Abstract
Intestinal homeostasis depends on the proper activity of the intestinal stem cells (ISCs) and an appropriate host response to the normal resident microbiota. The question on the effect of microbiota on ISCs behavior has not been addressed yet. Canonical Wnt pathway and ISC gene expression signature was compared in germfree vs. conventional and MyD88−/− vs. Myd88+/+ mice based on publicly available gene expression data sets. Microbiota and MyD88-dependent signaling have distinct effects on the Wnt pathway and ISC at gene expression level. In addition, the effect of microbiota and MyD88-dependent signaling on Wnt pathway and ISCs show regional variation. The net effect of microbiota on Wnt pathway and ISCs cannot be inferred from the available data. Nonetheless, the data are suggestive of a potential regulatory mechanism of the Wnt pathway by the microbiota and plausibly by any alteration in the microbiota composition.
Keywords: intestinal stem cells, microbiota, Lgr5, Wnt, MyD88, toll-like receptors, regional variation, colorectal cancer
A prominent difference between the small intestine and colon is the number and composition of the normal resident bacteria. A relatively lower number and fewer species reside in the stomach and upper small intestine due to the specific composition of luminal constituents and the propulsive motion of the region. However, the distal part of the small intestine and colon is habitat to a diverse and densely populated microbiota.1 The highest microbiota content is found in distal ileum and colon where the bacterial concentration reaches 1012–1014 cells/ml of the luminal content.2
Intestinal homeostasis depends on the proper activity of the intestinal stem cells (ISCs) and an appropriate host response to the normal resident microbiota. ISCs constitute a small proportion of intestinal epithelium which are defined by demonstrating self-renewal capacity and multipotency. They are responsible for intestinal epithelium rapid physiologic turnover and regeneration following injury. ISCs are identified at the base of the crypt and at the position 4 from the base of the crypt in the murine small intestine. The crypt base columnar (CBC) cells are Lgr5 positive and their cycling rate is estimated to be once every 24 h.3 The position 4 cells, on the other hand, are positive for Bmi1 and mTert and are quiescent and slow cycling.4,5
There is a constant interaction between intestinal epithelium and microbiota via pattern recognition receptors including toll-like receptors (TLRs). Lgr5+ ISCs express TLR4 which is shown to affect the crypt dynamic in vivo.6 However, the effect of microbiota and TLR signaling on ISCs is largely unknown. MyD88 is the main adaptor protein downstream of TLRs.7 MyD88 signaling is required for intestinal tissue homeostasis and response to injury as MyD88−/− mice are more susceptible to acute dextran sodium sulfate (DSS)-induced colitis and develop a more severe disease.8 However, MyD88-deficiency does not affect the crypt size and proliferative status under physiologic condition.9
The effect of the entire microbiota and MyD88-dependent signaling on the ISCs is not yet elucidated. Crypt size and proliferative status are indistinguishable in germfree wildtype, conventional MyD88−/−, and conventional wildtype mice.9 An intestinal pathogen, i.e., Salmonella typhimurium, has been shown to activate intestinal Wnt signaling pathway both in vitro and in vivo.10-15 Unlike mice, the intestinal epithelium proliferation rate is decreased in germfree zebrafish larvae compared with conventionally-reared. The effect of microbiota on the intestinal epithelium proliferation is mediated via MyD88 and is not dependent on microbiota-induced inflammatory response. The crosstalk between microbiota and Wnt pathway was investigated in axin1 mutated larvae. Axin1 mutant larvae have an increased level of intestinal epithelial proliferation which is significantly inhibited when reared germfree or treated with MyD88 inhibitor. Furthermore, microbiota affects the Wnt pathway activity as evident by reduced level of cytoplasmic β-catenin in germfree animal.16 In order to gain insight into the effect of microbiota and MyD88-dependent signaling on ISCs, I queried the publicly available gene expression data at http://microbiota.wall.gu.se/ (access date 1 July 2013) for the ISC gene signature and canonical Wnt pathway components. Larsson et al. have conducted a systematic gene expression analysis along the intestinal tract in mice. They assessed the effect of microbiota and MyD88-dependent signaling on intestinal whole tissue gene expression in different segments of the small intestine and colon by cDNA microarray.17 Both microbiota and ISCs are required for physiologic intestinal homeostasis. It is plausible that microbiota and MyD88-dependent signaling exert their effect on epithelial maintenance via their effect on ISCs. To assess this hypothesis, the ISC signature and canonical Wnt pathway gene expression were compared in germfree vs. conventional and MyD88−/− vs. MyD88+/+ mice. A set of 28 genes identified in two separate reports of the gene expression signature of ISC in mice was selected as ISC signature.18,19 The genes which were significantly altered (P < 0.05) were noted. Wnt pathway and ISCs are affected by both microbiota and MyD88-dependent signaling. However, it seems that the effect of microbiota is not totally mediated by MyD88-dependent pathway. Furthermore, an interesting regional variation of the Wnt pathway and ISC gene expression signature in response to microbiota and MyD88 signaling is observed along the intestinal tract. Lgr5 is the best-studied single ISC marker. It is found to be downregulated in duodenum, unchanged in jejunum, and increased in ileum and colon in germfree mice compared with conventional whereas it is unchanged in duodenum, jejunum, and colon; and is increased in ileum in MyD88−/− vs. MyD88+/+ mice. Among the ISC gene signature, Ascl2, Cd44, Cenpf, Cyp2e1, and Phgdh are co-regulated in germfree vs. conventional mice; the relevance of which to ISC biology remains unclear. It is noteworthy that while some genes are consistently increased or decreased in an experimental condition, some are distinctly regulated in different regions of the intestinal tract (Figs. 1 and 2). The inherent limitations of cDNA microarray as well as assessing the whole intestinal tissue rather than isolated epithelial cells or sorted ISCs are the major caveats of Larsson et al. report. Nonetheless, the data are suggestive of the effect of microbiota and MyD88 signaling on Wnt pathway and ISC gene signature in mice with potentially important regional variations.
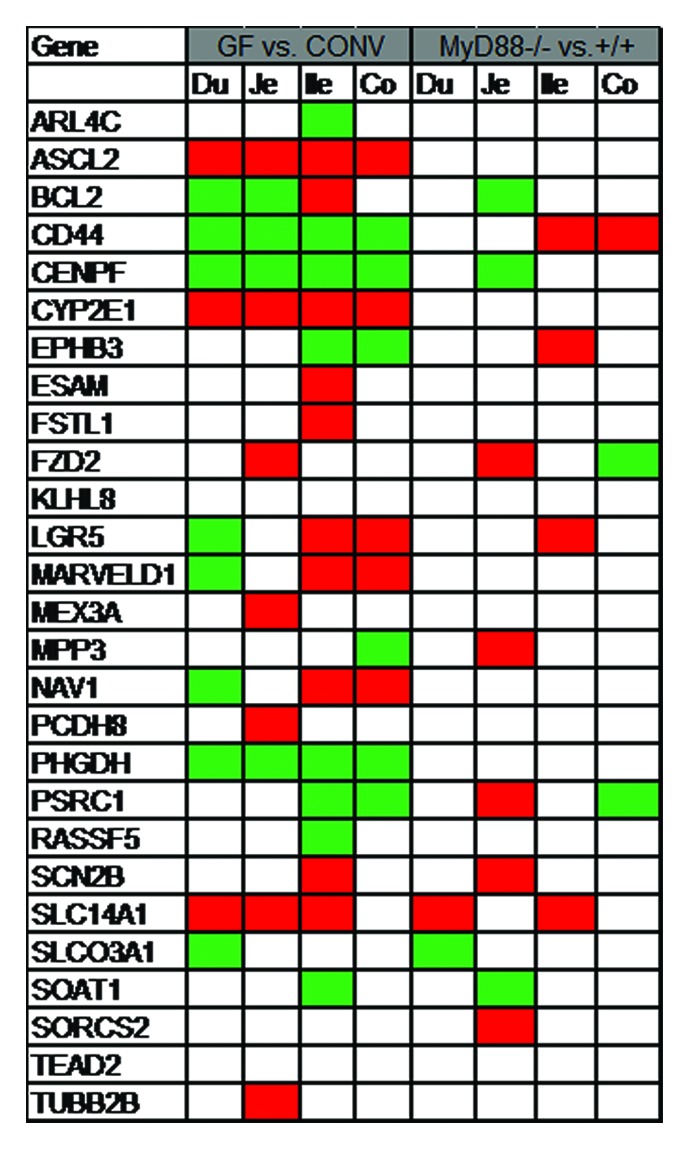
Figure 1. Intestinal stem cell gene expression signature alteration in Germfree (GF) vs. conventional (CONV) and MyD88−/− vs. MyD88+/+ mice. A set of 28 genes were selected as ISC mRNA signature based on previous reports.18,19 Publicly available database from Larsson et al. was queried for the genes of interest. Genes with significant alteration (P value < 0.05) were noted. There was no gene expression data available for PTPRO. Green, decreased; red, increased; white, unchanged; Du, duodenum; Je, jejunum; Ile, ileum; co, colon.
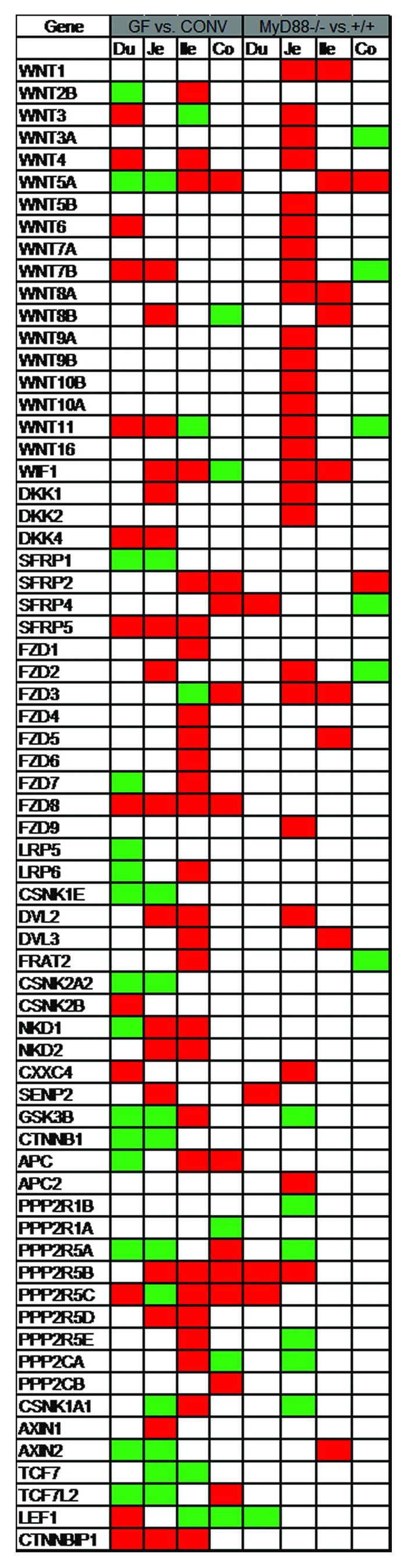
Figure 2. Wnt pathway in Germfree (GF) vs. conventional (CONV) and MyD88−/− vs. MyD88+/+ mice. Publicly available database from Larsson et al. was queried for the genes of interest. Genes with significant alteration (P value < 0.05) were noted. Green, decreased; red, increased; white, unchanged; Du, duodenum; Je, jejunum; Ile, ileum; co, colon.
Variation of the crypt dynamic and ISCs along the intestinal tract is poorly appreciated. Crypt structure is an indirect measure of the ISC activity. Regional differences in the crypt size, proliferative index, and distribution of proliferative cells along the crypt axis has been reported. The crypt length was found to be significantly shorter in the ascending colon compared with transverse and descending colon. Moreover, the total number of proliferative cells was significantly lower in ascending colon compared with other colon segments. Other suggested variations include the cell cycle time, duration of mitosis, and apoptotic index post-irradiation.20,21 King et al. also observed a regional difference in the expression pattern of a putative, yet controversial ISC marker along the crypt axis in proximal vs. distal colon in mice.22 In an elegant experiment by Leedham et al., it was observed that the total number of Lgr5+ stem cells significantly varies in different segments of the small intestine as well as between the small intestine and colon in mouse and human. There is also a regional difference in the gene expression level of stem-cell related genes and Wnt pathway modulators along the intestinal tract.23 ISCs are conceptually regarded as one entity. However, organizational differences exist in the small intestine compared with colon which plausibly dictates a distinct stem cell dynamic in the two organs. For example, two groups of stem cells are identified in the murine small intestine whereas in the colon only one group exists.24,25 Moreover, intestinal diseases show significant regional variation in disease predilection. It is conceivable that ISCs response to their environmental signals is different in distinct regions of the intestine. However, the exact mechanism of cross-regulation of ISCs by microbiota and potential regional variation requires further investigation which includes but is not limited to selectively comparing the sorted ISCs in terms of their transcriptome and in vitro organoid formation capacity from different segments of the intestinal tract in germfree and MyD88 −/− mice. Moreover, more robust techniques including RNA-seq chould be employed to allow for detection of the minute alterations across experiments.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Glossary
Abbreviations:
- CBC
crypt base columnar
- CONV
conventional
- GF
germfree
- ISC
intestinal stem cell
- TLR
toll-like receptor
References
- 1.Guarner F, Malagelada J-R. Gut flora in health and disease. Lancet. 2003;361:512–9. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 2.Carroll IM, Threadgill DW, Threadgill DS. The gastrointestinal microbiome: a malleable, third genome of mammals. Mamm Genome. 2009;20:395–403. doi: 10.1007/s00335-009-9204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 4.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–84. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neal MD, Sodhi CP, Jia H, Dyer M, Egan CE, Yazji I, Good M, Afrazi A, Marino R, Slagle D, et al. Toll-like receptor 4 is expressed on intestinal stem cells and regulates their proliferation and apoptosis via the p53 up-regulated modulator of apoptosis. J Biol Chem. 2012;287:37296–308. doi: 10.1074/jbc.M112.375881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Araki A, Kanai T, Ishikura T, Makita S, Uraushihara K, Iiyama R, Totsuka T, Takeda K, Akira S, Watanabe M. MyD88-deficient mice develop severe intestinal inflammation in dextran sodium sulfate colitis. J Gastroenterol. 2005;40:16–23. doi: 10.1007/s00535-004-1492-9. [DOI] [PubMed] [Google Scholar]
- 9.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Hobert ME, Rao AS, Neish AS, Madara JL. Bacterial activation of β-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004;287:G220–7. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, Hobert ME, Duan Y, Rao AS, He T-C, Chang EB, Madara JL. Crosstalk between NF-kappaB and beta-catenin pathways in bacterial-colonized intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2005;289:G129–37. doi: 10.1152/ajpgi.00515.2004. [DOI] [PubMed] [Google Scholar]
- 12.Duan Y, Liao AP, Kuppireddi S, Ye Z, Ciancio MJ, Sun J. β-Catenin activity negatively regulates bacteria-induced inflammation. Lab Invest. 2007;87:613–24. doi: 10.1038/labinvest.3700545. [DOI] [PubMed] [Google Scholar]
- 13.Ye Z, Petrof EO, Boone D, Claud EC, Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171:882–92. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu X, Lu R, Wu S, Sun J. Salmonella regulation of intestinal stem cells through the Wnt/β-catenin pathway. FEBS Lett. 2010;584:911–6. doi: 10.1016/j.febslet.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Lu R, Wu S, Zhang YG, Xia Y, Sartor RB, Sun J. Wnt2 inhibits enteric bacterial-induced inflammation in intestinal epithelial cells. Inflamm Bowel Dis. 2012;18:418–29. doi: 10.1002/ibd.21788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4570–7. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsson E, Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–31. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Muñoz P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–24. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 19.Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo B-K, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–91. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunter JP, Appleton DR, Dé Rodriguez MS, Wright NA, Watson AJ. A comparison of cell proliferation at different sites within the large bowel of the mouse. J Anat. 1979;129:833–42. [PMC free article] [PubMed] [Google Scholar]
- 21.Gândara RMC, Mahida YR, Potten CS. Regional differences in stem and transit cell proliferation and apoptosis in the terminal ileum and colon of mice after 12 Gy. Int J Radiat Oncol Biol Phys. 2012;82:e521–8. doi: 10.1016/j.ijrobp.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 22.King JB, von Furstenberg RJ, Smith BJ, McNaughton KK, Galanko JA, Henning SJ. CD24 can be used to isolate Lgr5+ putative colonic epithelial stem cells in mice. Am J Physiol Gastrointest Liver Physiol. 2012;303:G443–52. doi: 10.1152/ajpgi.00087.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leedham SJ, Rodenas-Cuadrado P, Howarth K, Lewis A, Mallappa S, Segditsas S, Davis H, Jeffery R, Rodriguez-Justo M, Keshav S, et al. A basal gradient of Wnt and stem-cell number influences regional tumour distribution in human and mouse intestinal tracts. Gut. 2013;62:83–93. doi: 10.1136/gutjnl-2011-301601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–71. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes KR, Gândara RMC, Javkar T, Sablitzky F, Hock H, Potten CS, Mahida YR. Heterogeneity in histone 2B-green fluorescent protein-retaining putative small intestinal stem cells at cell position 4 and their absence in the colon. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1188–201. doi: 10.1152/ajpgi.00080.2012. [DOI] [PubMed] [Google Scholar]


