/or detection can be difficult to achieve in online systems.31,33 In recent years the wealth of bioinformatics-based data emerging from genome studies has enabled researchers to take a fresh look at the evolution of TCSs, revealing modular protein designs displaying a limited range of structural arrangements.7,17 Such developments have enabled researchers to refocus recombinant approaches to exploit these features in design strategies for the optimization of TCS applications.8
In tandem with the contribution of bioinformatic inputs, functional characterisations of TCSs also have an important role to play, particularly with TCS systems involving multiple or duplicate domains, such as the TodS system, which has been well characterized in recent years.5,34 Amino acid sequence alignment of StyS from P. putida CA-3 with the TodS sensor kinase revealed a similar multidomain structure with five distinct, conserved domains within the protein: two histidine kinase (HK 1 and HK2) domains separated by a receiver domain, together with two input domains19,27,36,41 (Fig. 1A). HK1/2 both contain the characteristic amino acid blocks H, N, G1, F, and G2; while the receiver domain of StyS contains the D, D, S, K amino acid residues typical of bacterial response regulators of the RA2 receiver subfamily.13 In addition both sensory input regions contain blocks of conserved amino acids typical of PAS domains, previously linked with sensing oxygen, light, redox potential, and in the binding of small hydrophobic aromatic compounds.3,37 An unusual feature of styrene degradation in P. putida CA-3 is the involvement of an ABC-transporter protein, StyE, in actively transporting this relatively small, non-polar molecule into the cytosol.23 Overexpression of the styE gene in styrene grown cultures results in an 8-fold increase in transcriptional activation of the operon, presumably through some altered interaction with the TCS regulatory apparatus. No such association has however been reported for the TodS system for toluene degradation. As a result, we sought to functionally characterize the key domains within the StyS sensor and assess whether StyE overexpression revealed domain specific impacts. We report that HK1 does not appear to be essential for StyS functionality and that input 1 and input 2 display discrete roles in extracellular and intracellular styrene sensing, respectively. In addition, we report increased reporter system activation mediated via input 2 following StyE co-expression.
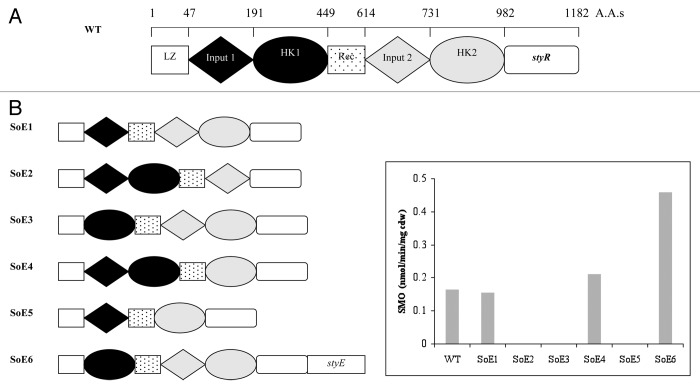
Figure 1. (A) Domain organization of the StyS sensor protein in Pseudomonas putida CA-3; LZ, Rec, and HK represent Leucine zipper, Receiver, and Histidine Kinase domains, respectively. styR indicates the position of the co-expressed, response regulator gene. (B) Diagrammatic representations of each SoE construct illustrating the deleted functional domain(s). A bar chart presents the styrene monoxygenase (SMO) activities from the indole to indigo reporter assay system harboring SoE constructs (1–6), respectively.
Results
Identification of functional domains in StyS
Figure 1A and B present graphical summaries of the different StyS deletion mutants, each deficient in a single domain, co-expressed with the transcriptional activator StyR. Each construct was assayed for indole to indigo transformation ability, indicative of the induced transcription of the pPstyABC reporter construct in the presence of styrene (Fig. 1B and 2). Deletion of the input 1 region (SOE3 Δ48–190) or the histidine kinase 2 domain (SOE2 Δ732–982) resulted in a complete loss of StyR dependent styABC transcription, (Fig. 1B). These findings are in keeping with previous work on the autophosphorylation capacities of truncated forms of the TodS sensor kinase.5 In contrast, constructs harboring a deletion of input 2 (SOE4 Δ568–730), or histidine kinase 1 (SOE1 Δ192–399), respectively, achieved reporter system activities comparable with the StySR wild type construct, suggesting the non-essential nature of these domains in response to extracellular styrene presence (Fig. 1B). Interestingly, construct SOE5 (Δ192–399 and Δ568–730), which lacked the apparently non-essential input 2 and HK1 domains, failed to induce transcription of the styABC genes in our system (Fig. 1B), suggesting that the overall structural and/or spatial integrity of the StyS protein domains may be critical.
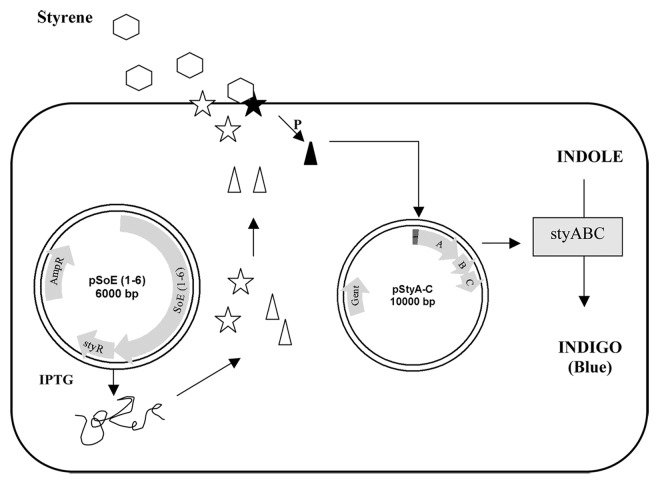
Figure 2. A diagrammatic representation of the indole to indigo assay reporter system. StyS, (star), detects styrene at membrane and activates StyR (triangle) by phosphorelay, which initiates transcription of styABC reporter genes.
Analysis of hydropathicity plots for StyS (Kyte-Doolittle model) revealed differing potential membrane associations for input domains 1 and 2, respectively. We thus investigated the possibility that input 2 provided an intracellular or inner membrane associated role in styrene detection. The SOE 6 construct (Δ48–190), harboring input 2 as the sole sensory domain, was therefore co-expressed with StyE, an ABC-transporter whose overexpression been previously shown to enhance transcriptional activation of the sty operon genes in P. putida CA-3.23 It was observed that this construct, in the presence of styrene, resulted in a 2.8-fold increase in indole to indigo activity from the reporter system when compared with wild type StySR construct (Fig. 1B). It would appear therefore that input 2 lacks an extracellular sensing role, but may interact strongly with styrene upon entry of the aromatic compound into the cell.
Identification of key amino acids in the input domains of StyS
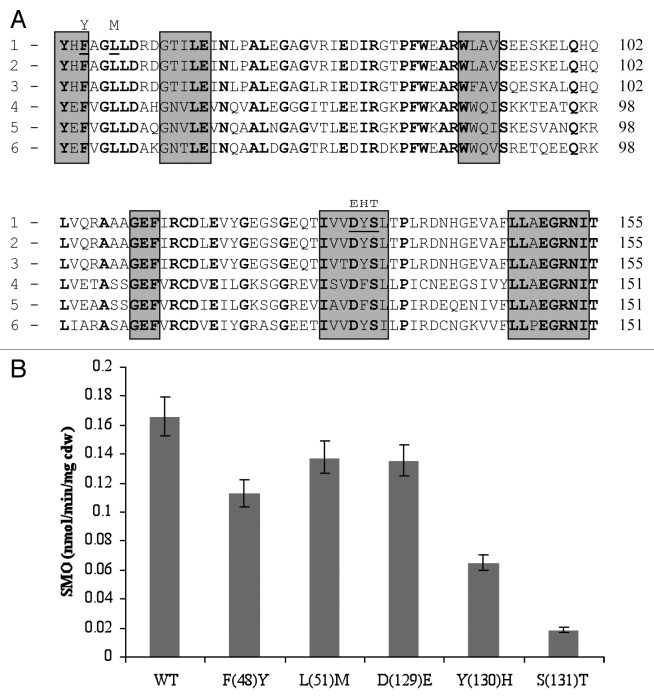
As both input domains appeared to have important, albeit differing, roles in styrene sensing we sought to examine key amino acids within these regions important in mediating styrene detection. PAS sensing domains have previously been shown to function by a variety of mechanisms, including the binding of specific compounds within a hydrophobic core, e.g., small aromatic compounds or ligands,3,42 and also numerous co-factors such as FAD, FMN, and Heme groups.10,37 Sequence alignments of the input 1 region of StyS with known HK associated PAS domain sequences from several Pseudomonas species identified a number of highly conserved amino acids (Fig. 3A). The following amino acid substitutions were introduced via site-directed mutagenesis—F(48)Y, L(51)M, D(129)E, Y(130)H, and S(131)T—and the impacts of the Input 1 mutations was assessed via the indole to indigo reporter system used earlier (Fig. 3B). From the data obtained it would appear that both Y130 and S131 play important roles in styrene detection by input 1, as substitutions of these amino acids resulted in 62% and 87% mean reductions in reporter system activity, respectively, when compared with wild type StyS. It was noted that these residues lie within a potential hydrophobic pocket in the predicted 3-D structure of the input 1 region of StyS generated using 3D-PSSM (www.sbg.bio.ic.ac.uk) (data not shown). It should be noted that many of the conserved amino acids present within the hydrophobic core of the input 1 PAS domain, e.g., glycine, proline and blocks of aromatic amino acids were not deemed suitable for mutational analysis due to the specific structural role known to be associated with them.4 Substitutions of F48, L51, and D129, had less significant negative impacts with mean reductions of approximately 29%, 14%, and 16%, respectively (Fig. 3B).
Figure 3. (A) Comparsion of the predicted amino acid sequence of the StyS Input 1 domain of (1) Pseudomonas putida CA-3 with PAS domains of: (2) Pseudomonas species Y2 (CAA03998), (3) P. fluorescens (AAC06271), (4) P. putida F1 (CAB43735), (5) P. mendocina (AAL13332), and (6) T. aromatica (AAD12184). Conserved amino acids are in bold, shaded boxes indicate hydrophobic regions. Amino acids targeted for substitution in StyS from P. putida CA-3 are underlined with the proposed substitution indicated by the letter above. (B) SMO: styrene monooxygenase reporter system activity (nmol min−1 mg cell dry weight−1) of StyS wild type WT and StyS Input 1 mutants; F(48)Y, L(51)M, D(129)E, Y(130)H, and S(131)T, respectively.
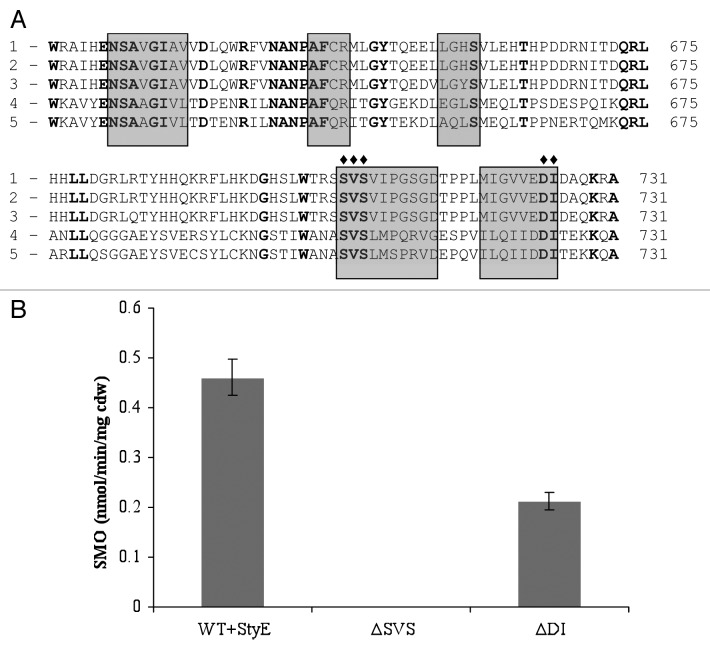
A similar strategy was applied to the Input 2 region of StyS and, following a sequence alignment of the amino acid sequence with input 2 domains of other TCS systems, a number of conserved amino acids regions, (with a differing architecture to those in input 1), were identified, namely S709, V710, and S711, together with D729 and I730 (Fig. 4A). Amino acid deletions and reporter system assays revealed that the removal of S709, V710, or S711, resulted in the complete inability of the StyS mutant construct to activate transcription of the reporter system in the presence of styrene (Fig. 4B). Furthermore, SMO activity decreased by >50% when either D729 or L730 were deleted from the input 2 region of StyS (Fig. 4B). The latter two residues also appeared to be located within a potential hydrophobic pocket identified in a hypothetical 3-D structure of the input 2 region of StyS generated using 3D-PSSM (www.sbg.bio.ic.ac.uk), (data not shown). The data suggest that both these regions (709–711 and 729–730) have key roles within the input 2 domain of StyS, in styrene induced activation of the sty ABCD operon in response to styrene entry into the cell. However, further work is required to determine the nature of the interactions of these regions with styrene and potential conformational shifts that may impact on signal transduction efficacy.
Figure 4. (A) Comparsion of the predicted amino acid sequence of the StyS Input 2 domain of (1) Pseudomonas putida CA-3 with PAS domains of: (2) Pseudomonas species Y2 (CAA03998), (3) P. fluorescens (AAC06271), (4) P. putida F1 (CAB43735), and (5) P. mendocina (AAL13332). Conserved amino acids are in bold, shaded boxes indicate hydrophobic regions. Amino acids targeted for deletion substitution are indicated by the ♦. (B) SMO: styrene monooxygenase reporter activity (nmol min−1 mg cell dry weight−1) with StyS wild type WT, and StyS Input 2 mutants ∆SVS and ∆DI, respectively.
Discussion
Previous characterization of TodS domains5 reveal certain overlapping similarities with the workings of StyS, e.g., the critical nature of the C-terminal HK2. The localization of kinase activity to the carboxy terminus has been well documented among other sensor kinase proteins also, including EnvZ,12 VirA,15 and FixL.22 The presence of a potentially redundant, duplicate HK domain, i.e., HK1 in StyS, has indeed been reported as a relatively common feature of bacterial TCSs, possibly arising from domain shuffling during their evolution.17,43 Silva-Jimenez and colleagues have recently tried to exploit this feature of TodS by designing a simplified, prototype TodS sensor, comprising only one HK2 domain.34 While toluene sensing was not improved, the half-life of the autophosphorylated sensor protein in response to other inducers was significantly increased. Such findings, while not readily amenable to bioinformatic prediction, may well contribute in the future to the design of increasingly robust sensor systems.
With respect to the PAS domain structures and key residues investigated in StyS, the substitutions of Y(130)H and S(131)T in input 1 lead to 62% and 87% reductions respectively, in styrene monooxygenase reporter activity (Fig. 3B). It is not possible as yet to predict the precise mechanisms associated with these observations, however one possibility involves the reported function of Ser and Tyr residues in anchoring cell membrane lipid components by covalent attachment.4 A StyS input 1 domain hydropathicity plot identified hydrophobic regions with a high probability of membrane association; thus, substitution of these amino acids may affect the ability of this protein region to position itself correctly in the membrane, reducing its availability to interact with styrene and initiate signal transduction. In relation to the PAS domain of input 2, serine residues are quite common in protein functional centers due to their reactive hydroxyl group,6,35 while valine residues have previously been reported to be involved in the binding and recognition of hydrophobic ligands.4,24 Thus the essential function associated with the S709, V710, and S711 triad may well involve styrene binding within the predicted hydrophobic niche.
A significant, novel finding of this study is the potential link between ligand active transport and TCS sensor efficacy in the activation of StyS. In a relatively recent review,38 Testch and Jung presented a small but growing body of evidence that bacterial transport systems, secondary and primary, can function as veritable co-sensors of histidine kinases. Indeed, bioinformatic analyses of sequenced members of the Firmicutes revealed 70 instances of TCS genes being co-located with related ABC-transporter genes, suggesting co-evolution and co-operative physiological roles. To our knowledge, this study is the first to report a functional link of this type in relation to aromatic hydrocarbon detection, although the specificities of interaction require further characterization. This may have implications for some of the challenges facing bioreporter strain design such as detection sensitivity and reporter signal strength. Traditional biosensor design has in the past largely focused on the integration of a reporter gene within the regulatory network, or “swapping” of sensor kinase domains to expand signal ranges.31,33 However, future iterations of these strategies could benefit considerably by investigating potential transport protein associations and the impacts of their co-expression in the optimization of such systems.
Materials and Methods
Bacterial strains, plasmids and growth conditions
E. coli XL-1 Blue (Promega) cells were used for routine transformation and maintenance of plasmids. Cells were grown at 37 °C in Luria-Bertani medium and aerated via continuous agitation on an orbital shaker at 120 rpm. E. coli cells containing both plasmids were grown in M9 medium with glucose (1% w/v), at the reduced temperature of 30 °C, to assist in plasmid maintenance. Styrene was supplied in the gaseous phase, as previously described.25 Cell growth was monitored by measuring optical density at 540 nm with a Beckman DU640 spectrophotometer. Cells were harvested at mid-log phase (OD540 0.6) for whole cell enzyme assays. Antibiotics, when required, were added at the following concentrations: ampicillin (100 μg/ml) and chloroamphenicol (25 μg/ml). TOPO pCR 2.1® (Invitrogen) was used for cloning of PCR products (according to the manufacturer’s instructions).
PCR amplifications and DNA cloning
Genomic DNA was isolated from P. putida CA-3 as previously described28 and used as a template for PCR. Gene constructs of styS harboring domain deletions were generated using a modification of the Splicing Overlap Extension (SOEing) PCR technique described by Horton and colleagues13,14 using the primers detailed in Table 1. The SOEing reaction involved the following steps: (1) sequences on either side of the area to be spliced out were amplified in two separate PCR reactions using the primer pairs A + B, and C + D; to generate fragments AB and CD, (note: primer C was designed so that the 5′ end contained a region complementary to the 3′ end of primer B); (2) the purified AB and CD fragments were both added to a single standard PCR reaction with primers A and D; (3) after an initial denaturation step, the reaction was held at 72 °C for 3 min to allow annealing of the complementary fragment termini and subsequent extension to generate full-length template for successive rounds of amplification with primers A and D; and (4) the mis-primed incorporation of the restriction enzyme sites XbaI and HindIII facilitated the subsequent cloning of each construct. All amplifications were performed in a MJ thermal cycler using 1 unit of VentR DNA Polymerase, 1X thermoPol buffer, 1.5 mM MgSO4 (New England Biolabs), 100 ng of each primer, and 1.25 mM of each dNTP and equimolar concentrations of AB and CD fragments as the template. The final volume of each reaction was adjusted to 50 μl using sterile distilled water. The PCR cycling conditions were as standard: 94 °C × 4 min, followed by 30 cycles of: 94 °C × 60 s, annealing × 60 s (see temperatures below), 72 °C × 60 s. In the amplification of AD full-length templates, the extension time was increased to 1.5 min and also the number of cycles was increased to 34. Annealing temperatures used for each primer pair are presented in Table 1. PCR products were separated by electrophoresis on a 1% agarose gel, purified (Qiagen Gel Extraction), cloned into TOPO pCR 2.1® vector, and transformed into E. coli Top10 cells according to the manufacturer’s instructions (Invitrogen). Transformants were provisionally identified via blue-white screening on LB agar containing X-gal/IPTG. Positive clones were subsequently tested with the primer pair StyRF and StyRR using the PCR conditions outlined above. Sequencing of relevant PCR amplicons was performed via Big DyeTM Terminator cycling and analyzed on 5.75% and 4.75% Long RangerTM gels for ABI 377 (Lark Technologies Inc). Sequence data was assembled and processed using DNASTAR software to confirm that the relevant domain had been deleted, and the remaining gene fragment was translationally in frame.
Table 1. Complementary regions between the respective B and C primers for splicing overlap extension are in italics.
|
Name |
Primer sequence 5′ – 3′ |
Tm (°C) |
Primer pair Annealing (°C) |
|---|---|---|---|
| StyS (A) | 1TCTAGAATGC CTGGAGCCTG GAACGTC | 75.6 | SA/SD: 60 |
| StyS (D) | 2AAGCTTTCAG TGGCCCCTTT CAAACGATTC ATA | 76.9 | “ |
| StyS (D2) | 3GAATTCTCAG TGGCCCCTTT CAAACGATTC ATA | 76.1 | SA/SD2: 60 |
| StyS (B1) | GAACCGATTC TTTTGCTCAT C | 62.4 | SA/SB1:55 |
| StyS (C1) | CGGTTCTTGG TGCGGAAGGG CTCG | 79.7 | SD/SC1: 65 |
| StyS (B2) | CAGCTCACCC ATCGCAGT | 65.4 | SA/SB2: 55 |
| StyS (C2) | GAGCTGATGA CCACAAAGCC ACA | 73.2 | SD/SC2: 65 |
| StyS (B3) | AAGCCCGGCG AAGTGATG | 65.6 | SA/SB3:55 |
| StyS (C3) | GGGCTTTCCA ACCTGAGTCA TCAG | 68.9 | SD/SC3:61 |
| StyS (B4) | CATCACCAGA TTACGGACCC T | 68.4 | SA/SB4:55 |
| StyS (C4) | GTGATGTCGA TCACCCATGA ACTG | 69.6 | SD/SC4:60 |
| StyEF(EcoRI) | 3GAATTCATGT TCTGCGCCTC GAGGGCT | 70.1 | EF/ER:51 |
| StyER(HindIII) | 2AAGCTTTCAG AAATTATGGG TATA | 58.7 | “ |
| pStyA F | 4ACTAGTTGCTC CTGGCCCTGT | 69.9 | pAF/CR:61 |
| StyC R | 2AAGCTTCGCA GCAGCGTGCG G | 79.4 | “ |
| StyAF | GATTTCCAGG TCGCTACC | 59.6 | AF/AR: 52 |
| StyAR | CATCTTGGCC TCTTCCTC | 59.9 | “ |
| StyRF | ATGACCACAA AGCCCACAGT A | 62.8 | RF/RR:51 |
| StyRR | CGCCCCTTTC AAACGATTCA T | 62.1 | “ |
Restriction enzyme sites are underlined; 1.XbaI 2.HindIII 3.EcoRI, and 4.SpeI.
Nucleic acid manipulations
All oligonucleotide primers used in this study were synthesized by Sigma-Genosys Ltd. Nucleic acid sequence determination was performed by Lark Technologies Inc, as above. Sequence data was assembled and processed using DNASTAR software, and amino acid database comparative analyses were performed using the BLAST algorithm of the National Centre for Biotechnological Information (http://www.ncbi.nlm.nih.gov). Hydropathicity plots of the StyS protein were generated using GeneJockey.
Construction of expression and reporter plasmids
E. coli transformants harboring styS deletion construct clones were inoculated into 10 ml LB-amp100 broths and incubated overnight at 37 °C to facilitate plasmid mini-prep isolations (Qiagen mini-kit). StyS deletion constructs were excised from TOPO pCR2.1® using XbaI and HindIII, the respective fragments were gel purified and ligated into pGEM-7Z (Promega) overnight at 16 °C using T4 DNA ligase (Promega) to produce constructs pSOE 1 to 5 (Fig. 1B and 2). The styABC styrene catabolic genes were co-amplified with the styA promoter region using primers PStyAF and StyCR (Table 1), which allowed mis-priming incorporation of SpeI and HindIII restriction sites at the 5′ and 3′ ends of the fragment, respectively. PCR reaction constituents and cycling conditions were as before; the annealing temperature was 61 °C. This reporter construct was subsequently cloned as a SpeI/HindIII fragment into the broad host range vector pBBR-1-MCS16 to generate pPstyABC (Fig. 2).
Construction of SOE5 and SOE6 constructs
The SOE1 construct was used as a PCR template in the generation of SOE5. Primers StyS A and B4 amplified the AB5 fragment, while primers StyS C4 and D produced the CD5 amplicon in a separate reaction (Table 1). The AB5 and CD5 fragments were purified, denatured, annealed and the full-length SOE5 construct amplified using the external 5′ and 3′ oligonucleotide pair, StyS A and D. Sequencing of the construct confirmed that the relevant domain had been deleted and that the SOE5 construct was translationally in frame.
Construct SOE3 was used as PCR template to generate SOE6, composed of styS harboring a deletion of Input 1, co-transcribed with styE; encoding the putative styrene transport protein gene, StyE23 (Fig. 1B). The full-length SOE3 construct was reamplified using primers StyS A and D2 (Table 1), in order to replace the 3′ HindIII site with an EcoRI restriction site, allowing the generation of an XbaI, EcoRI fragment upon restriction. In a separate reaction, styE was amplified from CA-3 genomic DNA using the primer pairs StyEF(EcoRI)/StyER(HindIII) (Table 1). The SOE3 and styE products were then visualized by 1% gel electrophoresis, purified (Qiagen Gel Extraction) and cloned into TOPO pCR 2.1® vector (Invitrogen). Positive transformants were identified as previously described, and SOE3 and styE were excised from the TOPO pCR 2.1® vector with XbaI, EcoRI and EcoRI, and HindIII, respectively (Promega). The fragments were purified (Qiagen Gel Extraction) and ligated to each other via the EcoRI restriction site, overnight at 16 °C using the T4 DNA ligase (Promega) to produce the StyS construct SOE6. SOE6 was gel purified and ligated into pGEM-7Z (Promega) to generate the construct pSOE6 (Fig. 1B). Sequencing across the SOE3, styE junction confirmed that both genes had been successfully spliced together and were translationally in frame.
Heterologous expression of constructs in E. coli XL-1 Blue
Each of the six pSOE constructs were transformed into chemically competent E. coli XL-1 Blue cells.32 The presence of the expression plasmids was in each case verified by PCR using the primer pairs StyRF and StyRR (as previously described above) and positive transformants were then made chemically competent, and transformed with the reporter plasmid pPstyA-C (Fig. 2). The presence of the plasmid was verified by PCR using the primers pairs StyAF and StyAR. Expression of each modified StyS from its respective construct (pSOE1-6) was induced by the addition of 1.0 mM IPTG.
Enzyme assays
The styrene monooxygenase assay (SMO) activity was monitored using the indole to indigo assay as previously described.26,27
Site directed mutagenesis in the characterization of input domains 1 and 2
Site directed mutagenesis of highly conserved, hydrophobic region amino acids within the input 1 PAS domain of StyS (Fig. 3A), was also achieved via a SOEing PCR based strategy. The mutagenic B and C primers listed in Table 2 provided the site-directed function, when combined with primers A and D (Table 1). The SOE4 construct, lacking input 2 (Fig. 1B), acted as template. Annealing temperatures for each primer pair are also provided in Table 2. The constructs generated were spliced together using the full-length 5′- 3′ gene primers StyS (A) and StyS (D) as before. The following amino acid substitutions were performed; F48Y, K51M, D129E, Y130H, and S131T (Fig. 3A). Constructs were sequenced to confirm the amino acid substitutions. Finally, the constructs were ligated into the reporter system pSoE expression vector and the effects of the amino acid substitutions assessed via monitoring of styrene monooxygenase activity.
Table 2. Amino acids substitutions and locations within Input 1 are presented in brackets. Bases underlined and in bold represent the incorporated point mutations. Complementary regions between the respective B and C primers for splicing overlap extension are in italics. A and D primers are the same as for Table 1.
| Name | Primer sequence 5′ – 3′ | Tm (°C) | Primer pair* Annealing (°C) |
|---|---|---|---|
| B(F48Y) | GGCGTAGTGA TACATGCG | 60.3 | SA/B(F48Y): 55 |
| C(F48Y) | TACGCCGGGC TTTTGGAC | 66.9 | SD/C(F48Y): 62 |
| B(L51M) | CAACATCCCG GCGAAGTG | 67.8 | SA/B(L51M): 55 |
| C(L51M) | ATGTTGGACC GTGATGGC | 64.6 | SD/C(L51M): 59 |
| B(D129Q) | GTATTCCACA ACATAGGT | 51.8 | SA/B(D129Q): 45 |
| C(D129Q) | GAATACTCGC TCACCCCG | 63.3 | SD/C(D129Q): 57 |
| B(Y130H) | CGAGTGGTCA CAACCGAT | 62.3 | SA/B(Y130H): 55 |
| C(Y130H) | CACTCGCTCA CCCCGTTG | 68.2 | SD/C(Y130H): 62 |
| B(S131T) | GAGCGTGTAG TCCACAAC | 56.5 | SA/B(S131T): 49 |
| C(S131T) | ACGCTCACCC CGTTGCGA | 72.7 | SD/C(S131T): 64 |
Similarly, conserved, hydrophobic amino acids in the PAS domain of input 2 (Fig. 4A), S709, V710, S711, D729, and I 730 were deleted using the primers in Table 3. Primers StyS (A) and StyS (D2) were used to splice the fragments together and the resulting constructs were fused to styE to generate SOE6 as previously described. Construct SOE6 was used as the template from which the input 2 mutants were generated. All amplicons were subjected to internal sequencing reactions to verify the success of the deletion mutagenesis and to confirm that each deletion mutant was translationally in frame.
Table 3. Complementary regions between the respective B and C primers for splicing overlap extension are in italics. A and D primers are the same as for Table 1.
| Name | Primer sequence 5′ – 3′ | Tm (°C) | Primer Pair Annealing (°C) |
|---|---|---|---|
| B(SVS) | TCGAGCGTGT CCATAGGC | 58.8 | SA/B(SVS): 54 |
| C(SVS) | GCTCGATGAT TCCGGGCAGT C | 78.3 | SD/C(SVS):63 |
| B(DI) | TCTTGCTTCG ACAACCCCGA T | 67.2 | SA/B(DI):55 |
| C(DI) | CGAAGACAAA AACGTGCCGA G | 76.2 | SD/C(DI):66 |
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
References
- 1.Achong GR, Rodriguez AM, Spormann AM. Benzylsuccinate synthase of Azoarcus sp. strain T: cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol. 2001;183:6763–70. doi: 10.1128/JB.183.23.6763-6770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso S, Bartolomé-Martín D, del Alamo M, Díaz E, García JL, Perera J. Genetic characterization of the styrene lower catabolic pathway of Pseudomonas sp. strain Y2. Gene. 2003;319:71–83. doi: 10.1016/S0378-1119(03)00794-7. [DOI] [PubMed] [Google Scholar]
- 3.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–61. doi: 10.1016/S0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 4.Betts MJ, Russell RB. 2003. Amino acid properties and consequences of substitutions. In Bioinformatics for Geneticists, pp. 289-316. Edited by M. R. Barnes & I. C. Gray. London: Wiley. [Google Scholar]
- 5.Blanco J, Moore RA, Faehnle CR, Viola RE. Critical catalytic functional groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase. Acta Crystallogr D Biol Crystallogr. 2004;60:1808–15. doi: 10.1107/S0907444904020104. [DOI] [PubMed] [Google Scholar]
- 6.Busch A, Guazzaroni ME, Lacal J, Ramos JL, Krell T. The sensor kinase TodS operates by a multiple step phosphorelay mechanism involving two autokinase domains. J Biol Chem. 2009;284:10353–60. doi: 10.1074/jbc.M900521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–47. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Checa SK, Zurbriggen MD, Soncini FC. Bacterial signaling systems as platforms for rational design of new generations of biosensors. Curr Opin Biotechnol. 2012;23:766–72. doi: 10.1016/j.copbio.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Coschigano PW, Young LY. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol. 1997;63:652–60. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coumailleau P, Poellinger L, Gustafsson JA, Whitelaw ML. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J Biol Chem. 1995;270:25291–300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- 11.Díaz E, Prieto MA. Bacterial promoters triggering biodegradation of aromatic pollutants. Curr Opin Biotechnol. 2000;11:467–75. doi: 10.1016/S0958-1669(00)00126-9. [DOI] [PubMed] [Google Scholar]
- 12.Forst S, Comeau D, Norioka S, Inouye M. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem. 1987;262:16433–8. [PubMed] [Google Scholar]
- 13.Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/S0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 14.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–35. [PubMed] [Google Scholar]
- 15.Huang Y, Morel P, Powell B, Kado CI. VirA, a coregulator of Ti-specified virulence genes, is phosphorylated in vitro. J Bacteriol. 1990;172:1142–4. doi: 10.1128/jb.172.2.1142-1144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–6. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 17.Krell T, Lacal J, Busch A, Silva-Jiménez H, Guazzaroni ME, Ramos JL. Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol. 2010;64:539–59. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 18.Labbé D, Garnon J, Lau PC. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium, Rhodococcus sp. strain M5. J Bacteriol. 1997;179:2772–6. doi: 10.1128/jb.179.8.2772-2776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau PC, Wang Y, Patel A, Labbé D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci U S A. 1997;94:1453–8. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leoni L, Rampioni G, Di Stefano V, Zennaro E. Dual role of response regulator StyR in styrene catabolism regulation. Appl Environ Microbiol. 2005;71:5411–9. doi: 10.1128/AEM.71.9.5411-5419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luengo JM, García JL, Olivera ER. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol. 2001;39:1434–42. doi: 10.1046/j.1365-2958.2001.02344.x. [DOI] [PubMed] [Google Scholar]
- 22.Monson EK, Weinstein M, Ditta GS, Helinski DR. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci U S A. 1992;89:4280–4. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mooney A, O’Leary ND, Dobson ADW. Cloning and functional characterization of the styE gene, involved in styrene transport in Pseudomonas putida CA-3. Appl Environ Microbiol. 2006;72:1302–9. doi: 10.1128/AEM.72.2.1302-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak WR, Wang PF, McLeish MJ, Kenyon GL, Babbitt PC. Isoleucine 69 and valine 325 form a specificity pocket in human muscle creatine kinase. Biochemistry. 2004;43:13766–74. doi: 10.1021/bi049060y. [DOI] [PubMed] [Google Scholar]
- 25.O’Connor K, Duetz W, Wind B, Dobson ADW. The effect of nutrient limitation on styrene metabolism in Pseudomonas putida CA-3. Appl Environ Microbiol. 1996;62:3594–9. doi: 10.1128/aem.62.10.3594-3599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connor KE, Dobson AD, Hartmans S. Indigo formation by microorganisms expressing styrene monooxygenase activity. Appl Environ Microbiol. 1997;63:4287–91. doi: 10.1128/aem.63.11.4287-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Leary ND, O’Connor KE, Dobson ADW. Biochemistry, genetics and physiology of microbial styrene degradation. FEMS Microbiol Rev. 2002;26:403–17. doi: 10.1016/S0168-6445(02)00126-2. [DOI] [PubMed] [Google Scholar]
- 28.O’Leary ND, O’Connor KE, Duetz W, Dobson ADW. Transcriptional regulation of styrene degradation in Pseudomonas putida CA-3. Microbiology. 2001;147:973–9. doi: 10.1099/00221287-147-4-973. [DOI] [PubMed] [Google Scholar]
- 29.O’Leary ND, O’Connor KE, Ward P, Goff M, Dobson ADW. Genetic characterization of accumulation of polyhydroxyalkanoate from styrene in Pseudomonas putida CA-3. Appl Environ Microbiol. 2005;71:4380–7. doi: 10.1128/AEM.71.8.4380-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olivera ER, Miñambres B, García B, Muñiz C, Moreno MA, Ferrández A, Díaz E, García JL, Luengo JM. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci U S A. 1998;95:6419–24. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park M, Tsai SL, Chen W. Microbial biosensors: engineered microorganisms as the sensing machinery. Sensors (Basel) 2013;13:5777–95. doi: 10.3390/s130505777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd edn. New York: Cols Spring Laboratory Press. [Google Scholar]
- 33.Shin HJ. Genetically engineered microbial biosensors for in situ monitoring of environmental pollution. Appl Microbiol Biotechnol. 2011;89:867–77. doi: 10.1007/s00253-010-2990-8. [DOI] [PubMed] [Google Scholar]
- 34.Silva-Jiménez H, Ramos JL, Krell T. Construction of a prototype two-component system from the phosphorelay system TodS/TodT. Protein Eng Des Sel. 2012;25:159–69. doi: 10.1093/protein/gzs001. [DOI] [PubMed] [Google Scholar]
- 35.Stephens AW, Siddiqui A, Hirs CH. Site-directed mutagenesis of the reactive center (serine 394) of antithrombin III. J Biol Chem. 1988;263:15849–52. [PubMed] [Google Scholar]
- 36.Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–90. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tetsch L, Jung K. The regulatory interplay between membrane-integrated sensors and transport proteins in bacteria. Mol Microbiol. 2009;73:982–91. doi: 10.1111/j.1365-2958.2009.06847.x. [DOI] [PubMed] [Google Scholar]
- 39.Tropel D, van der Meer JR. Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol Mol Biol Rev. 2004;68:474–500. doi: 10.1128/MMBR.68.3.474-500.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Meer JR, Tropel D, Jaspers M. Illuminating the detection chain of bacterial bioreporters. Environ Microbiol. 2004;6:1005–20. doi: 10.1111/j.1462-2920.2004.00655.x. [DOI] [PubMed] [Google Scholar]
- 41.Velasco A, Alonso S, García JL, Perera J, Díaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–71. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu YZ, Hogenesch JB, Bradfield CA. The PAS superfamily: sensors of environmental and developmental signals. Annu Rev Pharmacol Toxicol. 2000;40:519–61. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, Shi L. Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology. 2005;151:2159–73. doi: 10.1099/mic.0.27987-0. [DOI] [PubMed] [Google Scholar]