Abstract
Renewable lignocellulosic plant biomass is a promising feedstock from which to produce biofuels, chemicals, and materials. One approach to cost-effectively exploit this resource is to use consolidating bioprocessing (CBP) microbes that directly convert lignocellulose into valuable end products. Because many promising CBP-enabling microbes are non-cellulolytic, recent work has sought to engineer them to display multi-cellulase containing minicellulosomes that hydrolyze biomass more efficiently than isolated enzymes. In this review, we discuss progress in engineering the surfaces of the model microorganisms: Bacillus subtilis, Escherichia coli, and Saccharomyces cerevisiae. We compare the distinct approaches used to display cellulases and minicellulosomes, as well as their surface enzyme densities and cellulolytic activities. Thus far, minicellulosomes have only been grafted onto the surfaces of B. subtilis and S. cerevisiae, suggesting that the absence of an outer membrane in fungi and Gram-positive bacteria may make their surfaces better suited for displaying the elaborate multi-enzyme complexes needed to efficiently degrade lignocellulose.
Keywords: lignocellulose, consolidated bioprocessing, cellulase, minicellulosome, cell surface display, biofuels, Bacillus subtilis, Saccharomyces cerevisiae, Escherichia coli
Introduction
Dwindling supplies of petroleum and the need to reduce net carbon emissions have driven the search for innovative and cost-effective methods to produce biofuels, chemicals, and materials from lignocellulosic biomass.1 In the United States alone, it is estimated that over 1 billion tons of non-food lignocellulosic biomass can be produced annually on a sustainable basis at costs of only $40–50 per ton.2,3 However, a major obstacle limiting the use of lignocellulose as feedstock is its recalcitrance to degradation.2,4 While a number of technologies are being explored in industry to degrade lignocellulose, enzyme-based methods predominate, and are currently used to produce cellulosic ethanol (Fig. 1A).4 In this hydrolytic method, plant biomass is degraded in a two-step process in which it is first pretreated with various chemicals (e.g., acids or ionic liquids) to expose and partially degrade the cellulose and hemicellulose sugar polymers, and then hydrolyzed by adding a consortium of purified cellulase enzymes.4-9 Yeast then ferments the sugars into ethanol. To produce biomass-derived commodities cost-effectively, several groups are developing consolidated bioprocessing (CBP) microbes that combine cellulase production, cellulose hydrolysis, and fermentation into a single process (Fig. 1B). In principle, their use would significantly lower costs, as it would circumvent the need for adding purified cellulase enzyme cocktails and hydrolysate separation procedures.10-13 Avoiding the use of purified enzyme cocktails would be particularly advantageous as it is currently the single largest contributor to overall costs ($0.68–1.47 per gallon of cellulosic ethanol).14 An ideal CBP-enabling microbe would catabolize biomass efficiently and completely, utilize all of the sugars released from the biomass, and generate products at good yields, rates, and titers. It would also require minimal nutrient supplementation, be tolerant to low pH and high temperatures, and possess generally regarded as safe (GRAS) status.2,13 Many promising CBP-enabling microbes possess several of these characteristics, but they are unable to degrade and use biomass as a nutrient. To overcome this limitation, several groups have devised methods to create recombinant cellulolytic microbes that deconstruct plant biomass using surface displayed cellulases.
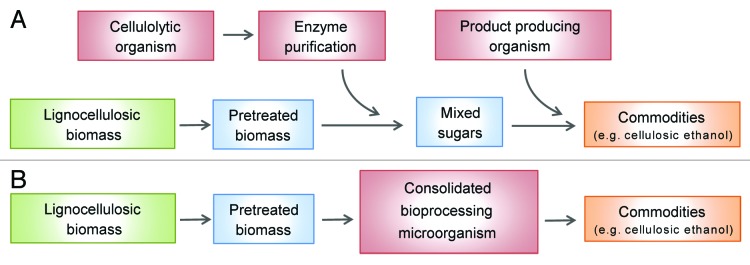
Figure 1. Consolidated bioprocessing of lignocellulosic biomass by cellulase displaying microbes. (A) The current steps involved in the industrial processing of plant biomass into ethanol using cellulase enzymes. Biomass degradation involves thermochemical pretreatment to expose its cellulose polymers, followed by exposure to purified cellulases to degrade cellulose into its component sugars. This is followed by a fermentation step in which yeast convert the sugars into ethanol. In principle, many other biocommodities can be produced from plant biomass using similar methods. (B) Steps in the consolidated bioprocessing (CBP) of biomass. CBP-enabling microbes would produce cellulase enzymes that degrade the cellulose and hemicellulose components of biomass, and then convert the resultant sugars into useful biocommodities. Microbes that naturally or recombinantly display cellulase enzymes are well suited for this process, as they are highly cellulolytic.
In order to degrade the complex structure of plant biomass, naturally cellulolytic microbes produce an array of cellulases that have different substrate specificities. Although a variety of plants are being considered as industrial feedstocks (corn stover, straw, Miscanthus, switchgrass, poplar, sugarcane bagasse, etc.), their cell walls all contain lignocellulose which is comprised of varying amounts of cellulose (25–55%), hemicellulose (8–30%), and lignin (18–35%) (Fig. 2).15 The most abundant component, cellulose, is a homopolymer of β-1,4-linked glucose molecules that are hydrogen bonded with other cellulose polymers to form both amorphous and crystalline regions, the latter of which is particularly recalcitrant to degradation.16 Naturally cellulolytic microorganisms produce three main types of cellulases that function synergistically: endoglucanases, exoglucanases, and β-glucosidases.17 Endoglucanases hydrolyze internal β-1,4-glucosidic bonds in the polymer, creating reducing and non-reducing ends that are further hydrolyzed by exoglucanases.18 Working together, the enzymes create shorter cellodextrins, including the disaccharide cellobiose, which is degraded into its component sugars by β-glucosidases.18 The hemicellulose component of lignocellulose is a heterogeneous polymer of pentose and hexose sugars.19 To liberate these sugars, microbes employ a variety of hemicellulases that have distinct substrate specificities, including exoxylanases, endoxylanases, arabinases, and mannanases, among others.20 Finally, the cellulose and hemicellulose carbohydrate polymers are embedded in lignin, a complex polymer containing a mix of phenolic compounds connected by a variety of linkages.21 Microbial lignin degradation remains poorly understood, but in white-rot fungi, it is mediated by a combination of extracellular peroxidases and laccases.22
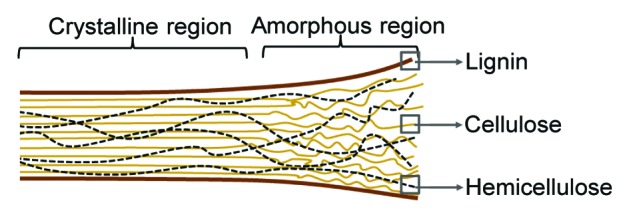
Figure 2. Schematic showing the structure of lignocellulose. Lignocellulose is composed of three components, including cellulose (solid tan lines), hemicellulose (dotted black lines), and lignin (solid brown lines). Cellulose is composed of β-1,4-linked glucose polymers, while hemicellulose is composed of a variety of pentoses. Lignin, which provides structural support for lignocellulose, coats these polymers.
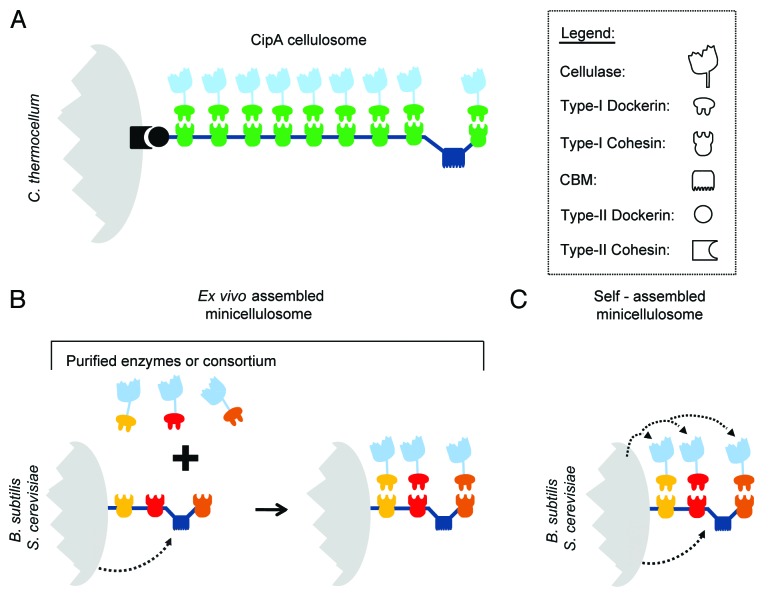
Recent work has engineered microorganisms to display multi-cellulase containing complexes called minicellulosomes (Fig. 3). These complexes are miniaturized versions of the cellulosomes used by naturally cellulolytic anaerobes to degrade plant biomass. Native cellulosomes contain a variety of cellulases that function synergistically to degrade biomass more efficiently than isolated enzymes. The cellulosome from the cellulolytic thermophile Clostridium thermocellum is archetypal (Fig. 3A). It contains a central scaffoldin protein, CipA, which coordinates the binding of nine cellulases.23 Binding is mediated by type-I cohesin modules within CipA that interact with sub-nanomolar affinity with type-I dockerin modules present in the cellulases.24 CipA also contains a carbohydrate-binding module (CBM) that tethers the cellulosome complex to its substrate, as well as a type-II dockerin module located at its C-terminus that anchors the cellulosome complex to cell wall associated proteins.25 Other species of anaerobic bacteria also display cellulosomes, which can adopt more elaborate structures that contain as many as 96 enzymes.24
Figure 3. The prototypical CipA cellulosome and methods used to recombinantly display miniaturized cellulosomes (minicellulosomes). (A) Architecture of the prototypical CipA cellulosome produced by C. thermocellum. It houses 9 cellulases enzymes that are bound to the central scaffoldin protein, CipA.23 Binding is mediated by type-I cohesin modules within CipA that interact with sub-nanomolar affinity with type-I dockerin modules present in the cellulases. CipA also contains a carbohydrate-binding module (CBM) that tethers the cellulosome complex to its substrate, as well as a type-II dockerin module located at its C-terminus that anchors the cellulosome complex to the cohesin module of cell wall associated proteins. (B) Ex vivo approach used to display minicellulosomes on the surfaces of B. subtilis or S. cerevisiae. The microbes secrete and display a scaffoldin protein that is displayed on their surface. Cellulase enzymes containing the appropriate type-1 dockerin module are incubated with the cells to construct the minicellulosome. The enzymes that are added to the cells are either purified enzymes or secreted by other microbes as part of a microbial consortium. Distinct colors are used to indicate species-specific type-1 dockerin and cohesin domains that selectively interact with one another to construct the minicellulosome. (C) Self-assembled approach used to display minicellulosomes on the surfaces of B. subtilis or S. cerevisiae. All of the components of the minicellulosome (scaffoldin and enzymes) are produced by the microbe and spontaneously assemble on the cell surface.
Surface displayed minicellulosomes exhibit enhanced cellulolytic activity. Studies have shown that co-localizing cellulases with different substrate preferences into a cellulosome facilitates enzyme-enzyme synergism; the enzymes in the complex collectively exhibit greater cellulolytic activity than the sum of the activities of the isolated enzymes.26 Synergy occurs because the enzymes have complementary activities, and their spacing and relative abundance is presumably optimal. The presence of both hemicellulolytic and cellulolytic activities in the cellulosome is also advantageous, since by working together these enzymes can remove “physical hindrances” blocking substrate access (e.g., hemicellulolytic enzymes degrade hemicellulose polysaccharides that might otherwise block access to cellulose). The displayed cellulosomes also tether the microbe to the biomass, thereby promoting cellulose-enzyme-microbe (CEM) synergistic interactions that increase the rate of hydrolysis.27 CEM interactions minimize the distance over which the hydrolysis products must diffuse to the cell, facilitating more efficient sugar uptake and preventing the build-up of potential enzyme inhibitors (e.g., cellobiose and glucose).28 It may also facilitate biomass degradation by promoting favorable substrate channeling of long-chain hydrolysis products to proximally bound cells. Thus, CBP-enabling microbes that display minicellulosomes should degrade biomass more rapidly and thoroughly than microbes that only secrete cellulases.
There have been many excellent reviews describing efforts to create cellulolytic and consolidated bioprocessing microorganisms.2,10,13,29 In this review, we focus solely on recent synthetic biology efforts to construct microbes that display cellulases and miniaturized cellulosomes (minicellulosomes). Specifically, we review progress in engineering three microorganisms: Saccharomyces cerevisiae, Escherichia coli, and Bacillus subtilis. Because they are well studied and robust genetic tools are available to manipulate them, they serve as model organisms for eukaryotes, and Gram-negative and Gram-positive eubacteria, respectively. Here we discuss the distinct approaches used to display cellulase complexes on their structurally unique surfaces, and we compare the cellulolytic activities that have been thus far achieved. This exciting work may lead to the direct use of these microbes in consolidated bioprocessing and it promises to facilitate the engineering of other industrially useful microbes.
Engineering Yeast to Display Cellulase Enzymes
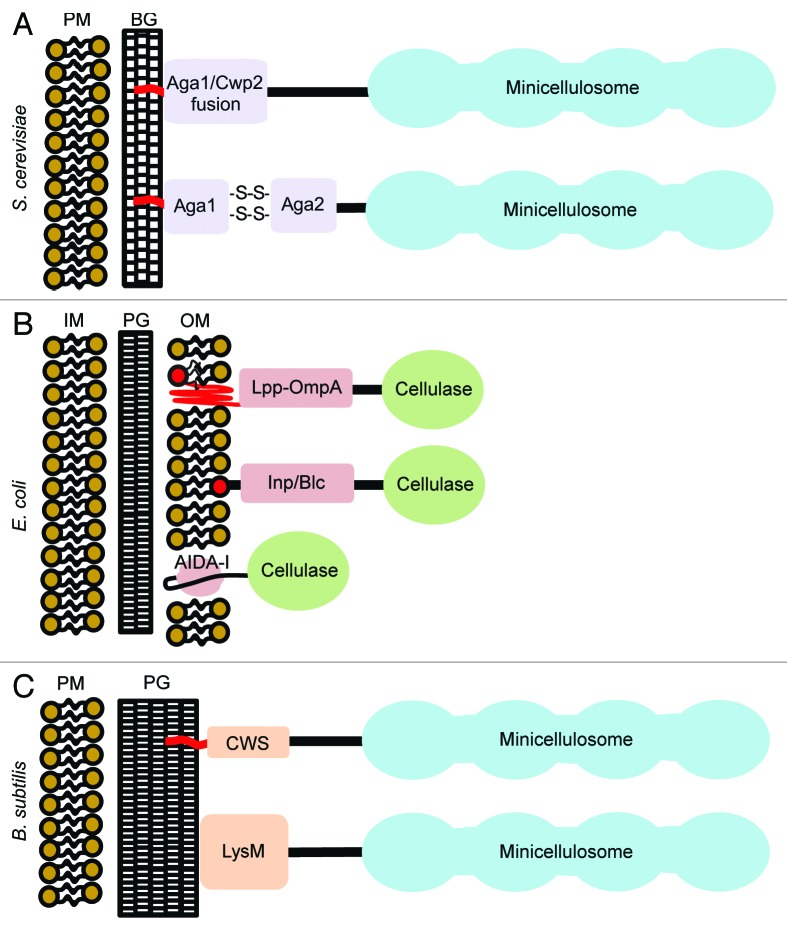
Since S. cerevisiae is already used industrially to produce ethanol from corn, considerable effort is being put forth to create recombinant cellulolytic strains that can degrade and utilize non-edible lignocellulose as a nutrient. While cellulase secreting yeast strains have been constructed, recent work is focused on generating strains that display cellulases and minicellulosomes in order to obtain improved cellulolytic activity. Cellulases are displayed on the cell surface using two related approaches. In the first approach, they are expressed as fusion proteins that contain a glycosylphosphatidylinositol (GPI) anchor signal sequence that is typically derived from the yeast Aga1 or Cwp2 proteins (Fig. 4A). After protein synthesis, the GPI anchor is added to the signal sequence’s ω-site amino acid by the GPI transamidase complex in the endoplasmic reticulum.30-32 GPI attachment initially targets the protein to the lipid bilayer, however, the protein is subsequently processed so as to become covalently linked to outer cell wall β-1,6-glucan, resulting in its display.32-34 In a second related approach, proteins are expressed as fusions to the yeast Aga2 protein, which associates with the endogenous Aga1 protein naturally displayed on the cell surface. Using these display systems, 1 × 104–1 × 105 proteins can be displayed per cell.35
Figure 4. Approaches used to display cellulases and minicellulosomes on different types of microbes. (A) S. cerevisiae: Proteins are displayed on the cell surface by embedding them into the lipid bilayer via a covalently attached C-terminal glycosylphosphatidylinositol (GPI) molecule. Heterologous proteins are displayed by either appending a GPI anchor signal sequence to their C-terminus (typically derived from the Aga1 or Cwp2 proteins) or by expressing them as fusion proteins with Aga2, a yeast protein that associates with the Aga1 protein naturally displayed on the cell surface. ~1 × 104–1 × 105 proteins are attached per cell.35 This approach has been used to display a minicellulosome. (B) E. coli: Heterologous proteins are expressed as fusion proteins with lipoproteins (e.g., Lpp-OmpA, Inp, and Blc) or to the autotransporter AIDA-I. This results in the protein being embedded in the outer membrane (OM). Sixty thousand proteins are attached per cell using the Lpp-OmpA display system.53 At present, only non-complexed cellulases have been displayed using this approach. (C) B. subtilis: Two methods are used to display proteins in this microbe. They are displayed non-covalently by expressing them as fusions with the LysM protein that interacts with cell wall N-acetylmuramic acid and N-acetyl-D-glucosamine.64 Alternatively, proteins containing a C-terminal cell wall sorting signal (CWS) are covalently linked to the peptidoglycan cross-bridge by the sortase transpeptidase enzyme.66 The LysM and sortase approaches are estimated to result in the display of 1.2 × 107and 2.4–3 × 105 proteins per cell, respectively.63,67,68 Both approaches have been used to display minicellulosomes. Key: PM, plasma membrane. BG, β-glucan. IM, inner membrane. PG, peptidoglycan. OM, outer membrane.
Displaying non-complexed enzymes
Initially, the Aga1 fusion system was used to display non-complexed cellulases. In pioneering work by Tanaka and colleagues, non-complexed endoglucanases and β-glucosidases from Aspergillus aculeatus were displayed by expressing each as a fusion protein containing a C-terminal GPI-anchor signal sequence.36 Cells displaying these enzymes could degrade cellodextrins, soluble glucose polymers that are more readily degraded by enzymes than the insoluble cellulose present in lignocellulose.36 During the past decade, Kondo and colleagues created cells with significantly improved cellulolytic activity and explored their ability to ferment cellulose into ethanol. They initially constructed strains that displayed two cellulases via a C-terminal GPI molecule, the T. reesei EGII endoglucanase and the A. aculeatus BGL1 β-glucosidase enzymes.37 After pre-culturing in nutrient-rich media, these strains fermented soluble β-glucan into ethanol. Later, the investigators improved activity by adding a third enzyme and several cellulose binding modules (CBM).38,39 These cells are capable of fermenting amorphous phosphoric acid swollen cellulose (PASC) into ethanol, which is a better lignocellulose mimic than soluble cellodextrins. The authors also demonstrated the industrial utility of the cells by showing that they could produce ethanol from acid pretreated rice straw using a simultaneous saccharification and fermentation (SSF) process.40 Although ethanol production still required the addition of a cellulase cocktail, as compared with native yeast strains that do not display cellulases, the amount of purified enzymes that needed to be added to hydrolyze the lignocellulose in the SSF process was reduced 10-fold. In addition, 1.4-fold more ethanol was produced (43.1 g/L ethanol from 200 g/L cellulosic material). The cellulase displaying cells could also be recycled between lignocellulose digestions, further demonstrating their practicality.41 Most recently, improved ethanol production from PASC was achieved by co-expressing a cellodextrin transporter, an intracellular β-glucosidase and three non-complexed displayed enzymes (endoglucanase, cellobiohydrolase, and β-glucosidase).42 Although the amount of ethanol produced was still low compared with industrial production levels (4.3 g/L ethanol from 20 g/L PASC), the results of this study highlight the benefits of optimizing both cellulase display and product import.
Displaying minicellulosomes
Because enzymes in cellulosome complexes degrade cellulose more efficiently than non-complexed enzymes, several groups have created yeast strains that display minicellulosomes. These complexes resemble the CipA cellulosome from C. thermocellum and are composed of a surface-displayed scaffoldin that contains cohesin modules that non-covalently bind to dockerin-cellulase fusion proteins (Fig. 3A). In 2009, two groups independently demonstrated that it was possible to display minicellulosomes on the surface of S. cerevisiae using an ex vivo assembly method in which yeast cells displaying a scaffoldin are incubated with a solution of purified cellulase-dockerin fusion proteins produced in E. coli (Fig. 3B).43,44 In these complexes, the scaffoldin is either directly fused to a GPI anchor signal sequence or it is fused to the Aga2 protein.43,44 Chen and colleagues constructed a minicellulosome that contained three enzymes targeted to specific sites within the complex via species-specific cohesin-dockerin interactions.44 By incorporating endoglucanase, exoglucanase, and β-glucosidase enzymes into the complex, the investigators generated yeast cells that could produce ethanol from insoluble PASC, a notable improvement over older-generation yeast strains that displayed non-complexed enzymes.44 In their systematic analysis, they demonstrated that the enzymes acted synergistically to hydrolyze cellulose, one of the first times enzyme-enzyme synergy was demonstrated in a cell surface displayed complex. Later, to eliminate the need to add purified enzymes to their cells, the investigators constructed surface-displayed minicellulosomes using a consortium of four yeast strains.45,46 In this ex vivo assembly method, cells displaying a scaffoldin are co-cultured with strains capable of secreting cellulase-dockerin fusion proteins, eliminating the need to add purified enzymes.46 After finding the optimal ratio of strains to maximize ethanol production, they demonstrated that ~1.87 g ethanol/L could be produced from PASC. Interestingly, in this system, the mechanism of scaffoldin anchoring appears to affect the efficiency of display, as an increased population of cells displaying minicellulosomes were observed when the scaffoldin was directly modified with a GPI anchor signal sequence instead of the Aga1-Aga2 anchoring system.45 Very recently, Hahn and colleagues used a similar consortium approach to construct cells that display ex vivo assembled minicellulosomes and they showed that the cells produce similar amounts of ethanol from PASC (1.8 g/L ethanol).47
Ex vivo assembly may be impractical for industrial applications, because it requires that purified enzymes be added to cells or that a consortium of different strains is used to construct the cellulosome. Two research groups have overcome this problem by constructing yeast cells that spontaneously assemble minicellulosomes on their surface (Fig. 3C).26,48 Zhao and colleagues were the first to achieve this milestone by constructing yeast cells that displayed a spontaneously assembling three-enzyme minicellulosome.26 This strain produced all of the components of the complex, including an Aga1-Aga2 tethered scaffoldin derived from C. thermocellum CipA and the T. reesei endoglucanase EGII, cellobiohydrolase CBHI, and A. aculeatus β-glucosidase BGLI enzymes. By systematically comparing the activities of uni-, bi-, and tri-functional minicellulosomes, they demonstrated that enzyme-enzyme synergistic interactions improved activity up to 1.6-fold. Moreover, cells displaying tri-functional minicellulosomes exhibited improved growth on PASC and could use it as a carbon source to produce 1.8 g/L of ethanol.
Tan and colleagues have displayed the largest and most complex self-assembling minicellulosome reported to date.48 To avoid having to display a long scaffoldin protein, the minicellulosome was constructed using two scaffoldins. The Aga1-Aga2 attached scaffoldin II protein is associated with the cell wall and coordinates the binding of scaffoldin I, mimicking nested architectures found in nature. Scaffoldin II contains four type-II cohesin modules from C. thermocellum, which coordinate the binding of four scaffoldin I proteins via its type-II dockerin. Scaffoldin I also contains a CBM and three type-I cohesin modules from different bacterial species that enable species-specific placement of dockerin fused enzymes.48 The cellulases and scaffoldin I are secreted using α-factor, and therefore their assembly and attachment to the cell presumably occurs extracellularly, avoiding export problems that would occur if the complex were assembled intracellularly. This approach enabled up to 12 enzymes to be displayed, four copies each of the C. cellulolyticum celCCA endoglucanase, celCCE cellobiohydrolase, and Ccel_2454 β-glucosidase. However, there may be a limit to the size of the scaffoldin that can be attached to the cell wall, as the investigators discovered that the percentage of cells displaying scaffoldin II decreased when longer scaffoldin II polypeptides were expressed. In the end, they chose to work with yeast displaying only 6 enzymes, two copies each of the endoglucanase, exoglucanase, and β-glucosidase, which produced 1.4 g/L of ethanol from insoluble Avicel. Very recently, Chen and colleagues used a similar “adaptive assembly” strategy to display a four enzyme containing minicellulosome that required ex vivo assembly.49 It also contains two scaffoldins enabling the display of four enzymes; two copies each of an endoglucanase and a β-glucosidase. Although the requirement for ex vivo assembly limited the ability of these cells to grow using cellulose as a nutrient, they could produce 1.9 g ethanol/L from PASC, which was double the amount of ethanol produced by cells displaying a related minicellulosome that contained a total of only two enzymes.
Recombinant Cellulolytic Eubacteria
Many species of eubacteria are promising consolidated bioprocessors because they are already used industrially to produce chemicals (e.g., amino acids, vitamins, solvents, etc.). They can be divided into Gram-negative and Gram-positive groups, whose distinct cell surface architectures necessitate that different approaches be used to display proteins. Work thus far has concentrated on the model Gram-negative and Gram-positive microorganisms Escherichia coli and Bacillus subtilis, respectively. They are not naturally cellulolytic, but contain robust genetic systems that enable their genetic manipulation. The most progress has been made with B. subtilis, leading to the display of minicellulosomes that enable it to grow on untreated lignocellulose, while only non-complexed cellulases have been displayed on the surface of E. coli. Below we describe this work, which could lead to their direct use in the consolidated bioprocessing of biomass, and facilitate the introduction of cellulolytic activity into less well studied industrially useful eubacteria.
Single Cellulase Display on the Surface of E. coli
The cell wall in Gram-negative bacteria consists of inner and outer membranes separated by peptidoglycan. In the model Gram-negative organism E. coli, a variety of approaches have been developed to display heterologous proteins in the outer membrane (Fig. 3B).50 Two general approaches have been used to display cellulase enzymes on the surface of E. coli. In the first approach, the cellulase is expressed as a fusion protein with an E. coli lipoprotein. For example, display has been achieved by fusing to the N-terminus of a cellulase the signal sequence and the first nine amino acids from the major outer membrane lipoprotein (Lpp) and the transmembrane domain from the outer membrane protein (OmpA).51 The Lpp component targets and anchors the protein to the outer membrane, while the OmpA segment is required for surface expression of the passenger cellulase.52 Using an Lpp-OmpA display system, ~6 x 104 proteins can be displayed per cell.53 Similar display strategies fuse cellulases to either the Ice nucleation protein (Inp) or the bacterial lipocalin (Blc) lipoproteins that are in turn embedded in the outer membrane.54-58 In the second approach, the cellulase is displayed on the bacterial surface using the type V secretion system.59 Specifically, the enzyme is expressed such that it contains an N-terminal signal sequence and the C-terminal translocator domain derived from the autotransporter AIDA-I protein which is embedded in the outer membrane.
At present, only non-complexed cellulases have been displayed on the surface of E. coli.51,54-59 As only a single type of enzyme was displayed, these cells exhibit limited cellulolytic activity. Although multi-enzyme display is desirable to maximize cellulolytic activity, E. coli cells that secrete cellulases can also degrade biomass. Recently, Keasling’s group engineered a consortium of enzyme-secreting E. coli cells that can degrade ionic liquid (IL) pretreated switchgrass and produce a range of chemicals (butanol, fatty acid ethyl esters, and pinene).60 After 48 h, cell densities of 140 × 107 CFU/mL were obtained, which is ~50% lower than E. coli cells grown in minimal media containing glucose as a carbon source. The rate of lignocellulosic degradation may be growth limiting, as the consortium digested only ~5–6% of the cellulose and hemicellulose. An endoglucanase was also displayed on the surface of the Gram-negative bacterium Zymobacter palmae by fusing it to the ice nucleation protein from Pseudomonas syringae. However, the activity of these cells was only verified using soluble CMC as a substrate.61
Cellulolytic Bacillus subtilis
Gram-positive bacteria typified by B. subtilis contain a single membrane surrounded by a thick peptidoglycan cell wall. Two approaches are used to display cellulases and minicellulosomes on its surface (Fig. 4C). Proteins are displayed non-covalently by expressing them as fusions with the LysM protein that contains binding modules that interact with N-acetylmuramic acid and N-acetyl-D-glucosamine within the cell wall, or they are covalently attached to the peptidoglycan using sortase transpeptidase enzymes.62-65 In the latter procedure, the protein is expressed as a fusion protein that contains a C-terminal cell wall sorting signal (CWS), which is then is covalently linked to the peptidoglycan cross-bridge by the sortase.66 Although direct comparisons have not been made, Chen et al. concluded that 1.2 × 107 proteins are attached to each cell using the LysM fusion approach, whereas it is estimated that 2.4–3 × 105 proteins can be displayed per cell using sortase transpeptidases.63,67,68
Two research groups reported the construction of B. subtilis cells that display an ex vivo assembled minicellulosome (Fig. 3B).63,65 Zhang and colleagues created cells that display a minicellulosome that is non-covalently associated with the cell wall. These cells secrete the mini-CipA scaffoldin, which associates with the cell wall via its LysM domain. Mini-CipA also contains three cohesin modules and a CBM derived from C. thermocellum CipA.65 To construct the minicellulosome, cells displaying mini-CipA were incubated with three E. coli purified cellulases (B. subtilis endoglucanase Cel5, C. phytofermentans exoglucanase Cel48, and C. thermocellum endoglucanase Cel9). Interestingly, similar to C. thermocellum that naturally displays a cellulosome, the recombinant B. subtilis cells exhibited cellulolytic CEM synergy, as the activity of the surface-displayed complex was superior to a purified minicellulosome that contained the same enzymes; the cells degraded RAC and microcrystalline cellulose 2.3- and 4.5-fold better than the isolated complex, respectively. It was proposed that interactions between cells might also contribute to CEM synergy as presumably adjacent cells can assimilate long-chain hydrolysis products before they diffuse into the bulk phase, which prevents product inhibition of the cellulases and cellulosomes. Independently, Anderson and colleagues engineered B. subtilis cells to display an ex vivo assembled minicellulosome that is covalently attached to the cell wall.63 In this system, a scaffoldin (called Scaf) is joined via a peptide bond to the cross-bridge peptide of the peptidoglycan by a heterologous sortase enzyme.66 Tri-functional minicellulosomes are then assembled when the Scaf displaying cells are incubated with the appropriate purified cellulase-dockerin fusion proteins (the C. thermocellum endoglucanase Cel8A and the C. cellulolyticum exoglucanase Cel9E and endoglucanase Cel9G enzymes). These cells degraded RAC, and exhibited enzyme-enzyme synergism that increased cellulolytic activity 1.3-fold.63 Importantly, the sortase displayed minicellulosome exhibited stable activity for up to 70 h when the WprA cell wall protease was genetically eliminated, presumably because the complex is covalently linked to the cell wall. A sortase-utilizing system was also used to display miniaturized scaffoldins on the cell surface of L. lactis, which could then bind purified β-glucuronidase UidA.69
B. subtilis cells displaying a self-assembled minicellulosome grow on untreated lignocellulose
To overcome the requirement for ex vivo assembly, we recently engineered B. subtilis cells that display a covalently attached minicellulosome that assembles spontaneously (Fig. 3C).62 The minicellulosome was constructed by co-expressing five proteins: the SrtA sortase from B. anthracis, a chimeric scaffoldin (Scaf) composed of three cohesin modules that are covalently attached to the cell wall by SrtA, and three dockerin-cellulase fusion proteins that bind to the scaffoldin non-covalently via species-specific dockerin-cohesin interactions. Three enzymes derived from the mesophile C. cellulolyticum were displayed (endoglucanase/xylanase Cel5A, exoglucanase/endoglucanase Cel9E, and the processive endoglucanase Cel48F) and based on immunoblot analyses, they are present in the complex at saturating levels. The cells exhibit potent cellulolytic activity enabling growth on dilute acid-pretreated corn stover to densities similar to those achieved by cells cultured in minimal media containing glucose. Recombinant azide-treated B. subtilis cells required ~96 h at 37 °C to degrade 62% of a 5 g/L solution of pretreated corn-stover biomass. Azide-killed cells supplemented with β-glucosidase released 21% and 33% of the glucose and xylose found in corn stover after 48 h. This result is promising, since when assayed under similar conditions at 37 °C, cells displaying a tri-functional minicellulosome exhibit ~1/3 the cellulolytic activity of a Ctec2/Htec2 enzyme cocktail (Novozymes) that contains dozens of enzymes. Importantly, bacteria displaying the tri-functional minicellulosome could also grow on untreated plant biomass (corn stover, straw, or switchgrass) to high cell densities. To the best of our knowledge, this is the first such demonstration of this capability by a recombinant organism. The specific growth rates of cells cultured in minimal media containing 0.5% wt/volume of glucose, acid-treated corn stover, and untreated corn stover were ~0.17, ~0.08, and ~0.05 per hour, respectively. Additional improvements in their activity are needed if they are to rival naturally cellulolytic microbes such as C. thermocellum, which at 60 °C degrades 2 g/L of microcrystalline Avicel in 20 h.70 Creating recombinant B. subtilis that display more than three types of enzymes can be expected to lead to even more potent cellulolytic microbes with better growth properties.
Comparing Activities of Recombinant Cellulolytic B. subtilis, E. coli, and S. cerevisiae
Direct comparisons of the cellulolytic activities of the microbes discussed in this review are not possible because different experimental approaches and cellulose substrates have been used to assess their activities. The methods range from detailed analyses of the amount of biomass degraded and the sugars produced from hydrolysis, to less informative approaches that monitored only microbial growth or ethanol production. Further hindering direct comparisons, the identities and numbers of enzymes displayed can vary between each study and there are differences in the abilities of each microbe to import and metabolize the lignocellulosic degradation products. A variety of substrates have been used to assess the cellulolytic activity of recombinant microbes. They vary in their recalcitrance to enzymatic degradation because they differ in their crystallinity, degree of polymerization, fraction of reducing ends, and presence of hemicellulose and lignin.29 Based on these properties, the substrates range from easy to difficult to degrade as follows: PASC/RAC < microcrystalline cellulose < pretreated lignocellulose < untreated lignocellulose. With these considerations in mind, below we compare the cellulolytic activity of the recombinant microbes that have been discussed in this review.
The data shown in Table 1 compares the cellulolytic activities of the recombinant microbes discussed in this review. For simplicity, only strains capable of degrading insoluble forms of cellulose without the need for adding purified enzyme cocktails are considered. B. subtilis cells displaying a covalently attached self-assembling minicellulosome have the highest demonstrated activity.62 They degrade the most complex forms of cellulose, both untreated and acid-treated lignocellulose. Notably, these bacteria grow in minimal media containing industrially relevant forms of untreated lignocellulosic biomass as a primary nutrient source (corn stover, hatched straw, and switchgrass) to densities that are similar to those achieved by cells that are cultured in glucose. B. subtilis cells displaying an ex vivo assembled complex can degrade microcrystalline cellulose, but the need to add purified enzymes to construct the complex limits their ability to replicate using cellulose as a nutrient.62,65 The most cellulolytic yeast strains reported to date display a complex that contains 6 enzymes (two copies of three types of enzymes).48 These cells degrade microcrystalline cellulose, but their ability to metabolize more complex lignocellulose has not been tested. In addition, growth of these cells on microcrystalline cellulose required supplementation with rich nutrients unlike recombinant B. subtilis grown on biomass. Notably, the same group reported cells that display 12 enzymes, but these microbes were less cellulolytic because fewer enzyme complexes were displayed per cell, presumably because of the increased metabolic burden of displaying this large complex. Thus far, the surface of E. coli has only been engineered to display non-complexed cellulases, with only a single type of enzyme displayed on each cell.51,55,56,71 As expected, these cells have limited degradative capacity, as they only demonstrated significant activity on CMC. The limited progress that has been made thus far in engineering the surface of E. coli to display multi-enzyme complexes may be due to difficulties associated with exporting proteins across its two membranes and may explain why cellulosomes are predominantly found in Gram-positive species.
Table 1. Cellulolytic Activities of Recombinant Cellulase Displaying Microbes.
| Recombinant Microorganism | Cellulase(s)a Displayed | Insoluble Cellulose Substrate | |||
|---|---|---|---|---|---|
| PASC/ RAC |
Microcrystalline Cellulose |
Pretreated Lignocellulose |
Untreated Lignocellulose |
||
| B. subtilis | |||||
| Anderson et al. 2011 | Ex vivo mini.a | + | - | - | - |
| You et al. 2012 | Ex vivo mini. | + | + | - | - |
| Anderson et al. 2013 | Self-assembled mini.a | - | - | + | + |
| S. cerevisiae | |||||
|---|---|---|---|---|---|
| Tsai et al. 2009 | Ex vivo mini. | +, Eb | - | - | - |
| Yanase et al. 2010 | Non-complexed cellulase.a | +, E | - | - | - |
| Tsai et al. 2010 | Ex vivo mini. Consortium. a | +, E | - | - | - |
| Wen et al. 2010 | Self-assembled mini. | +, E | - | - | - |
| Goyal et al. 2011 | Ex vivo mini. Consortium. | +, E | - | - | - |
| Fan et al. 2012 | Self-assembled mini. | +, E | +, E | - | - |
| Tsai et al. 2013 | Ex vivo mini. | +, E | - | - | - |
| Kim et al. 2013 | Ex vivo mini. Consortium. | +, E | - | - | - |
a Ex vivo mini: minicellulosome assembled ex vivo. Self-assembled mini: minicellulosome assembled spontaneously. Non-complexed cellulase: individually displayed enzyme. Consortium: multiple strains required to assemble minicellulosome. bE, ethanol.
Because these model organisms are still under development, the cellulolytic activity that can ultimately be obtained by engineering their surfaces remains unknown. However, B. subtilis may have the greatest potential for further development because a higher density of enzyme complexes can presumably be displayed on its surface. This may result in increased rates of cellulolysis, as more enzymes will be available to degrade the cellulose fibers bound to each cell. Specifically, based on documented levels of heterologous protein display, B. subtilis can display ~60–3.2 × 104 times more proteins per micron2 of surface area as compared with S. cerevisiae (B. subtilis and S. cerevisiae display ~2.4 × 105–1.2 × 107 and ~1 × 104–1 × 105 heterologous proteins per cell resulting in surface densities of ~1.9 × 104–9.6 × 105 and ~30–300 proteins/micron,2 respectively).35,67,68 The surface of E. coli can also be densely coated with proteins (~6.4 × 103 proteins/micron2 assuming 6 × 104 proteins displayed per cell), but as of yet, only non-complexed cellulases have been displayed.53 In order to develop improved minicellulosome display methods, it will be important to rigorously quantify the number of complexes displayed per cell. Thus far, the number of minicellulosome complexes displayed on the surface of S. cerevisiae has not been quantified, while in B. subtilis, experimental measurements have shown that 2 × 104 and 1.5 × 105 minicellulosomes can be attached to each cell using LysM and sortase-mediated approaches, respectively.62,65 These studies suggest that the number of minicellulosomes displayed per cell may decrease as the size of the complex increases. For example, in B. subtilis, quantitative studies have shown that it is possible to display 1.2 × 107 individual LysM fusion proteins, but only 2 × 104 LysM anchored minicellulosomes.65,68 Similar decreases in anchoring efficiency also occur in S. cerevisiae with increasing protein size, suggesting that this is a general problem.48
Future Directions
Surface engineering efforts thus far have created recombinant microbes with potent cellulolytic activities. However, even more elaborate structures will need to be grafted onto their surfaces if their activities are to rival those of naturally cellulolytic organisms or cellulase cocktails that are currently used in industry. The composition and structure of lignocellulose varies depending upon its source and the method of pretreatment. Therefore, the number, type, relative abundance, and positioning of the surfaced displayed enzymes will need to be optimized to degrade different types of plant feedstocks. Displaying minicellulosomes that more closely resemble native cellulosomes is an obvious strategy to improve cellulolytic activity, as it will presumably maximize enzyme-enzyme and CEM synergy. However, constructing these large self-assembling complexes may prove difficult as the results of recent work in S. cerevisiae and B. subtilis indicate that the surface density of displayed complexes decreases as they become larger and more complex. Overcoming this problem may require minicellulosome construction using adaptive assembly strategies and/or using protein components produced from a microbial consortium. The activity of these displayed complexes may be further improved using directed evolution approaches using growth on biomass to select for cells that display minicellulosomes possessing the best enzyme compositions and architectures. Combined, this work promises to yield potent recombinant surface-engineered microbes that can degrade biomass. Concurrently, many research groups are using metabolic engineering, synthetic biology, and systems biology approaches to construct microorganisms capable of producing next generation biofuels and useful chemicals.72-75 When paired with the novel lignocellulosic degrading platforms described in this review, these microbes could significantly reduce the world’s dependency on oil by directly producing biofuels and other useful biocommodities from renewable and abundant plant biomass.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was supported by Department of Energy grant DE-FC-03–87ER60615. Huang GL was supported by a Ruth L. Kirschstein National Research Service Award GM007185.
Glossary
Abbreviations:
- CBP
consolidated bioprocessing
- CEM
cellulose-enzyme-microbe
- PASC
phosphoric acid swollen cellulose
- RAC
regenerated amorphous cellulose
- IL
ionic liquid
- GPI
glycosylphosphatidylinositol
- CWS
cell wall sorting signal
- CBM
cellulose binding module
References
- 1.Kerr RA. Energy. World oil crunch looming? Science. 2008;322:1178–9. doi: 10.1126/science.322.5905.1178. [DOI] [PubMed] [Google Scholar]
- 2.Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–83. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Perlack RD, Energy USDo, Agriculture USDo, Laboratory ORN. Biomass as feedstock for a bioenergy and bioproducts industry the technical feasibility of a billion-ton annual supply. Oak Ridge, Tenn.: Oak Ridge National Laboratory, 2005. [Google Scholar]
- 4.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–7. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 5.Wilson DB. Cellulases and biofuels. Curr Opin Biotechnol. 2009;20:295–9. doi: 10.1016/j.copbio.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Yeoman CJ, Han Y, Dodd D, Schroeder CM, Mackie RI, Cann IK. Thermostable enzymes as biocatalysts in the biofuel industry. Adv Appl Microbiol. 2010;70:1–55. doi: 10.1016/S0065-2164(10)70001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller PS, Blum PH. Extremophile-inspired strategies for enzymatic biomass saccharification. Environ Technol. 2010;31:1005–15. doi: 10.1080/09593330903536113. [DOI] [PubMed] [Google Scholar]
- 8.Hendriks AT, Zeeman G. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol. 2009;100:10–8. doi: 10.1016/j.biortech.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Zhao X, Cheng K, Liu D. Organosolv pretreatment of lignocellulosic biomass for enzymatic hydrolysis. Appl Microbiol Biotechnol. 2009;82:815–27. doi: 10.1007/s00253-009-1883-1. [DOI] [PubMed] [Google Scholar]
- 10.Olson DG, McBride JE, Shaw AJ, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X-Z, Zhang Y-HP. One-step production of biocommodities from lignocellulosic biomass by recombinant cellulolytic Bacillus subtilis: Opportunities and challenges. Eng Life Sci. 2010;10:398–406. doi: 10.1002/elsc.201000011. [DOI] [Google Scholar]
- 12.Lynd LR, Laser MS, Bransby D, Dale BE, Davison B, Hamilton R, Himmel M, Keller M, McMillan JD, Sheehan J, et al. How biotech can transform biofuels. Nat Biotechnol. 2008;26:169–72. doi: 10.1038/nbt0208-169. [DOI] [PubMed] [Google Scholar]
- 13.la Grange DC, den Haan R, van Zyl WH. Engineering cellulolytic ability into bioprocessing organisms. Appl Microbiol Biotechnol. 2010;87:1195–208. doi: 10.1007/s00253-010-2660-x. [DOI] [PubMed] [Google Scholar]
- 14.Klein-Marcuschamer D, Oleskowicz-Popiel P, Simmons BA, Blanch HW. The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol Bioeng. 2012;109:1083–7. doi: 10.1002/bit.24370. [DOI] [PubMed] [Google Scholar]
- 15.Zhao X, Zhang L, Liu D. Biomass recalcitrance. Part I: the chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels. Bioproducts and Biorefining. 2012;6:465–82. doi: 10.1002/bbb.1331. [DOI] [Google Scholar]
- 16.Zhao X, Zhang L, Liu D. Biomass recalcitrance. Part II: Fundamentals of different pre-treatments to increase the enzymatic digestibility of lignocellulose. Biofuels. Bioproducts and Biorefining. 2012;6:561–79. doi: 10.1002/bbb.1350. [DOI] [Google Scholar]
- 17.Mba Medie F, Davies GJ, Drancourt M, Henrissat B. Genome analyses highlight the different biological roles of cellulases. Nat Rev Microbiol. 2012;10:227–34. doi: 10.1038/nrmicro2729. [DOI] [PubMed] [Google Scholar]
- 18.Ghose T. Cellulase biosynthesis and hydrolysis of cellulosic substances. In: Ghose T, ed. Advances in Biochemical Engineering: Springer Berlin / Heidelberg, 1977. [Google Scholar]
- 19.Scheller HV, Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–89. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee G, Car S, Scott-Craig JS, Borrusch MS, Aslam N, Walton JD. Synthetic enzyme mixtures for biomass deconstruction: production and optimization of a core set. Biotechnol Bioeng. 2010;106:707–20. doi: 10.1002/bit.22741. [DOI] [PubMed] [Google Scholar]
- 21.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–46. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 22.Bugg TD, Ahmad M, Hardiman EM, Rahmanpour R. Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep. 2011;28:1883–96. doi: 10.1039/c1np00042j. [DOI] [PubMed] [Google Scholar]
- 23.Bayer EA, Shimon LJ, Shoham Y, Lamed R. Cellulosomes-structure and ultrastructure. J Struct Biol. 1998;124:221–34. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 24.Fontes CM, Gilbert HJ. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem. 2010;79:655–81. doi: 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 25.Leibovitz E, Ohayon H, Gounon P, Béguin P. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J Bacteriol. 1997;179:2519–23. doi: 10.1128/jb.179.8.2519-2523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wen F, Sun J, Zhao H. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl Environ Microbiol. 2010;76:1251–60. doi: 10.1128/AEM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y, Zhang YH, Lynd LR. Enzyme-microbe synergy during cellulose hydrolysis by Clostridium thermocellum. Proc Natl Acad Sci U S A. 2006;103:16165–9. doi: 10.1073/pnas.0605381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demain AL, Newcomb M, Wu JH. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev. 2005;69:124–54. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv. 2006;24:452–81. doi: 10.1016/j.biotechadv.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Fujita M, Kinoshita T. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett. 2010;584:1670–7. doi: 10.1016/j.febslet.2009.10.079. [DOI] [PubMed] [Google Scholar]
- 31.Ohishi K, Inoue N, Kinoshita T. PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J. 2001;20:4088–98. doi: 10.1093/emboj/20.15.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Lu CF, Kurjan J, Lipke PN. A pathway for cell wall anchorage of Saccharomyces cerevisiae alpha-agglutinin. Mol Cell Biol. 1994;14:4825–33. doi: 10.1128/mcb.14.7.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu CF, Montijn RC, Brown JL, Klis F, Kurjan J, Bussey H, Lipke PN. Glycosyl phosphatidylinositol-dependent cross-linking of alpha-agglutinin and beta 1,6-glucan in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1995;128:333–40. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–68. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 36.Murai T, Ueda M, Kawaguchi T, Arai M, Tanaka A. Assimilation of cellooligosaccharides by a cell surface-engineered yeast expressing beta-glucosidase and carboxymethylcellulase from aspergillus aculeatus. Appl Environ Microbiol. 1998;64:4857–61. doi: 10.1128/aem.64.12.4857-4861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita Y, Takahashi S, Ueda M, Tanaka A, Okada H, Morikawa Y, Kawaguchi T, Arai M, Fukuda H, Kondo A. Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Appl Environ Microbiol. 2002;68:5136–41. doi: 10.1128/AEM.68.10.5136-5141.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita Y, Ito J, Ueda M, Fukuda H, Kondo A. Synergistic saccharification, and direct fermentation to ethanol, of amorphous cellulose by use of an engineered yeast strain codisplaying three types of cellulolytic enzyme. Appl Environ Microbiol. 2004;70:1207–12. doi: 10.1128/AEM.70.2.1207-1212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito J, Fujita Y, Ueda M, Fukuda H, Kondo A. Improvement of cellulose-degrading ability of a yeast strain displaying Trichoderma reesei endoglucanase II by recombination of cellulose-binding domains. Biotechnol Prog. 2004;20:688–91. doi: 10.1021/bp034332u. [DOI] [PubMed] [Google Scholar]
- 40.Matano Y, Hasunuma T, Kondo A. Display of cellulases on the cell surface of Saccharomyces cerevisiae for high yield ethanol production from high-solid lignocellulosic biomass. Bioresour Technol. 2012;108:128–33. doi: 10.1016/j.biortech.2011.12.144. [DOI] [PubMed] [Google Scholar]
- 41.Matano Y, Hasunuma T, Kondo A. Cell recycle batch fermentation of high-solid lignocellulose using a recombinant cellulase-displaying yeast strain for high yield ethanol production in consolidated bioprocessing. Bioresour Technol. 2013;135:403–9. doi: 10.1016/j.biortech.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Yamada R, Nakatani Y, Ogino C, Kondo A. Efficient direct ethanol production from cellulose by cellulase- and cellodextrin transporter-co-expressing Saccharomyces cerevisiae. AMB Express. 2013;3:34. doi: 10.1186/2191-0855-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lilly M, Fierobe HP, van Zyl WH, Volschenk H. Heterologous expression of a Clostridium minicellulosome in Saccharomyces cerevisiae. FEMS Yeast Res. 2009;9:1236–49. doi: 10.1111/j.1567-1364.2009.00564.x. [DOI] [PubMed] [Google Scholar]
- 44.Tsai SL, Oh J, Singh S, Chen R, Chen W. Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2009;75:6087–93. doi: 10.1128/AEM.01538-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goyal G, Tsai SL, Madan B, DaSilva NA, Chen W. Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb Cell Fact. 2011;10:89. doi: 10.1186/1475-2859-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai SL, Goyal G, Chen W. Surface display of a functional minicellulosome by intracellular complementation using a synthetic yeast consortium and its application to cellulose hydrolysis and ethanol production. Appl Environ Microbiol. 2010;76:7514–20. doi: 10.1128/AEM.01777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S, Baek SH, Lee K, Hahn JS. Cellulosic ethanol production using a yeast consortium displaying a minicellulosome and β-glucosidase. Microb Cell Fact. 2013;12:14. doi: 10.1186/1475-2859-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan LH, Zhang ZJ, Yu XY, Xue YX, Tan TW. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci U S A. 2012;109:13260–5. doi: 10.1073/pnas.1209856109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai SL, DaSilva NA, Chen W. Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth Biol. 2013;2:14–21. doi: 10.1021/sb300047u. [DOI] [PubMed] [Google Scholar]
- 50.van Bloois E, Winter RT, Kolmar H, Fraaije MW. Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol. 2011;29:79–86. doi: 10.1016/j.tibtech.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Francisco JA, Stathopoulos C, Warren RA, Kilburn DG, Georgiou G. Specific adhesion and hydrolysis of cellulose by intact Escherichia coli expressing surface anchored cellulase or cellulose binding domains. Biotechnology (N Y) 1993;11:491–5. doi: 10.1038/nbt0493-491. [DOI] [PubMed] [Google Scholar]
- 52.Earhart CF. Use of an Lpp-OmpA fusion vehicle for bacterial surface display. Methods Enzymol. 2000;326:506–16. doi: 10.1016/S0076-6879(00)26072-2. [DOI] [PubMed] [Google Scholar]
- 53.Chen G, Cloud J, Georgiou G, Iverson BL. A quantitative immunoassay utilizing Escherichia coli cells possessing surface-expressed single chain Fv molecules. Biotechnol Prog. 1996;12:572–4. doi: 10.1021/bp960041s. [DOI] [PubMed] [Google Scholar]
- 54.Jung HC, Lebeault JM, Pan JG. Surface display of Zymomonas mobilis levansucrase by using the ice-nucleation protein of Pseudomonas syringae. Nat Biotechnol. 1998;16:576–80. doi: 10.1038/nbt0698-576. [DOI] [PubMed] [Google Scholar]
- 55.Liu W, Zhang XZ, Zhang Z, Zhang YH. Engineering of Clostridium phytofermentans Endoglucanase Cel5A for improved thermostability. Appl Environ Microbiol. 2010;76:4914–7. doi: 10.1128/AEM.00958-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim YS, Jung HC, Pan JG. Bacterial cell surface display of an enzyme library for selective screening of improved cellulase variants. Appl Environ Microbiol. 2000;66:788–93. doi: 10.1128/AEM.66.2.788-793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung HC, Park JH, Park SH, Lebeault JM, Pan JG. Expression of carboxymethylcellulase on the surface of Escherichia coli using Pseudomonas syringae ice nucleation protein. Enzyme Microb Technol. 1998;22:348–54. doi: 10.1016/S0141-0229(97)00224-X. [DOI] [PubMed] [Google Scholar]
- 58.Soma Y, Inokuma K, Tanaka T, Ogino C, Kondo A, Okamoto M, Hanai T. Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system. J Biosci Bioeng. 2012;114:80–5. doi: 10.1016/j.jbiosc.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 59.Muñoz-Gutiérrez I, Oropeza R, Gosset G, Martinez A. Cell surface display of a β-glucosidase employing the type V secretion system on ethanologenic Escherichia coli for the fermentation of cellobiose to ethanol. J Ind Microbiol Biotechnol. 2012;39:1141–52. doi: 10.1007/s10295-012-1122-0. [DOI] [PubMed] [Google Scholar]
- 60.Bokinsky G, Peralta-Yahya PP, George A, Holmes BM, Steen EJ, Dietrich J, Lee TS, Tullman-Ercek D, Voigt CA, Simmons BA, et al. Synthesis of three advanced biofuels from ionic liquid-pretreated switchgrass using engineered Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:19949–54. doi: 10.1073/pnas.1106958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kojima M, Akahoshi T, Okamoto K, Yanase H. Expression and surface display of Cellulomonas endoglucanase in the ethanologenic bacterium Zymobacter palmae. Appl Microbiol Biotechnol. 2012;96:1093–104. doi: 10.1007/s00253-012-4424-2. [DOI] [PubMed] [Google Scholar]
- 62.Anderson TD, Miller JI, Fierobe HP, Clubb RT. Recombinant Bacillus subtilis that grows on untreated plant biomass. Appl Environ Microbiol. 2013;79:867–76. doi: 10.1128/AEM.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Anderson TD, Robson SA, Jiang XW, Malmirchegini GR, Fierobe HP, Lazazzera BA, Clubb RT. Assembly of minicellulosomes on the surface of Bacillus subtilis. Appl Environ Microbiol. 2011;77:4849–58. doi: 10.1128/AEM.02599-10. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64.Frankel MB, Schneewind O. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J Biol Chem. 2012;287:10460–71. doi: 10.1074/jbc.M111.336404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.You C, Zhang XZ, Sathitsuksanoh N, Lynd LR, Zhang YH. Enhanced microbial utilization of recalcitrant cellulose by an ex vivo cellulosome-microbe complex. Appl Environ Microbiol. 2012;78:1437–44. doi: 10.1128/AEM.07138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82:1044–59. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen HD, Schumann W. Establishment of an experimental system allowing immobilization of proteins on the surface of Bacillus subtilis cells. J Biotechnol. 2006;122:473–82. doi: 10.1016/j.jbiotec.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 68.Chen CL, Wu SC, Tjia WM, Wang CL, Lohka MJ, Wong SL. Development of a LytE-based high-density surface display system in Bacillus subtilis. Microb Biotechnol. 2008;1:177–90. doi: 10.1111/j.1751-7915.2007.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wieczorek AS, Martin VJ. Engineering the cell surface display of cohesins for assembly of cellulosome-inspired enzyme complexes on Lactococcus lactis. Microb Cell Fact. 2010;9:69. doi: 10.1186/1475-2859-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lynd LR, Grethlein HE, Wolkin RH. Fermentation of Cellulosic Substrates in Batch and Continuous Culture by Clostridium thermocellum. Appl Environ Microbiol. 1989;55:3131–9. doi: 10.1128/aem.55.12.3131-3139.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kojima M, Akahoshi T, Okamoto K, Yanase H. Expression and surface display of Cellulomonas endoglucanase in the ethanologenic bacterium Zymobacter palmae. Appl Microbiol Biotechnol. 2012;96:1093–104. doi: 10.1007/s00253-012-4424-2. [DOI] [PubMed] [Google Scholar]
- 72.Dellomonaco C, Fava F, Gonzalez R. The path to next generation biofuels: successes and challenges in the era of synthetic biology. Microb Cell Fact. 2010;9:3. doi: 10.1186/1475-2859-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang YS, Park JM, Choi S, Choi YJ, Seung Y, Cho JH, Lee SY. Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol Adv. 2012;30:989–1000. doi: 10.1016/j.biotechadv.2011.08.015. [DOI] [PubMed] [Google Scholar]
- 74.Li H, Cann AF, Liao JC. Biofuels: biomolecular engineering fundamentals and advances. Annu Rev Chem Biomol Eng. 2010;1:19–36. doi: 10.1146/annurev-chembioeng-073009-100938. [DOI] [PubMed] [Google Scholar]
- 75.Rabinovitch-Deere CA, Oliver JW, Rodriguez GM, Atsumi S. Synthetic biology and metabolic engineering approaches to produce biofuels. Chem Rev. 2013;113:4611–32. doi: 10.1021/cr300361t. [DOI] [PubMed] [Google Scholar]