Abstract
The intestinal microbiota changes dynamically from birth to adulthood. In this study we identified γ-Proteobacteria as a dominant phylum present in newborn mice that is suppressed in normal adult microbiota. The transition from a neonatal to a mature microbiota was in part regulated by induction of a γ-Proteobacteria-specific IgA response. Neocolonization experiments in germ-free mice further revealed a dominant Proteobacteria-specific IgA response triggered by the immature microbiota. Finally, a role for B cells in the regulation of microbiota maturation was confirmed in IgA-deficient mice. Mice lacking IgA had persistent intestinal colonization with γ-Proteobacteria that resulted in sustained intestinal inflammation and increased susceptibility to neonatal and adult models of intestinal injury. Collectively, these results identify an IgA-dependent mechanism responsible for the maturation of the intestinal microbiota.
Keywords: IgA, microbiota, proteobacteria, colitis, necrotizing enterocolitis
Introduction
The intestinal microbiota is highly dynamic and alters with age. Humans demonstrate a distinct change in the composition of bacterial populations from birth to adulthood, after which the intestinal bacteria remain relatively stable and are characterized by two dominant phyla, Bacteroidetes and Firmicutes.1,2 The factors contributing to the establishment of the mature microbiota in adult humans and mice have largely been attributed to alterations in the diet and changes in the oxygen content of the intestinal lumen.3 In addition, the involvement of immune mechanisms in regulation of the gut bacterial composition is suggested by altered microbiota in genetically modified mice or during experimental intestinal infections.4,5 However, the precise innate and adaptive immune mechanisms regulating the composition of the microbiota from birth to adulthood are still poorly understood.
On one end of the spectrum, incomplete development of the immune system in preterm infants is associated with the development of necrotizing enterocolitis, a disease with high mortality strongly correlated with aberrant microbiota and disrupted immune regulation.6-8 Altered control of intestinal bacteria in adults can also result in intestinal inflammation and carcinogenesis.9 Mouse models for intestinal inflammation have revealed several key factors that are involved in maintenance of homeostatic interactions of the host with intestinal bacteria. Among the most prominent innate immune factors are TLRs, the TLR/IL-1R adaptor protein MyD88 and Nod-like receptors (NLR).10,11 Adult mice lacking these proteins harbor altered microbiota and are very susceptible to intestinal injury caused by chemicals or intestinal pathogens.5,12-14 In addition, both B and T cells are indispensable for the regulation of bacterial communities under steady-state and inflammatory conditions.15 By understanding how innate and adaptive mechanisms control the transition of the intestinal microbiota into adulthood, we can identify targets for potential therapies of related diseases. We therefore hypothesized that innate or adaptive mechanisms are responsible for regulating the intestinal bacterial composition from the neonate to the adult.
Results
Proteobacteria dominates in the “immature,” but not the “mature,” microbiota
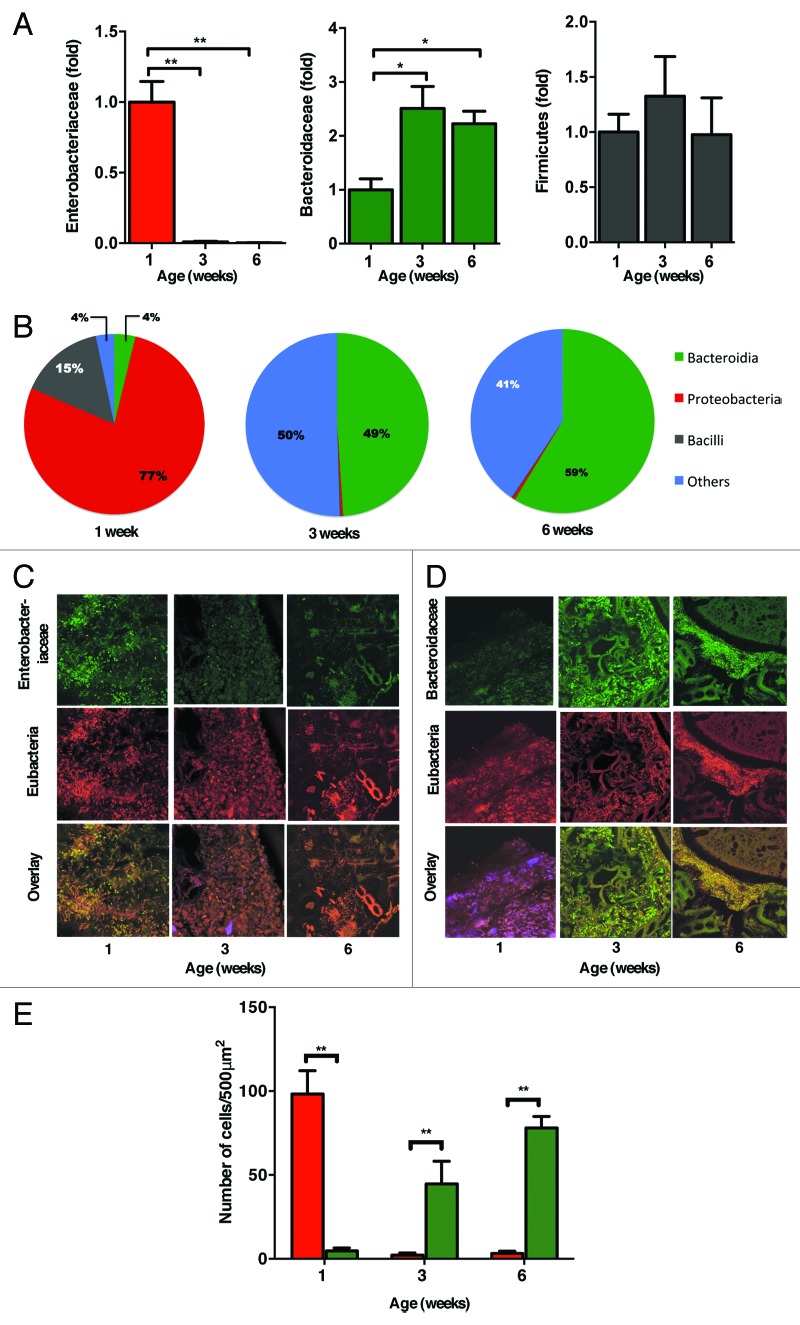
The newborn period is marked by neo-colonization of the gastrointestinal tract. The mature microbiota provides multiple beneficial effects to the host, but mechanisms determining composition of the microbiota are poorly understood.16,17 We first analyzed the dynamics of the microbiota by quantifying major bacterial phylogenetic groups in newborn and adult mice. Quantitative analysis of intestinal bacteria by RT-PCR revealed dramatic dominance of γ-Proteobacteria (Fig. 1A), in particular of Enterobacteriaceae. in newborn mice compared with adult mice. The progressive maturation of the intestinal microbiota was characterized by a significant diminution of the proportion of this genus over time. This was accompanied by the progressive expansion of Bacteroides, a phylum that together with Firmicutes dominates in the luminal colonic contents of adult mice. These results were confirmed by 454-based DNA pyrosequencing of 16S rRNA libraries from colonic contents (Fig. 1B; Fig. S1 and S2A and B). Fluorescence in situ hybridization (FISH) analysis of luminal colonic bacteria in 1-, 3- and 6-week old mice further demonstrated that Enterobacteriaceae are the major bacteria in neonatal mice (Fig. 1C and D), which comprised over 75% of the total commensal bacteria (Fig. 1E). Our results revealed that overwhelming dominance by Proteobacteria marks the neonatal, “immature” microbiota, which is in stark contrast to the composition of the mature microbiota, predominantly consisting of Bacteroidetes and Firmicutes.
Figure 1. Immature microbiota in newborn mice is characterized by a predominance of Proteobacteria. (A) The relative abundance of Proteobacteria (red), Bacteroidetes (green) and Firmicutes (black) present in colonic contents of 1, 3, and 6-week-old mice was analyzed by qRT-PCR. Data are represented as fold change relative to 1-week old mice. (B) The proportion of major bacterial phyla seen in 1, 3, and 6 weeks old mice was analyzed by 454-pyrosequencing. (C) Fluorescence in situ hybridization detection of Enterobactericeae (green) and Eubacteria (red) or (D) Bacteroidaceae (green) and Eubacteria (red) in colons of 1, 3, and 6 week old mice. (E) Quantification of the FISH positive cells per 500 μm2 section from a minimum of 10 sections from 3‒4 mice at each age group shown in (C) and (D). The data shown are representative of 3 experiments with 5‒7 mice in each group. Data are depicted as mean ± SEM (* p < 0.05, ** p < 0.01).
Immature microbiota is a cause of intestinal inflammation
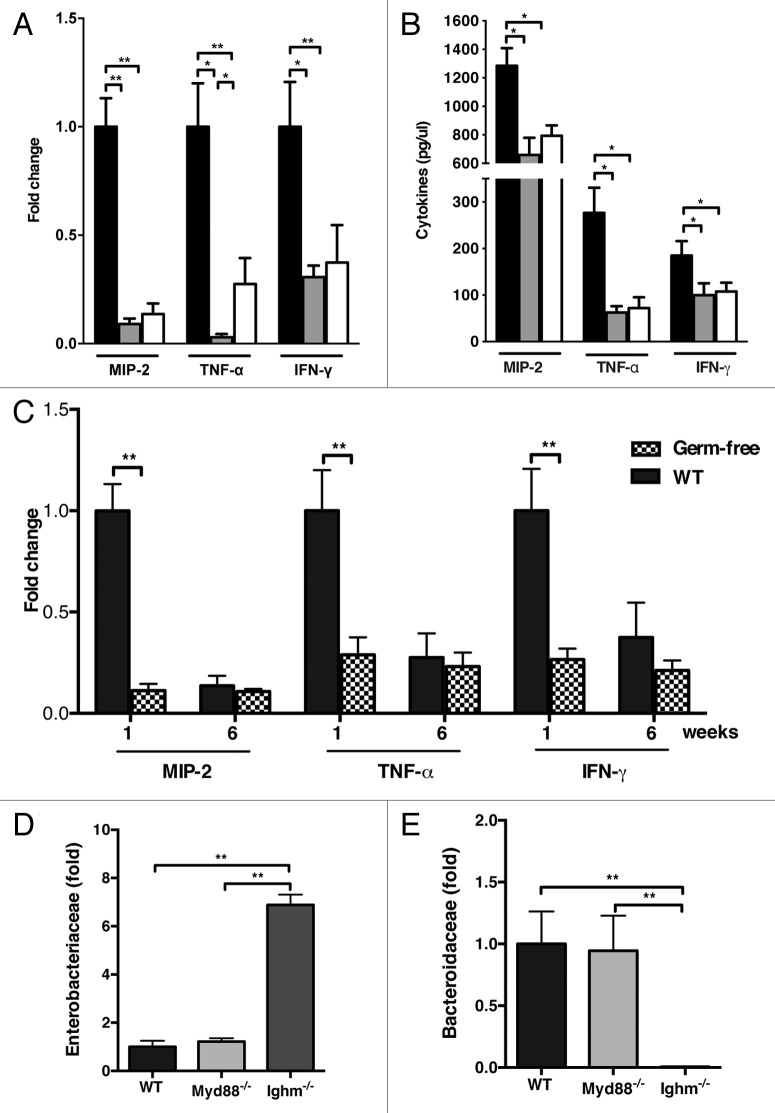
Clinical studies reveal that the developing intestine is more prone to inflammation.18 While the precise mechanisms behind this are incompletely understood, early aberrant bacterial colonization is thought to play a role.19 Our analysis of proinflammatory cytokines revealed dramatically elevated levels of MIP-2, TNF-α and IFN-γ in 1-week old mice when compared with adult controls (Fig. 2A). These results were observed at both the mRNA and protein levels in colonic tissue (Fig. 2B). To formally examine if the immature microbiota is responsible for early colonic inflammation, age-matched germ-free mice, which are microbiologically sterile, were studied in similar experiments. We found that the lack of intestinal bacteria in germ-free animals ameliorated the difference seen in newborn and adult mice (Fig. 2C).
Figure 2. B cells restrict the immature microbiota responsible for early intestinal inflammation. Quantification of MIP-2, TNF-α and IFN-γ by (A) qRT-PCR and (B) ELISA. Data represented as fold change relative to 1-week old mice. (C) Comparative expression of colonic inflammatory cytokines in conventional and GF mice was analyzed by qRT-PCR in 1 and 6 week old mice. (D) qRT-PCR-based quantification of Enterobacteriaceae and (E) Bacteroidaceae in adult WT, Myd88−/−, and Ighm−/− mice. Data represented as fold change relative to WT adult mice. The data shown are representative of 3 experiments for conventional and 2 experiments for GF mice, with 5‒7 mice in each group. Data are depicted as mean ± SEM (* p < 0.05, ** p < 0.01).
TLR and MyD88 play a limited role in regulation of intestinal microbiota
Toll-like receptors and the adaptor protein MyD88 play a major role in the regulation of host-microbial interactions.20 In addition, MyD88 is indispensable for control of commensal bacteria under steady-state and inflammatory conditions.10 We therefore investigated the involvement of MyD88 and major Toll-like receptors involved in bacterial recognition in limiting the expansion of Proteobacteria. While newborn mice lacking MyD88 showed an enhanced expression of Proteobacteria 16s rRNA, the difference was ameliorated by 3 weeks of age (data not shown). Furthermore, in agreement with previous results,21,22 no significant variation in composition of adult microbiota was observed in mice lacking MyD88 when compared with their wild-type controls (Fig. 2D). Thus, MyD88 is largely dispensable for limiting Enterobacteriaceae and establishing Bacteroidaceae in the mature microbiota.
In addition to Toll-like receptors, B-cells are also known to play a role in the regulation of the microbiota in adult mice.23-26 In order to investigate if B-cells are involved in establishing the mature microbiota, we profiled the composition of major bacterial phyla in the colon of newborn and adult Ighm−/− deficient mice that have arrested B-cell differentiation. Lack of B-cells in adult mice resulted in an altered phenotype more consistent with the immature microbiota seen in neonatal WT mice characterized by expansion of Proteobacteria spp. (Fig. 2D; Fig. S2C). These results indicate that B-cells play a role in the control of Proteobacteria by limiting their expansion in adult mice.
Induction of Proteobacteria-specific IgA in developing intestine
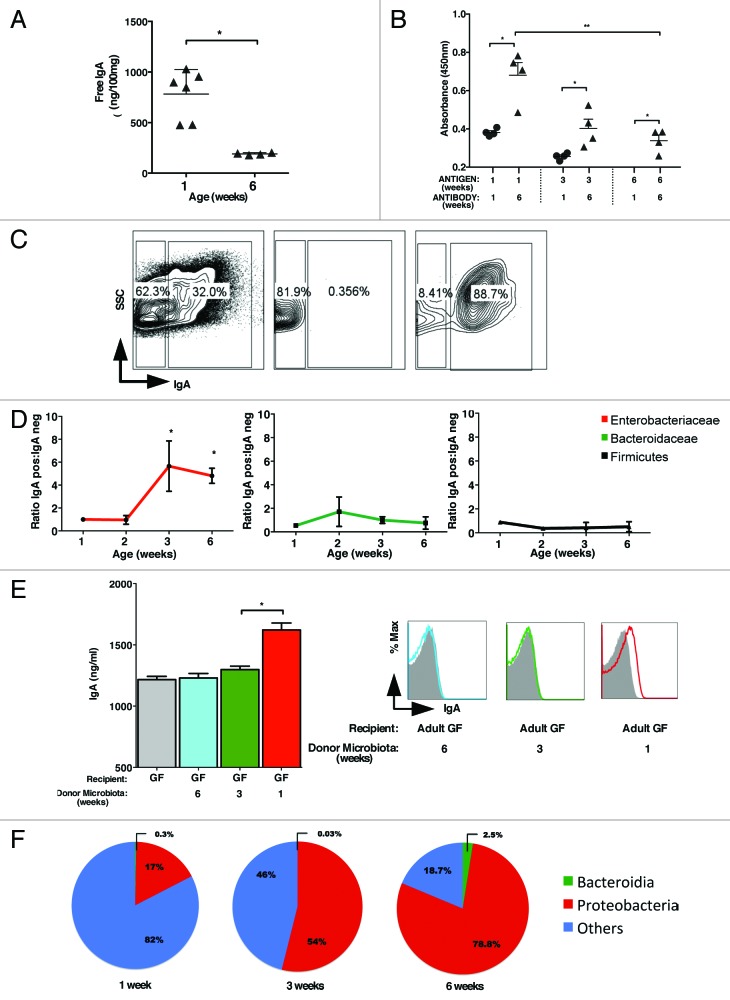
A major function for B-cells in mucosal tissues is regulation of commensal bacteria via production of IgA.27 To determine interaction of fecal IgA with bacterial antigens, we prepared antigens from 1-, 3- and 6-week old mice colonic contents. Whereas we detected greater quantities of total luminal IgA in colonic fecal contents of 1-week old mice vs. 6-week old mice (Fig. 3A), there was a limited ability of this antibody to interact with antigens prepared from intestinal bacteria (Fig. 3B). This was observed for bacterial antigens prepared from both the neonatal and adult mice. In contrast, higher antigen reactivity to commensal bacterial antigens was observed for IgA secreted in the lumen of adult mice, and that reactivity was highest to antigens obtained from 1-week old mice (Fig. 3B). To identify bacterial taxa targeted by B-cell mediated antibody responses, we sorted highly purified IgA-coated and non-coated bacteria from the same mice (Fig. 3C) and analyzed their 16S RNA by qPCR. Our experiments revealed that the majority of IgA-coated bacteria were Proteobacteria spp. (Fig. 3D), in particular Enterobacteriaceae. In contrast, Bacteroidetes and Firmicutes were nearly equally represented among IgA positive and negative bacteria. To further test if IgA preferentially targets Proteobacteria, we used a re-colonization model of GF mice utilizing colonic contents prepared from 1-, 3- and 6-week-old mice. We observed that GF mice reconstituted with the microbiota prepared from 1 week old, but not adult mice, triggered IgA secretion that interacted with Proteobacteria known to be enriched in newborn mice (Fig. 3E). These results provide an explanation for the higher reactivity of adult IgA with the immature microbiota. Furthermore, the induction of IgA by 1-wk-old but not adult commensal bacteria suggested that Proteobacteria are likely the major inducers of IgA production by B cells.
Figure 3. Preferential induction of Proteobacteria specific IgA. (A) Quantification of total colonic IgA in 1 and 6 week old mice by ELISA. (B) Detection of bacterial antigen specific IgA in 1 and 6 week old mice by ELISA. Bacterial antigens were prepared from colonic contents of 1, 3, and 6- week old mice by sonication as described in the Materials and Methods section. (C) Cell-sort based purification of IgA positive and IgA negative bacteria and their post sort analysis. The percentages shown indicate purity of IgA positive and negative populations achieved. (D) qRT-PCR analysis of IgA positive and negative Enterobacteriaceae (red), Bacteroidaceae (green) and Firmicutes (black) shown as ratio of IgA positive to IgA negative bacteria. (E) Total IgA measured in GF mice gavage fed with 1-, 3-, or 6-week microbiota measured on day 5 post-recolonization (left). Enterobacteriaceae-specific IgA in the recolonized GF mice was measured by flow-cytometry in the same sample (right). (F) Proportion of major bacterial phyla seen in pooled APCs from 4 mice per group isolated from lamina propria in 1, 3, and 6 week old mice were analyzed by 454-pyrosequencing. The data shown are representative of three experiments with 4–6 mice in each group. Data are depicted as mean ± SEM (* p < 0.05 and ** p < 0.01).
Antigen presenting cells (APCs), in particular DCs, play an important role in the induction of IgA by B-cells.28 We therefore investigated if APCs are responsible for guiding Proteobacteria-specific antibody responses. To address this possibility, we sort purified lamina propria APCs based on the CD11c cell surface marker highly expressed on dendritic cells and intestinal macrophages.29 Pooled APCs from 1-wk, 3-wk and 6-wk-old mice were analyzed by 454-based DNA pyrosequencing of 16S rRNA. We observed that despite the high frequency of Bacteroides and Firmicutes detected in the lumen of adult mice, CD11c+ cells were enriched for Proteobacterial specific 16S RNA (Fig. 3F; Fig. S3). At the same time, Bacteriodes specific signals remained similar in these APCs. Taken together, our results combined with previously established requirements for DCs30 in the regulation of antibody responses provide an explanation for the preferential induction of the Proteobacteria-specific IgA response.
IgA deficiency leads to increased colonization by Protebacteria and enhanced susceptibility to intestinal damage
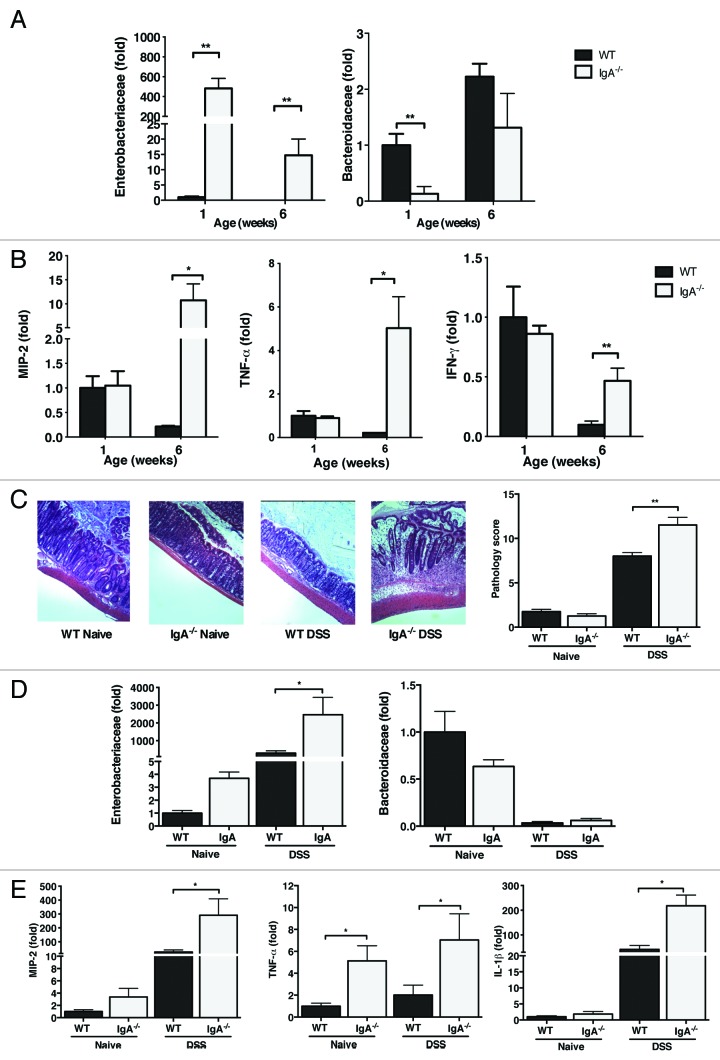
To assess the physiological importance of Proteobacteria-specific IgA seen in adult mice, the composition of the microbiota in IgA deficient mice was examined in comparison to their WT counterparts. Lack of IgA resulted in a significant expansion of Proteobacteria seen in colon of newborn mice (Fig. 4A). Furthermore, in contrast to WT mice, IgA deficient animals harbored increased numbers of Proteobacteria even as adults. These conclusions were established by qRT-PCR and further confirmed by FISH analysis (data not shown). This phenotype persisted after co-housing of WT and Igha−/− mice with no transmission between groups (Fig. S4A). It appears that Proteobacteria represents a persistent phylum in the absence of IgA. Because of the observed association between Proteobacteria and intestinal inflammation,4,31 we investigated if the Proteobacteria-enriched microbiota seen in IgA deficient mice result in elevated levels of colonic pro-inflammatory mediators. Both WT and Igha−/− had comparable and elevated expression of MIP-2, TNF-α and IFN-γ in their colons at 1-week of life likely due to the high frequency of Proteobacteria seen in both strains of mice (Fig. 4B). In addition, the persistence of Proteobacteria in 6-week-old Igha−/− mice resulted in sustained or enhanced expression of these pro-inflammatory factors. Thus lack of IgA resulted not only in expansion of Proteobacteria, but also triggered a persistent and exaggerated inflammatory phenotype.
Figure 4. IgA deficiency leads to persistence of Proteobacteria and enhanced susceptibility to DSS colitis. (A) Quantification of Enterobacteriaceae and Bacteroidaceae in the colonic contents of 1- and 6- week old IgA deficient mice by qRT-PCR in comparison to their WT controls. The data are represented as fold change from 1-week-old WT mice. (B) Expression of MIP-2, TNF-α, and IFN-γ in colon of 1- and 6-week old WT and IgA deficient mice. (C) WT and IgA deficient mice were treated with 2% DSS in their drinking water for 10 d. Representative images of colons from control and DSS treated WT and IgA- deficient mice (left) and their respective colitis scores (right) are shown. (D) Relative expression of Enterobacteriaceae and Bacteroidaceae in Naïve and DSS exposed mice. (E) Expression of pro-inflammatory cytokines from colon in DSS treated mice. The data shown are representative of three experiments with 4–6 mice in each group. Data are depicted as mean ± SEM (* p < 0.05 and ** p < 0.01).
To determine if the increased abundance of Proteobacteria seen in IgA deficient mice would result in increased susceptibility to inflammatory states, we first utilized a model of DSS-induced colonic injury in adult mice. IgA deficient mice showed more severe and extensive colitis than WT mice represented by higher pathology scores and higher expression of pro-inflammatory cytokines (Fig. 4C). Additionally, DSS colitis in IgA deficient mice was further associated with an expansion of Proteobacteria, in particular Enterobacteriaceae (Fig. 4D), and increased expression of the pro-inflammatory cytokines MIP-2, TNF-α and IL1β (Fig. 4E).
Using a different model in which PAF/LPS are administered to pre-weaned 10-d-old mice, Igha−/− pups were also found to be more susceptible to intestinal injury when compared with their WT controls (Fig. S4B). These results suggest that Proteobacteria and its persistence as seen in IgA deficient mice is dysbiotic and results in enhanced susceptibility to intestinal injury in neonatal and adult mice.
Discussion
In our studies here, we extensively characterized the dynamics of bacterial composition in newborn and adult mice. We established that the microbiota in newborn mice was dominated by γ-Proteobacteria, mostly consisting of Enterobacteriaceae. We observed that the transition of this “immature” to a more “mature” microbiota in adult mice was characterized by replacement of Enterobacteriaceae with Bacteroidetes and Firmicutes. Upon investigation of the mechanisms regulating this unique transition, we established that, while TLR/MyD88 played a role in the maturation of the intestinal microbiota, B cells, and in particular IgA, were indispensable for this process. Analysis of IgA specificity in conventional adult mice colonized with commensal intestinal bacteria revealed that IgA was largely directed toward Enterobacteriaceae. We found that lamina propria (LP) CD11c+ antigen-presenting cells preferentially sample Enterobacteriaceae. Furthermore, experiments with neocolonization of germ-free (GF) mice with immature and adult microbiota revealed a potent IgA response targeting Enterobacteriaceae. Finally, examination of IgA-deficient mice strongly supported a major role for IgA in restricting Enterobacteriaceae in adult mice. Lack of IgA resulted in incomplete transition to a mature microbiota and greatly enhanced susceptibility in both neonatal and adult models of intestinal injury. Taken together, these results reveal that a targeted IgA response against Enterobacteriaceae is crucial in regulating maturation of the intestinal microbiota and in limiting Proteobacteria-associated colonic inflammation.
The transition of the microbiota in humans from the neonate to the adult is unique and follows a consistent pattern, with slight variations based on diet and mode of delivery at birth.32-34 The role of immune mechanisms in the regulation of the microbiota has only recently become the subject of investigation. In this study, we identified an IgA-dependent mechanism responsible for the establishment of the “mature” microbiota in adult mice. While the microbiota of newborn mice, which we defined as “immature,” was dominated by γ-Proteobacteria, the intestinal bacteria of adult mice were dominated by Bacteroidetes and Firmicutes. We as well as others have shown that the pre-weaned murine intestine is pro-inflammatory.35,36 This proinflammatory environment has been shown to produce nitrate, which consequently confers a growth advantage to members of the Enterobacteriaceae genus, including E. coli.31,37 We show here that genus-specific IgA production is important in limiting expansion of Enterobacteriaceae. Failure to produce IgA resulted in abnormal composition of the intestinal bacteria in adult mice that was reminiscent of the immature microbiota, resulting in enhanced colonic inflammation. A major role for immature microbiota in driving inflammation is implicated by marked decreased in production of proinflammatory cytokines as Proteobacteria were progressively replaced by Bacteroidetes and Firmicutes in adult mice. The finding of non-age-dependent lower levels of intestinal pro-inflammatory cytokines during the neonatal period in germ-free mice strongly supports the concept that bacterial colonization is essential for colonic inflammation. Collectively, our findings argue in favor of a major role for Proteobacteria in driving colonic inflammation, which is particular relevant in neonatal mice colonized with the immature microbiota.
Our studies show that, in conventionally raised neonatal mice, the development of selective induction of IgA against Proteobacteria resulted in the gradual loss of this phylum. The lack of IgA specificity to Proteobacteria despite a significant amount seen in 1-week-old mice may be attributable to non-specific IgA from breast-milk. Whether specificity to Proteobacteria develops in breast milk as well or solely as an intrinsic response warrants investigation. We further showed that lack of IgA prevented the suppression of Proteobacteria and resulted in dysbiosis associated with increased intestinal inflammation. Previous studies examining mouse mutants lacking secretory immunoglobulins have identified an altered microbial profile in the small intestine.24-26 While these studies did now show an increase in Proteobacteria, the microbiota in the small and large intestine are known to be unique38 and differences in the mouse models can alter the microbiota based on the presence or lack of IgM and IgG. Of note, there appears to be a reciprocal decrease in Bacteroidetes in mice that show an expansion of Proteobacteria, we can speculate that this is due to competition or inhibition by Proteobacteria, however the precise mechanism is unknown and is being investigated. Neo-colonization experiments in germ-free mice with intestinal bacteria isolated from 1, 3 and 6 weeks old mice further support the role of Proteobacteria in inducing a specific IgA response. An equivalent response could potentially be triggered at a later time point in mice colonized with mature microbiota as a result of lower baseline IgA producing plasma cells. However, the fact that a robust response is seen at 5 d with the Proteobacteria-rich “immature” microbiota highlights the importance of this phylum in triggering the adaptive immune system.
It appears that IgA deficiency predisposed these mice to a high susceptibility to colonic damage. These results are of significance since IgA deficiency is one of the most common immunodeficiencies seen in humans and there is an increased susceptibility to colitis in these individuals.39 Of particular note, 10–15% of IgA deficient individuals are susceptible to inflammatory and autoimmune gastrointestinal disorders including Crohn disease and ulcerative colitis.40 Mouse models of immunoglobulin deficient mice have shown altered transcriptome profiles,26 indicating compensation by other immune mechanisms of defense. This may explain the mild baseline phenotype seen in IgA deficient mice but could also contribute to the enhanced susceptibility when challenged with DSS. It is also interesting that there is a reduced incidence of NEC in preterm infants supplemented with breast milk which is rich in IgA.41 An association of NEC with Proteobacteria42 suggests that IgA specific to Proteobacteria play a role in preventing NEC. In our studies, IgA deficiency did not result in spontaneous colitis, but there was an increased susceptibility to intestinal injury seen in both adult and newborn gut injury models. Furthermore, compared with control mice, some aging IgA deficient mice developed spontaneous rectal prolapse (our unpublished observation). Our data support the hypothesis that complete or relative insufficiency in IgA against Proteobacteria, as seen in newborns, results in a persistence of Proteobacteria and increased susceptibility to inflammation and injury.
Utilizing FlSH staining, Tsuruta et al. have shown that Enterobacteriaceae have preferential coating of IgA in 9-week-old mice.43 Our study further shows that this specificity develops over time and with exposure to Proteobacteria, as seen in the experiments with specific re-colonization of germ-free mice. The mechanism of production of Proteobacteria-specific IgA appears to be mediated by the selective uptake of these bacteria by DCs. The mechanisms by which DCs take up this more invasive phylum could include phagocytosis.4 Proteobacteria-specific IgA antibodies may then act to establish the normal microbiota in adult mice by steric hindrance or as a potent neutralizer of mitogenic activity.44 The precise molecular mechanism of IgA induction to Proteobacteria may be T-cell dependent or independent45 and warrants further investigation. The lack of histologically evident colonic inflammation in non-stressed IgA deficient mice may be due to compensation by other immunoglobulins, in particular IgM and IgG, since it has been found that IgM levels in IgA deficiency are increased46 and IgM plays a role in restricting colonic bacteria.5
In summary, our study highlights the importance of IgA in establishing a beneficial commensal bacterial population via selective suppression of Proteobacteria needed for reducing susceptibility to colonic injury and inflammation.
Materials and Methods
Mice
C57BL/6 mice were obtained from the University of Texas Southwestern (UTSW) Medical Center Mouse Breeding Core Facility. Germ-free C57BL/6 mice have been described previously.47,48 Igha−/− mice were initially purchased from Jackson Laboratory and bred for 3 generations in our animal facility prior to experimentation to ensure a stable microbiota composition within the environment of our animal facility. Myd88−/− mice were generously provided by S. Akira and have been described previously in experimental colitis studies.5 All mice were maintained at the American Association of Laboratory Animal Care-accredited animal facility at the UTSW Medical Center. All of the animals that were used were age and sex-matched. All mice except GF animals were maintained in the same animal room.
RT-PCR
Total RNA was purified using TRIZOL reagent and subjected to first-strand cDNA synthesis by using SuperScript III (Invitrogen). Real-time PCR was performed using Ssofast PCR Master Mix (Biorad) and the MyIQ real-time PCR machine according to the manufacturer’s instructions. For colonic cytokines, data were analyzed by the Ct threshold cycle method with normalization for starting template performed using a housekeeping gene, SRP-14. For bacterial 16S rRNA analysis, samples were normalized to Eubacteria. Primer sequences were used as follows: Murine SRP-14 5′-AAGTGTCTGTTGAGAGCCACGGAT-3′ and 5′-CTGTCACTGTGCTGGTTTGCTCTT-3′; MIP-2 5′-CTCTCAAGGGCGGTCAAAAAGTT-3′ and 5′-TCAGACAGCGAGGCACATCAGGTA-3′; TNF-α 5′-CCACCACGCTCTTCTGTCTAC-3′ and 5′-TGGGCTACAGGCTTGTCACT-3′; IFN-γ 5′-AACGCTACACACTGCATCT-3′ and 5′-GAGCTCATTGAATGCTTGG-3′; IL-1β 5′-CCTTCCAGGATGAGGACATGA-3′ and 5′-TGAGTCACAGAGGATGG-GCTC-3′. Bacterial primers used are as follows: Eubacteria 5′-ACTCCTACGGGAGGCAGCAGT-3′ and 5′-ATTACCGCGGCTGCTGGC-3′; γ-Proteobacteria 5′-TAACGCTTGGGAATCTGCCTRTT-3′ and 5′-CATCTRTTAGCGCCAGGCCTTGC-3′; Enterobacteriaceae 5′-GTGCCAGCMGCCGCGGTAA-3′ and 5′-GCCTCAAGGGCACAACCTCCAAG-3′; Bacteroidetes 5′-GGTTCTGAGAGGAGGTCCC-3′ and 5′-GCTGCCTCCCGTAGGAGT-3′; Firmicutes 5′-GGAGYATGTGGTTTAATTCGAAGCA-3′ and 5′-AGCTGACGACAACCATGCAC-3′.
454-based DNA pyrosequencing
Colonic contents were extracted and genomic DNA isolated using DNA stool kit (QIAGEN). Sort purified lamina propria CD11c+ cells pooled from 4 mice per age group were used to prepare bacterial DNA using DNeasy Blood and Tissue Kit (QIAGEN). DNA prepared was further purified and cleaned using a DNA clean-up kit (MoBio). Bacterial Tag-Encoded FLX 454-Pyroseqencing was performed using bar-coded primers 28F-519R for the V1–V3 region of the 16S rRNA gene by the Research and Testing Laboratory (Lubbock, TX). Three thousand reads per sample were obtained. FASTA formatted sequences were analyzed for quality and sequences that had low quality tags, primer, ends or that failed to be at least 250bp in length were excluded from the analysis.
Once FASTA formatted sequences pass quality-controlled checks as described above, the identity of each remaining sequence was first sorted such that the FASTA formatted file contained reads from longest to shortest. These sequences were then clustered into OTU clusters with 96.5% identity (3.5% divergence) using USEARCH. For each cluster the seed sequence was put into a FASTA formatted sequence file. This file was then queried against a database of high quality sequences derived from NCBI using a distributed .NET algorithm that utilizes BLASTN+ (KrakenBLAST, www.krakenblast.com). Using a .NET and C# analysis pipeline the resulting BLASTN+ outputs were compiled and data reduction analysis performed.
Based upon the above BLASTn+ derived sequence identity percentage the sequences were classified at the appropriate taxonomic levels based upon the following criteria. Sequences with identity scores, to well characterized 16S sequences, greater than 97% identity (< 3% divergence) were resolved at the species level, between 95% and 97% at the genus level, between 90% and 95% at the family and between 85% and 90% at the order level, 80 and 85% at the class, and 77% to 80% at phyla. Any match below this percent identity was discarded. In addition, the HSP was at least 75% of the query sequence or it was discarded, regardless of identity.
After resolving based upon these parameters, the percentage of each organism was individually analyzed for each sample providing relative abundance information within and among the individual samples based upon relative numbers of reads within each. Evaluations presented at each taxonomic level, including percentage compilations represent all sequences resolved to their primary identification or their closest relative.
ELISA
Each colon sample was weighed, cut longitudinally, washed extensively with PBS and incubated overnight in cell culture media (RPMI + 10% FBS). MIP-2, TNF-α, IL-1β and IFN-γ concentrations in the supernatants were determined with ELISA kits from eBioscience according to the manufacturer’s instructions.
To detect unbound IgA in colonic contents, colonic contents were obtained and normalized by weight of stool. Samples were then vortexed and passed through a 0.22 μm filter with a vacuum (Millipore, Steriflip). Samples were then spun at 13,000 RPM and the supernatant obtained. The amount of IgA was measured by ELISA (Bethyl Laboratories). To prepare antigens from colonic contents for ELISA, stool from 1-week, 3-week, and 6-week old mice was normalized by weight. Samples were initially vortexed for 5 min and then spun down. The pellet was then resuspended in PBS, sonicated for 10 min and spun down for 15 min at maximum RPM. Supernatant was taken and protein measured using the Bradford assay. ELISA plates were coated with 1 mg/ml of antigen for specific age groups. IgA from colonic contents were loaded at 1:10 dilution in triplicates. Anti-IgA Biotin and HRP-Conjugated Streptavidin were used to detect binding of IgA.
Isolation of IgA positive bacteria
Fecal colonic contents were disrupted in PBS by vortexing for 5 min, and insoluble material was removed by filtering through a 20 μm filter (Millipore). The bacteria were washed twice in PBS with 5 mM EDTA and 2% FBS before resuspension in Alexa 488-conjugated anti-IgA monoclonal antibodies (BD Biosciences). Commensal bacteria-specific IgA were assessed by staining luminal bacteria for 30 min on ice. Bacterial sorting was then performed using MoFlo High Speed Sorter (Beckman Coulter) for IgA positive and IgA negative bacteria.
Histopathology and in situ hybridization
Portions of the colon were fixed in the Carnoy fixative, embedded in paraffin and stained with hematoxylin and eosin (H&E). For in situ hybridization, fixed colon samples were deparaffinized and rehydrated in hybridization buffer. The colon sections utilizing specific bacterial probes conjugated to Alexa-532, EUB338 general bacteria probe for a bacterial 16S rRNA gene (5′-GCT GCC TCC CGT AGG AGT-3′) and Alexa-488 for Enterobacteriaceae, ENTBAC (5′-CAT GAA TCA CAA AGT GGT AAG CFC C-3′) or Bacteroides, BAC303 (5′-CCA ATG TGG GGG ACC TT-3′). Counting of bacterial cells was performed using Image J software with background correction for Eubacteria present. Histological changes were analyzed in a double-blind fashion using a 17-point scale as follows: For crypt integrity: 0, normal; 1, irregular crypts; 2, mild crypt loss; 3, severe crypt loss; 4, complete crypt loss with an intact epithelial cell layer; 5, complete loss of crypts and surface epithelium (< 10 crypt width); and 6, complete loss of crypts and surface epithelium (> 10 crypts). For infiltration of inflammatory cells into the mucosa: 0, normal; 1, mild; 2, modest; and 3, severe. For infiltration of the submucosa: 0, normal; 1, mild; 2, modest; and 3, severe. For infiltration of the muscle: 0, normal; 1, mild; 2, modest; and 3, severe. These scores were added, resulting in a total scoring range of 0 to 15.
Isolation of intestinal antigen presenting cells
Isolation of LP cells was performed as follows. The small intestine was removed and carefully cleaned of mesentery and the Peyer patches were excised. The intestine was then opened longitudinally, washed of fecal contents, cut into smaller sections and subjected to 2 sequential incubations in PBS with 5 mM EDTA and 1mM DTT at 37°C with agitation to remove epithelial cells. The solution was discarded between incubation steps and replaced. The remaining tissue was agitated in PBS and then filtered through a 100-μm strainer. The tissue was then incubated for 30 min with gentle agitation in 0.4 mg/ml of Collagenase D and 50 mg/ml of DNase I at 37C. The samples were then washed through a strainer (100 μm). LP cells were then stained with CD45, CD11c and CD11b and sort purified using MoFlo for CD45 and CD11c positivity. Consistent sort purification of approximately 93% was obtained.
Experimental design
Colonization of germ free (GF) mice with age-specific microbiota
Fecal colonic contents were vortexed for 5 min and then spun down. The pellet was then resuspended in PBS and insoluble material removed by filtering through a 20 μm filter (Millipore). The bacteria were washed twice in PBS and GF mice were colonized by gavaging 200μl (~107 CFUs) of colonic contents prepared as described above on day 0 from 1, 3 and 6-week-old conventional C57BL/6 mice. Mice were sacrificed after 5 d. Fecal colonic contents from recolonized and control GF mice were analyzed for the presence of total IgA by ELISA. To detect Enterobacteriaceae-specific IgA, E. coli were isolated from 1 wk old conventional C57BL/6 mice on selective Chromagar plates. Bacterial colonies were expanded and used as antigen in the FACS-based assay. Specifically, 3 × 106 of isolated bacteria were incubated with unbound IgA prepared from gavage fed recolonized and control GF mice for 30 min at 4°C. Samples were then spun at 1500 RPM for 5 min, washed twice and stained with FITC conjugated anti-IgA for analysis by flow cytometry. The data were analyzed with FlowJo software (TreeStar). Two experiments were performed with a total of 4–6 mice in each group.
Adult DSS-colitis model
Adult mice were treated with 2% (w/v) DSS (MP Biomedicals) in their drinking water for 10 d as described previously.5 Control animals were treated with regular water without DSS. After 10 d, mice were sacrificed and colonic samples obtained for RNA isolation, ELISA and histological analysis.5 3 experiments were performed with 4‒6 mice in each group.
Neonatal PAF/LPS model of intestinal injury
Gut mucosal injury was induced in 12‒14 d mouse pups by intraperitoneal administration of platelet-activating factor (PAF, 50 μg/kg) and LPS (1mg/kg).49-51 Mice were sacrificed 2 h after PAF and LPS administration and mucosal injury was graded in a blinded-fashion using a scale as described previously52 and below. Three experiments were performed with 4‒6 mice in each group.
Statistical analysis
Statistical differences were analyzed by ANOVA and t-test using Prism 6 GraphPad Software, Inc. (San Diego, CA). Values are expressed as mean ± SEM, with statistical significance identified as a p value < 0.05.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgements
We would like to thank Cassie Behrendt and Charmaine Clements for germ-free mouse husbandry. This research was supported by NIH R01 AI08263 and a Burroughs Wellcome Foundation Investigators in the Pathogenesis of Infectious Diseases award to FY, NIH R01 DK070855 and the Howard Hughes Medical Institute to LVH, NIH R01 HL093535, Children’s Medical Center Foundation Grant and the William Buchanan Chair to RCS and NIH K12 1K12HD068369 and Children’s Medical Center Foundation Grant to JM.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/gutmicrobes/article/26489
Glossary
Abbreviations:
- NLR
Nod-like receptor
- LP
lamina propria
- GF
germ free
- DSS
dextran sodium sulfate
- FISH
fluorescence in situ hybridization
- NEC
necrotizing enterocolitis
- APC
antigen presenting cells
- DC
dendritic cells
References
- 1.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirkland D, Benson A, Mirpuri J, Pifer R, Hou B, DeFranco AL, Yarovinsky F. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity. 2012;36:228–38. doi: 10.1016/j.immuni.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, Theriaque D, Li N, Sharma R, Hudak M, et al. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One. 2011;6:e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afrazi A, Sodhi CP, Richardson W, Neal M, Good M, Siggers R, Hackam DJ. New insights into the pathogenesis and treatment of necrotizing enterocolitis: Toll-like receptors and beyond. Pediatr Res. 2011;69:183–8. doi: 10.1203/PDR.0b013e3182093280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weitkamp JH, Koyama T, Rock MT, Correa H, Goettel JA, Matta P, Oswald-Richter K, Rosen MJ, Engelhardt BG, Moore DJ, et al. Necrotising enterocolitis is characterised by disrupted immune regulation and diminished mucosal regulatory (FOXP3)/effector (CD4, CD8) T cell ratios. Gut. 2013;62:73–82. doi: 10.1136/gutjnl-2011-301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tlaskalová-Hogenová H, Stěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, Rossmann P, Hrnčíř T, Kverka M, Zákostelská Z, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–20. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- 11.Cheesman SE, Guillemin K. We know you are in there: conversing with the indigenous gut microbiota. Res Microbiol. 2007;158:2–9. doi: 10.1016/j.resmic.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Salcedo R, Worschech A, Cardone M, Jones Y, Gyulai Z, Dai RM, Wang E, Ma W, Haines D, O’hUigin C, et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J Exp Med. 2010;207:1625–36. doi: 10.1084/jem.20100199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupaul-Chicoine J, Yeretssian G, Doiron K, Bergstrom KS, McIntire CR, LeBlanc PM, Meunier C, Turbide C, Gros P, Beauchemin N, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32:367–78. doi: 10.1016/j.immuni.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Macpherson AJ, Geuking MB, McCoy KD. Homeland security: IgA immunity at the frontiers of the body. Trends Immunol. 2012;33:160–7. doi: 10.1016/j.it.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chinen T, Rudensky AY. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol Rev. 2012;245:45–55. doi: 10.1111/j.1600-065X.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- 18.Neu J, Walker WA. Necrotizing enterocolitis. N Engl J Med. 2011;364:255–64. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bury RG, Tudehope D. Enteral antibiotics for preventing necrotizing enterocolitis in low birthweight or preterm infants. Cochrane Database Syst Rev. 2001:CD000405. doi: 10.1002/14651858.CD000405. [DOI] [PubMed] [Google Scholar]
- 20.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–56. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–8. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reikvam DH, Derrien M, Islam R, Erofeev A, Grcic V, Sandvik A, Gaustad P, Meza-Zepeda LA, Jahnsen FL, Smidt H, et al. Epithelial-microbial crosstalk in polymeric Ig receptor deficient mice. Eur J Immunol. 2012;42:2959–70. doi: 10.1002/eji.201242543. [DOI] [PubMed] [Google Scholar]
- 25.Sait L, Galic M, Strugnell RA, Janssen PH. Secretory antibodies do not affect the composition of the bacterial microbiota in the terminal ileum of 10-week-old mice. Appl Environ Microbiol. 2003;69:2100–9. doi: 10.1128/AEM.69.4.2100-2109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, Orandle M, Mayer L, Macpherson AJ, McCoy KD, et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med. 2011;17:1585–93. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagarasan S, Honjo T. Regulation of IgA synthesis at mucosal surfaces. Curr Opin Immunol. 2004;16:277–83. doi: 10.1016/j.coi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Rescigno M. Intestinal dendritic cells. Adv Immunol. 2010;107:109–38. doi: 10.1016/B978-0-12-381300-8.00004-6. [DOI] [PubMed] [Google Scholar]
- 29.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–55. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swiatczak B, Rescigno M. How the interplay between antigen presenting cells and microbiota tunes host immune responses in the gut. Semin Immunol. 2012;24:43–9. doi: 10.1016/j.smim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–11. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salminen S, Gibson GR, McCartney AL, Isolauri E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53:1388–9. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guaraldi F, Salvatori G. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol. 2012;2:94. doi: 10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlisle EM, Poroyko V, Caplan MS, Alverdy J, Morowitz MJ, Liu D. Murine gut microbiota and transcriptome are diet dependent. Ann Surg. 2013;257:287–94. doi: 10.1097/SLA.0b013e318262a6a6. [DOI] [PubMed] [Google Scholar]
- 35.Mirpuri J, Brazil JC, Berardinelli AJ, Nasr TR, Cooper K, Schnoor M, Lin PW, Parkos CA, Louis NA. Commensal Escherichia coli reduces epithelial apoptosis through IFN-alphaA-mediated induction of guanylate binding protein-1 in human and murine models of developing intestine. J Immunol. 2010;184:7186–95. doi: 10.4049/jimmunol.0903116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirpuri J, Yarovinsky F. IL-6 signaling SOCS critical for IL-12 host response to Toxoplasma gondii. Future Microbiol. 2012;7:13–6. doi: 10.2217/fmb.11.147. [DOI] [PubMed] [Google Scholar]
- 37.Spees AM, Wangdi T, Lopez CA, Kingsbury DD, Xavier MN, Winter SE, Tsolis RM, Bäumler AJ. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. MBio. 2013;4 doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–40. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang N, Shen N, Vyse TJ, Anand V, Gunnarson I, Sturfelt G, Rantapää-Dahlqvist S, Elvin K, Truedsson L, Andersson BA, et al. Selective IgA deficiency in autoimmune diseases. Mol Med. 2011;17:1383–96. doi: 10.2119/molmed.2011.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yel L. Selective IgA deficiency. J Clin Immunol. 2010;30:10–6. doi: 10.1007/s10875-009-9357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleary TG. Human milk protective mechanisms. Adv Exp Med Biol. 2004;554:145–54. doi: 10.1007/978-1-4757-4242-8_14. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, Antonopoulos DA, Chang EB, Claud EC. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. ISME J. 2009;3:944–54. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuruta T, Inoue R, Iwanaga T, Hara H, Yajima T. Development of a method for the identification of S-IgA-coated bacterial composition in mouse and human feces. Biosci Biotechnol Biochem. 2010;74:968–73. doi: 10.1271/bbb.90801. [DOI] [PubMed] [Google Scholar]
- 44.Norrby-Teglund A, Ihendyane N, Kansal R, Basma H, Kotb M, Andersson J, Hammarström L. Relative neutralizing activity in polyspecific IgM, IgA, and IgG preparations against group A streptococcal superantigens. Clin Infect Dis. 2000;31:1175–82. doi: 10.1086/317423. [DOI] [PubMed] [Google Scholar]
- 45.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–6. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 46.Plebani A, Mira E, Mevio E, Monafo V, Notarangelo LD, Avanzini A, Ugazio AG. IgM and IgD concentrations in the serum and secretions of children with selective IgA deficiency. Clin Exp Immunol. 1983;53:689–96. [PMC free article] [PubMed] [Google Scholar]
- 47.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raetz M, Hwang SH, Wilhelm CL, Kirkland D, Benson A, Sturge CR, Mirpuri J, Vaishnava S, Hou B, Defranco AL, et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-γ-dependent elimination of Paneth cells. Nat Immunol. 2013;14:136–42. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsueh W, González-Crussi F, Arroyave JL. Platelet-activating factor: an endogenous mediator for bowel necrosis in endotoxemia. FASEB J. 1987;1:403–5. doi: 10.1096/fasebj.1.5.3678700. [DOI] [PubMed] [Google Scholar]
- 50.Sun X, Rozenfeld RA, Qu X, Huang W, Gonzalez-Crussi F, Hsueh W. P-selectin-deficient mice are protected from PAF-induced shock, intestinal injury, and lethality. Am J Physiol. 1997;273:G56–61. doi: 10.1152/ajpgi.1997.273.1.G56. [DOI] [PubMed] [Google Scholar]
- 51.Maheshwari A, Kelly DR, Nicola T, Ambalavanan N, Jain SK, Murphy-Ullrich J, Athar M, Shimamura M, Bhandari V, Aprahamian C, et al. TGF-β2 suppresses macrophage cytokine production and mucosal inflammatory responses in the developing intestine. Gastroenterology. 2011;140:242–53. doi: 10.1053/j.gastro.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirpuri J, Sotnikov I, Myers L, Denning TL, Yarovinsky F, Parkos CA, Denning PW, Louis NA. Lactobacillus rhamnosus (LGG) regulates IL-10 signaling in the developing murine colon through upregulation of the IL-10R2 receptor subunit. PLoS One. 2012;7:e51955. doi: 10.1371/journal.pone.0051955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.