Abstract
Due to the recent rapid expansion in our understanding of the composition of the gut microflora and the consequences of altering that composition the question of how bacteria colonise mucus layers and interact with components of mucus, such as mucin, is now receiving widespread attention. Using a combination of mucus secreting cells, and a novel mucin microarray platform containing purified native mucins from different sources we recently demonstrated that two gastrointestinal pathogens, Helicobacter pylori and Campylobacter jejuni, colonise mucus by different mechanisms. This result emphasizes the potential for even closely related bacteria to interact with mucus in divergent ways to establish successful infection. Expanding the use of the mucin arrays described in the study to other microorganisms, both pathogenic and commensal, should lead to the discovery of biologically important motifs in bacterial-host interactions and complement the use of novel in vitro cell models, such as mucus secreting cell lines.
Keywords: Helicobacter pylori, Campylobacter jejuni, mucus, mucin, microarray
Introduction
Mucosal surfaces serve as a portal of entry for the majority of infections that occur in humans and animals. All mucosal surfaces are covered in a layer of mucus. In the gastrointestinal tract this mucus is colonised by tens of trillions of commensal organisms, essential for the development and health of the host. Mucus in the murine colon and the stomach has been shown to consist of an inner closely adherent mucus layer and an outer less dense, loosely adherent layer.1,2 The small intestine has a single loosely adherent layer.2 In the colon the outer mucus layer is colonised by commensal bacteria while the inner mucus layer is devoid of microbiota.1 The protective function of the inner mucus layer can be disrupted in disease, allowing access of organisms to the underlying epithelium. For example, the mucus layer has been shown to be depleted in animal models of colitis, and also in humans with ulcerative colitis resulting in bacterial penetration of the inner layer.3
A complex, mutually dependent relationship is established between the gut microbiota and the human host from birth which persists throughout life.4 Initial cross talk between the host and gut microbiota influences postnatal intestinal epithelial differentiation by altering gene expression and improves immune tolerance.5-7 The microbiota also provides other benefits to the host including priming of the immune system and protection of the mucosal surface by competing with invading pathogens for space, nutrients, and binding receptors.6 Recent advances in sequencing technology and metagenomics have revealed changes in the microbial population of the gut in both health and disease. Strikingly, these studies have shown that alterations in the composition of the gut microflora, sometimes referred to as dysbiosis, are associated with a number of chronic diseases including obesity, diabetes and inflammatory disease.8-10
The predominant proteins found in mucus are mucins, high molecular weight and heavily O-glycosylated proteins. The high proportion of O-glycans present (usually 50–80%) confer viscoelastic properties on mucin molecules and result in the formation of mucus gels. There are four human gel forming mucins, MUC2, MUC5AC, MUC5B, and MUC6, which are expressed in a site specific fashion throughout the human body.11 In the intestinal tract mucus contains almost exclusively MUC2, whereas MUC5AC and MUC6 are found in gastric mucus. The oligosaccharide structures present on the surface of mucins vary depending on cell lineage, tissue location and developmental stage and can also be influenced by infection, inflammation, and neoplasia.12
The mucus gel can act as a reservoir for mucus-adapted pathogens. Pathogens which infect mucosal surfaces share two main goals: (1) to overcome the mucus barrier and (2) to interact with the underlying epithelial cells and cause disease. Glycans are often exploited by pathogens to facilitate colonization and disease13-15 and, as heavily glycosylated molecules, mucins play a central role during bacterial colonisation of the intestinal tract of humans and animals. The oligosaccharide structures on mucins display a high degree of heterogeneity between species and among different anatomic locations within the body, resulting in very varied endogenous microbial populations. Glycan epitopes act as binding receptors for many bacterial adhesins.16,17 Differences in mucin glycosylation may account for the diverse disease outcome between species and individuals.
Organisms have devised a number of strategies to enable penetration and/or colonisation of the mucus layer, including production of proteases and glycosidases to degrade the mucins.18-21 In addition, bacteria may signal to the host cell to either downregulate expression of mucin genes or to alter the glycosylation profile of the mucin.22 Recent studies which show that bacterial gene expression can be altered upon binding to mucin,23,24 suggests that mucus can prime bacterial pathogens for interaction with epithelial cells and to promote colonization and/or virulence. A transcriptomic analysis of the intestinal tract of flies shows that genes linked to mucus production are regulated by the pathogen Erwinia carotovora, suggesting that the mucus barrier is remodelled during infection.25 It is clear that the interaction of bacteria with mucins is an important first step in the colonisation of mucosal surfaces. Knowledge of how bacteria interact with mucus could lead to novel strategies to prevent infection, either directly or by promoting colonisation by “beneficial” bacteria, such as probiotics. However, despite the obvious importance of elucidating how microorganisms, both pathogens and commensals, interact with mucins and other components of mucus, it is only relatively recently that this subject has received widespread attention. In part this is due to the recent rapid expansion in our understanding of the composition of the gut microflora and the consequences of altering that composition. However, it is also due in no small part to the development of new improved tools that make such studies much more feasible than previously.
Use of Novel Tools to Assess the Interactions of Bacteria with Mucus and Mucins
In a recent paper we exploited cell lines that secrete mucins and form an adherent mucus layer together with novel mucin microarrays that contain natural mucins from different animal species to examine the interaction of two gastrointestinal pathogens Campylobacter jejuni and Helicobacter pylori with mucus.26 We showed that despite being closely related, these two bacteria have very divergent mechanisms of interaction with mucus and mucins. Our results also highlight the role of mucin in promoting infection and indicate that the tissue tropism exhibited by different bacteria may be mediated by the glycans present on mucins.
Studies of host bacterial interactions traditionally have relied heavily on the use of cultured cell lines. While these studies have been invaluable in advancing our knowledge on how bacteria interact with and signal to epithelial cells in order to subvert their function and cause disease, there is now a recognition that intestinal cell lines commonly used for such studies do not accurately reflect conditions encountered in the gut. The development of gut–derived epithelial cells that secrete mucins into supernatants has helped develop our understanding of bacterial interactions with, and responses to, mucin.27 However, in recent studies we have started to explore the interactions of bacteria with cells that harbor an overlying adherent mucus layer, a state that more accurately mimics conditions in vivo. We used the non mucin secreting intestinal HT29 cell line and two of its derivatives, the methotrexate adapted cell line HT29-MTX which secretes mucins into the culture supernate and a subclone of those cells HT29-MTX-E12 (E12) cells which form an adherent mucus layer to assess the effect of mucus and mucins on the interaction of C. jejuni and H. pylori with cells. These results clearly show that the presence of mucus enhances C. jejuni infection of the cells a finding that is in agreement with a previous study which showed that both adhesion to and internalization of C. jejuni were enhanced in E12 cells harboring mucus compared with parental cells without mucus.28 Interestingly, it has also recently been reported that Salmonella virulence is enhanced in the presence of mucus.29 On the other hand, H. pylori did not interact with HT29 cells and while it did interact with the mucin secreting HT29MTX cells, infection was markedly enhanced for E12 cells. Thus, either H. pylori increases expression of factors that mediate infection upon finding itself in an environment of mucus or, alternatively, the mucus layer offers an enhanced number of receptors that enables effective infection. In contrast to C. jejuni, H. pylori was unable to bind to the mucin purified from E12 cells. Rather, H. pylori bound to the glycolipid fraction of the mucus which expressed both sialyl Lewisx and the Lewisb blood group antigen, two well characterized receptors for this organism.17,30
Another promising model of intestinal infection relies on the use of polarized in vitro organ culture. This has been used to examine both enteropathogenic Escherichia coli and C. jejuni pathogenicity.31,32 In addition several groups have made recombinant proteins consisting of mucin domains.33-36 While recombinant production of full-length gel-forming human mucins has not been reported to date, murine Muc5ac has been cloned in its entirety and a murine model of MUC5AC overexpression established37 suggesting that this approach should be feasible in the future to modulate cellular mucin production.
Studies on how microbes interact directly with mucins have been hampered previously due to difficulties in obtaining mucus from animals and humans, variation in mucin glycosylation in different individuals, the cost and length of time that mucin purification takes and typically low yields of purified mucin. We overcame some of these hurdles through the use of a novel high throughput mucin microarray platform containing mucins from different animal species.38 This array enabled high throughput and quantitative analysis of the interaction of fluorescently labeled bacteria with the mucins. Other advantages include efficient use of limited mucin quantities enabling optimization of the number of binding experiments that can be done and optimal presentation of the glycans, thereby maximizing the access of the bacteria to potential glycan receptors. Glycan arrays which contain single glycans have been used previously for the study of bacterial glycan interactions. However, mucins harbor hundreds of heterogeneous glycans. In addition, as protein-glycan interactions tend to have low binding energies the multimeric presentation of the glycans of mucin molecules maximizes the potential for high affinity binding.
The mucin arrays were interrogated with C. jejuni and H. pylori organisms which had been stained with Syto 82, a fluorescent vital dye. Results show that, despite being closely related organisms, C. jejuni and H. pylori each bound to different mucin subsets. Strikingly, C. jejuni displayed a distinct tropism for mucins from the chicken intestinal tract compared with mucins from other animals. In addition, the strength of the interaction with avian mucins was dependent on the site of origin of the mucin (in descending order of interaction: large intestine, proximal small intestine, cecum). We previously showed that chicken mucin attenuates virulence of C. jejuni by inhibiting the organism from invading epithelial cells.39 The gradient of inhibition mirrored that seen with mucin arrays (greatest with mucin from the large intestine) indicating that the strong binding of C. jejuni to chicken mucin may explain why this organism acts as a commensal in chickens but causes disease when ingested by humans. Further exploration of this area should include a comparison of the relative strength of interaction of C. jejuni with human mucin.
The strength of the interaction of H. pylori with mucins was not as great as that seen with C. jejuni and chicken mucin. This finding was not unexpected given that natural H. pylori infection only occurs in humans and non human primates. However, it was surprising that wild type strains of H. pylori, known to differ in expression of two well characterized outer membrane adhesins BabA and SabA, (which interact with Lewisb blood group antigen and sialyl Lewisx respectively), all bound to the same extent to each of the mucins. This suggests that H. pylori expresses other lectins that have not yet been described and highlights the potential of mucin arrays to explore novel adhesin-receptor interactions in mucosal pathogens.
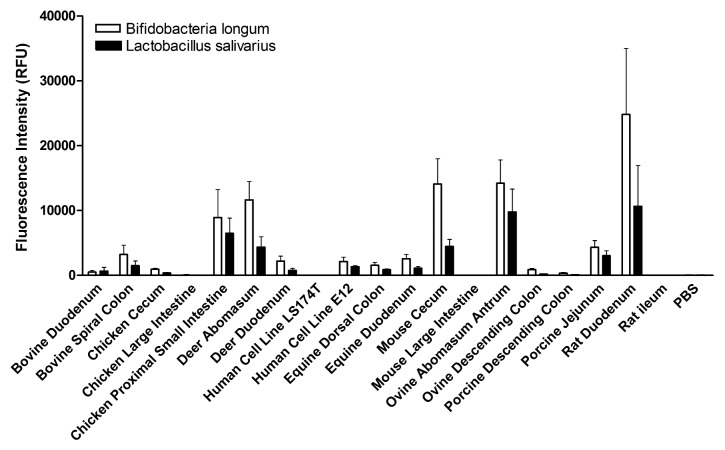
We now have started to expand our study of the interaction of bacteria with mucin arrays to include commensal organisms. Mucin binding is one of the key characteristics used to identify potential probiotic species. Using the mucin arrays we have assessed the binding of two commensal bacteria Lactobacillus salivarius AH102 and Bifidobacteria longum AH1205, both previously shown to influence C. jejuni virulence using the E12 infection model described above.28 Binding of the bacteria to a subset of mucins present on the array is presented in Figure 1. The two organisms displayed overlapping but distinct mucin binding signatures. While each bacterium demonstrated a preference for a specific subset of mucins, tropism was not determined by either the species or anatomical site of origin of the mucin highlighting the importance of specific glycosylation patterns of individual mucins.
Figure 1. The interaction of Lactobacillus salivarius AH102 and Bifidobacteria longum AH1205 with animal mucins. Mucin microarrays were probed with fluorescently-labeled L. salivarius and B. longum organisms. L. salivarius and B. longum were cultured at 37 °C in MRS broth under microaerophillic conditions. Cultures were harvested, washed, and resuspended in PBS to an OD600 of 1.0. Bacteria were stained with Syto82, washed 7 times in low salt TBS-tween, and incubated with the animal mucin microarrays for 1 hr at 37 °C at an OD600 of 1.0. Histogram represents the mean fluorescence intensity from triplicate subarrays on three replicate microarray slides, values for each subarray consisting of the median of six feature replicates. Error bars are the standard deviation of the mean of three microarray slides.
Future Perspectives
While the manuscript focuses on a comparative analysis of H. pylori and C. jejuni, this work will be of interest to those studying a wide range of mucosal pathogens. Expanding the use of mucin arrays to different organisms, both pathogenic and commensal, will continue the discovery of biologically important motifs in bacterial-host interactions and can complement the use of novel in vitro cell models, such as mucus secreting cell lines described above. Future developments should include the addition of human mucin samples to arrays. Mucins isolated from various anatomical sites of healthy individuals and from patients with a variety of diseases can be screened on a single chip enabling binding between different mucins directly comparable. As our knowledge of the organisms that colonise mucus expands so does the potential of the tools described in this paper. A case in point is the recent recognition that phage to bacteria ratios may be increased relative to the adjacent environment, on all mucosal surfaces.40 In vitro studies of tissue culture cells with and without surface mucus demonstrated that this increase in phage abundance is mucus dependent and protects the underlying epithelium from bacterial infection. Thus improved tools to assess mucus interactions could be used to screen not just bacteria but also phage, viruses and purified proteins that interact with mucins. The success of research programmes examining microbial organisms and their interaction with mucus, both gastro-intestinal and at other mucosal surfaces, requires a truly multidisciplinary approach encompassing teams of researchers with expertise in microbiology, mucin biology, chemistry, and glycobiology.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
This work was supported by a grant from Science Foundation Ireland (08/SRC/B1393).
References
- 1.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–9. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ermund A, Schütte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am J Physiol Gastrointest Liver Physiol. 2013;305:G341–7. doi: 10.1152/ajpgi.00046.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson ME, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2013 doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanthakumar NN, Dai D, Meng D, Chaudry N, Newburg DS, Walker WA. Regulation of intestinal ontogeny: effect of glucocorticoids and luminal microbes on galactosyltransferase and trehalase induction in mice. Glycobiology. 2005;15:221–32. doi: 10.1093/glycob/cwi004. [DOI] [PubMed] [Google Scholar]
- 6.Leser TD, Mølbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol. 2009;11:2194–206. doi: 10.1111/j.1462-2920.2009.01941.x. [DOI] [PubMed] [Google Scholar]
- 7.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. 1997;159:1739–45. [PubMed] [Google Scholar]
- 8.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 9.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 11.Longman RJ, Douthwaite J, Sylvester PA, Poulsom R, Corfield AP, Thomas MG, Wright NA. Coordinated localisation of mucins and trefoil peptides in the ulcer associated cell lineage and the gastrointestinal mucosa. Gut. 2000;47:792–800. doi: 10.1136/gut.47.6.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corfield AP, Myerscough N, Longman R, Sylvester P, Arul S, Pignatelli M. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut. 2000;47:589–94. doi: 10.1136/gut.47.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–8. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 14.Corfield AP, Wiggins R, Edwards C, Myerscough N, Warren BF, Soothill P, Millar MR, Horner P. A sweet coating--how bacteria deal with sugars. Adv Exp Med Biol. 2003;535:3–15. doi: 10.1007/978-1-4615-0065-0_1. [DOI] [PubMed] [Google Scholar]
- 15.Moran AP, Gupta A, Joshi L. Sweet-talk: role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut. 2011;60:1412–25. doi: 10.1136/gut.2010.212704. [DOI] [PubMed] [Google Scholar]
- 16.Bouckaert J, Berglund J, Schembri M, De Genst E, Cools L, Wuhrer M, Hung CS, Pinkner J, Slättegård R, Zavialov A, et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol Microbiol. 2005;55:441–55. doi: 10.1111/j.1365-2958.2004.04415.x. [DOI] [PubMed] [Google Scholar]
- 17.Borén T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–5. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 18.Corfield AP, Wagner SA, Clamp JR, Kriaris MS, Hoskins LC. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun. 1992;60:3971–8. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corfield AP, Wagner SA, O’Donnell LJ, Durdey P, Mountford RA, Clamp JR. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj J. 1993;10:72–81. doi: 10.1007/BF00731190. [DOI] [PubMed] [Google Scholar]
- 20.Haider K, Hossain A, Wanke C, Qadri F, Ali S, Nahar S. Production of mucinase and neuraminidase and binding of Shigella to intestinal mucin. J Diarrhoeal Dis Res. 1993;11:88–92. [PubMed] [Google Scholar]
- 21.Homer KA, Whiley RA, Beighton D. Production of specific glycosidase activities by Streptococcus intermedius strain UNS35 grown in the presence of mucin. J Med Microbiol. 1994;41:184–90. doi: 10.1099/00222615-41-3-184. [DOI] [PubMed] [Google Scholar]
- 22.Cooke CL, An HJ, Kim J, Canfield DR, Torres J, Lebrilla CB, Solnick JV. Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori. Gastroenterology. 2009;137:1061–71, e1-8. doi: 10.1053/j.gastro.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Skoog EC, Sjöling A, Navabi N, Holgersson J, Lundin SB, Lindén SK. Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PLoS One. 2012;7:e36378. doi: 10.1371/journal.pone.0036378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu QV, McGuckin MA, Mendz GL. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J Med Microbiol. 2008;57:795–802. doi: 10.1099/jmm.0.47752-0. [DOI] [PubMed] [Google Scholar]
- 25.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5:200–11. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Naughton JA, Mariño K, Dolan B, Reid C, Gough R, Gallagher ME, Kilcoyne M, Gerlach JQ, Joshi L, Rudd P, et al. Divergent mechanisms of interaction of Helicobacter pylori and Campylobacter jejuni with mucus and mucins. Infect Immun. 2013;81:2838–50. doi: 10.1128/IAI.00415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Favre-Bonté S, Licht TR, Forestier C, Krogfelt KA. Klebsiella pneumoniae capsule expression is necessary for colonization of large intestines of streptomycin-treated mice. Infect Immun. 1999;67:6152–6. doi: 10.1128/iai.67.11.6152-6156.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alemka A, Clyne M, Shanahan F, Tompkins T, Corcionivoschi N, Bourke B. Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect Immun. 2010;78:2812–22. doi: 10.1128/IAI.01249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gagnon M, Zihler Berner A, Chervet N, Chassard C, Lacroix C. Comparison of the Caco-2, HT-29 and the mucus-secreting HT29-MTX intestinal cell models to investigate Salmonella adhesion and invasion. J Microbiol Methods. 2013;94:274–9. doi: 10.1016/j.mimet.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 30.Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–8. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schüller S, Lucas M, Kaper JB, Girón JA, Phillips AD. The ex vivo response of human intestinal mucosa to enteropathogenic Escherichia coli infection. Cell Microbiol. 2009;11:521–30. doi: 10.1111/j.1462-5822.2008.01275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corcionivoschi N, Alvarez LA, Sharp TH, Strengert M, Alemka A, Mantell J, Verkade P, Knaus UG, Bourke B. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe. 2012;12:47–59. doi: 10.1016/j.chom.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Godl K, Johansson ME, Lidell ME, Mörgelin M, Karlsson H, Olson FJ, Gum JR, Jr., Kim YS, Hansson GC. The N terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J Biol Chem. 2002;277:47248–56. doi: 10.1074/jbc.M208483200. [DOI] [PubMed] [Google Scholar]
- 34.Lidell ME, Johansson ME, Mörgelin M, Asker N, Gum JR, Jr., Kim YS, Hansson GC. The recombinant C-terminus of the human MUC2 mucin forms dimers in Chinese-hamster ovary cells and heterodimers with full-length MUC2 in LS 174T cells. Biochem J. 2003;372:335–45. doi: 10.1042/BJ20030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lidell ME, Hansson GC. Cleavage in the GDPH sequence of the C-terminal cysteine-rich part of the human MUC5AC mucin. Biochem J. 2006;399:121–9. doi: 10.1042/BJ20060443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci U S A. 2006;103:9298–303. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehre C, Worthington EN, Liesman RM, Grubb BR, Barbier D, O’Neal WK, Sallenave JM, Pickles RJ, Boucher RC. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc Natl Acad Sci U S A. 2012;109:16528–33. doi: 10.1073/pnas.1206552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilcoyne M, Gerlach JQ, Gough R, Gallagher ME, Kane M, Carrington SD, Joshi L. Construction of a natural mucin microarray and interrogation for biologically relevant glyco-epitopes. Anal Chem. 2012;84:3330–8. doi: 10.1021/ac203404n. [DOI] [PubMed] [Google Scholar]
- 39.Alemka A, Whelan S, Gough R, Clyne M, Gallagher ME, Carrington SD, Bourke B. Purified chicken intestinal mucin attenuates Campylobacter jejuni pathogenicity in vitro. J Med Microbiol. 2010;59:898–903. doi: 10.1099/jmm.0.019315-0. [DOI] [PubMed] [Google Scholar]
- 40.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, Stotland A, Wolkowicz R, Cutting AS, Doran KS, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci U S A. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]