Abstract
It is known that the gastrointestinal tract (GIT) microbiota responds to different antibiotics in different ways and that while some antibiotics do not induce disturbances of the community, others drastically influence the richness, diversity, and prevalence of bacterial taxa. However, the metabolic consequences thereof, independent of the degree of the community shifts, are not clearly understood. In a recent article, we used an integrative OMICS approach to provide new insights into the metabolic shifts caused by antibiotic disturbance. The study presented here further suggests that specific bacterial lineage blooms occurring at defined stages of antibiotic intervention are mostly associated with organisms that possess improved survival and colonization mechanisms, such as those of the Enterococcus, Blautia, Faecalibacterium, and Akkermansia genera. The study also provides an overview of the most variable metabolic functions affected as a consequence of a β-lactam antibiotic intervention. Thus, we observed that anabolic sugar metabolism, the production of acetyl donors and the synthesis and degradation of intestinal/colonic epithelium components were among the most variable functions during the intervention. We are aware that these results have been established with a single patient and will require further confirmation with a larger group of individuals and with other antibiotics. Future directions for exploration of the effects of antibiotic interventions are discussed.
Keywords: antibiotic therapy, human gut microbiota, metagenomic, metaproteomic, metabolomic, metatranscriptomic
Introduction
The human gastrointestinal tract (GIT) contains trillions of finely tuned bacteria,1-6 referred to as the GIT microbiota, which function as an organ with its own metabolism.7-12 A number of investigations have innovatively employed state-of-the-art technologies to explore, primarily, the bi-directional communication between the GIT and human health13-18 and, second, the way in which external factors such as nutrition and antibiotics modulate this loop.1,19-32
The use of antibiotics to treat bacterial infections in children, adolescents, and adults living mainly in developed countries is increasing substantially. While some of the treatments are temporally targeted, others are constantly provided over long periods, and this may ultimately cause the emergence of antibiotic-resistant bacteria. The consequences of two sets of factors, namely, the target, dose, and duration of the antibiotic treatment and the emergence of resistant bacteria, are beginning to be understood. In this context, antibiotics could play a key role in the bi-directional communication between the gut and the host, and accordingly, this communication could also be modulated. There have been reports of microbial shifts associated with a number of antibiotics, including the following: (1) combined intravenous therapy with ampicillin, sulbactam, and cefazolin;30 (2) penicillin, vancomycin, and chlortetracycline;33 (3) ciprofloxacin;19,24 (4) streptomycin;34 (5) ASP250 (chlortetracycline, sulfamethazine and penicillin);35 (6) streptomycin;36 (7) amoxicillin and metronidazole;20 (8) metronidazole alone;37 and (9) vancomycin and imipenem,38 to cite just a few. Our group recently published omic results that demonstrated the potential of antibiotics to affect the energy metabolism of the GIT microbiota and its capacity to metabolize molecules known to be produced by the host (such as bile acids and sterols) or exclusively produced by bacteria (such as vitamins essential for neuronal development) during follow-up treatment.30 The treatment of a 68-y-old male with β-lactam therapy was used as a proof-of-concept; fecal samples were examined at days 0 (FS-0), 3 (FS-3), 6 (FS-6), 11 (FS-11) and 14 (FS-14) after the initiation of therapy and at 40 d after cessation of therapy. Here, we aimed to extend our knowledge regarding time-span-specific bacterial lineages and anabolic activity blooms occurring as a consequence of an antibiotic treatment through an in-depth examination of previously published data.30
Bacterial Blooms Occurring at Defined Stages of β-lactam Treatment
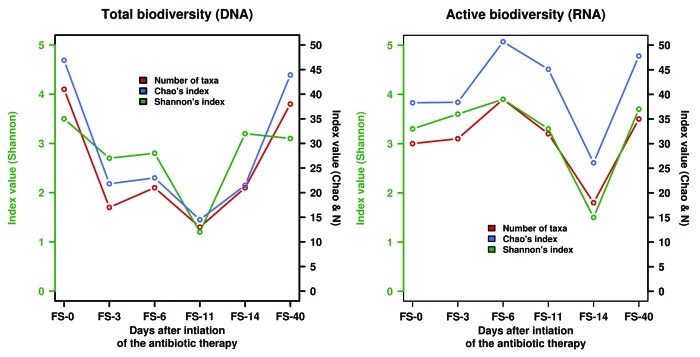
Our recent article showed that during an intervention, the administration of β-lactam intravenous therapy consisting of ampicillin, sulbactam, and cefazolin exerts changes both at the level of total (Fig. 1, left) and active (Fig. 1, right) bacterial taxa. Additionally, we have now observed a division-wide bloom of the active Firmicutes. An active Enterococcus lineage (E. durans) in the Firmicutes expands dramatically only at day 3 (~14.6% of the total active community) (Table 1). Its contribution accounted for ≤ 0.5% in other sampled populations, and no active members were found before or after the intervention. Cultivated members of this lineage have been shown to produce tyramine, a metabolite associated with tyrosine metabolism that has been found to be involved in the enhancement of bacterial adhesion.39,40 Although we did not measure the exact levels of tyramine in the sampled populations, an inspection of tyramine mass spectrometry (MS) adduct signatures ([M+H]+, [M+Na]+ and [M+K]+) in the HPLC-ESI-QTOF-MS chromatograms30 tentatively identified one or several such signatures in sampled populations at days 3 and 6 (in at least two replicates) but not in any other samples. This suggests that during β-lactam therapy, E. durans-like bacteria may survive in the intestinal environment and synthesize tyramine in the colon, drawing on this ability as a survival and colonization mechanism to enhance adhesion to the colonic mucosa.40 Additionally, a bloom of active bacteria associated with unclassified Firmicutes sequences (~10.6%) occurred at day 6, while accounting for 1.4% at the beginning of the therapy and for ≤ 2.7% in other sampled populations (Table 1); unfortunately, because there are no cultured representatives related to these sequences, the physiology of the represented bacteria remains unknown. A contrasting scenario was found for other Firmicutes members, such as those belonging to the genus Blautia, which were most active (up to ~27.8%) in the absence of antibiotic (FS-0 and FS-14) while contributing ≤ 3.1% in populations subjected to antibiotics (Table 1). Finally, bacteria assigned to the Faecalibacterium and Ruminococcus genera were most active after the end of the antibiotic treatment (Table 1). Thus, they were from 8.3- to 192-fold more active (in terms of the total number of sequences assigned to these genera) at the end of the therapy than before. This indicates that Clostridiales have an increased fitness relative to all other Firmicutes identified in the community when the intervention ended. It is noteworthy that the significant decline in the diversity of Blautia and Faecalibacterium lineages has been suggested to play a role in improved intestinal permeability because these bacteria influence mucin synthesis and composition.41-43 This may be a reason for the active Enterococcus bloom that uses tyramine as a strategy to colonize a “modified” colonic mucosa.39,40
Figure 1. Biodiversity measurements of the total (left) and active (right) GIT microbiota from a patient receiving β-lactam therapy. The number of observed taxa (N), the biodiversity index value (Shannon), and the richness estimator (Chao1) are adapted from reference 30.
Table 1. Active bacterial composition based on 16S rRNA analyses in the follow-up study.
| Relative percentage of active bacteria (%) | ||||||
|---|---|---|---|---|---|---|
| FS-0 | FS-3 | FS-6 | FS-11 | FS-14 | FS-40 | |
| Blautia (Firmicutes) | 27.84 | 3.13 | 0.51 | 0.14 | 0 | 14.02 |
| Gemmiger (Betaproteobacteria) | 4.44 | 0.06 | 0 | 0.05 | 0 | 0.70 |
| Uncl. Actinobacteria | 2.21 | 0.40 | 0 | 0.00 | 0.05 | 0.15 |
| Enterococcus (Firmicutes) | 0 | 14.56 | 0.51 | 0.42 | 0.05 | 0.00 |
| Uncl. Bacteroidetes | 3.99 | 13.37 | 11.34 | 4.66 | 4.44 | 3.68 |
| Bifidobacterium (Actinobacteria) | 1.44 | 4.38 | 0.13 | 0 | 0.02 | 0.73 |
| Uncl. Firmicutes | 1.42 | 2.10 | 10.57 | 2.68 | 0.52 | 2.34 |
| Akkermansia (Verrucomicrobia) | 0.53 | 0 | 6.62 | 1.98 | 0 | 0 |
| Alistipes (Bacteroidetes) | 0 | 0.63 | 4.20 | 2.59 | 0.52 | 0.58 |
| Parabacteroides (Bacteroidetes) | 1.49 | 4.84 | 4.33 | 17.36 | 7.94 | 2.51 |
| Bacteroides (Bacteroidetes) | 23.15 | 24.74 | 10.83 | 36.12 | 74.24 | 20.98 |
| Faecalibacterium (Firmicutes) | 1.37 | 0 | 0.51 | 1.32 | 0.07 | 11.35 |
| Uncl. Ruminococcaceae (Firmicutes) (Firmicutes) | 0.82 | 0.17 | 3.06 | 2.30 | 0.17 | 9.69 |
| Ruminococcus (Firmicutes) | 0.03 | 0 | 0 | 0 | 0 | 6.59 |
Samples FS-0, FS-3, FS-6, FS-11, and FS-14 correspond to the materials collected on days 0, 3, 6, 11, and 14 of the antibiotic treatment, respectively. The FS-40 sample corresponds to the materials collected 40 days after cessation of the antibiotic treatment. Only bacteria with abundance level higher than 1% were included.
A division-wide bloom of naturally antibiotic-resistant Bacteroidetes44 was also observed. While species of Alistipes become most active at day 6 (~6.6%), the maximum bloom of active Parabacteroides species (P. merdae and P. distasonis) occurred at day 11 (~17.4%), and that of active Bacteroides species (B. fragilis, B. dorei, B. uniformis and B. ovatus) occurred at day 14 (~74.2%) (Table 1). This time-delay effect has also been observed after gentamicin and ceftriaxone treatment in mice.45 Indeed, bacteria belonging to the Bacteroides lineage have been shown to acquire and metabolize a wide range of sugars, using a carbohydrate acquisition strategy based on outer membrane proteins that confers a competitive growth advantage.46,47 This suggests that the bloom of these active bacteria may have metabolic consequences at the level of carbohydrate metabolism. In agreement with this, among all sampled populations, protein extracts from the fecal sample at day 14 have been recently shown to be the most active in terms of sugar hydrolysis.48 As observed for the Firmicutes, a bloom of bacteria affiliated with unclassified active Bacteroidetes sequences occurred at days 3 (~11.3%) and 6 (~13.4%) while accounting for ≤4.7% in other sampled populations (Table 1); unfortunately, because there are no cultured representatives related to these sequences, the physiology of the represented bacteria remains unknown. Finally, it is noteworthy that in contrast to Firmicutes members, no significant bloom of any active Bacteroidetes occurred after the end of the antibiotic therapy. This indicates that the Bacteroidetes have an increased fitness only when the end of the treatment is approaching (days 6 to 14), most likely because these microorganisms are able to develop resistance to several antimicrobial drugs due to the production of endogenous β-lactamases.44
Within Actinobacteria inhabitants, the Bifidobacterium spp. become most active at day 3 (4.4%) while accounting for 1.4% before the intervention and for ≤ 0.73% in other sampled populations (Table 1). Additionally, bacteria affiliated with unclassified active Actinobacteria sequences (~2.2%) were strongly affected by the antibiotic, becoming strongly depleted during and after the intervention (≤0.4%); a similar situation was observed for Betaproteobacteria assigned to the Gemmiger genus, which accounted for 4.4% before the treatment and less than 0.7% in other sampled populations (Table 1). Finally, we observed a significant activation of mucin-degrading commensal bacteria assigned to the Akkermansia genus, which are members of the Verrucomicrobia, at day 6 (~6.62%) and, to a much lesser extent, at day 11 (~2.0%) (Table 1). In other sampled populations, no sequences assigned to active Akkermansia were detected, and this genus only accounted for 0.5% of the total active community before the intervention. The activation of this bacterial group at days 6 and 11 agrees with our experimental evidence that demonstrated that mucin degradation increased at these time points.48 These data also agree with the results of previous investigations that suggest a link between mucin degradation, gut inflammation, and the abundance of Akkermansia muciniphila.49
Time-Span Prevalence of Active Proteins and Associated Functions
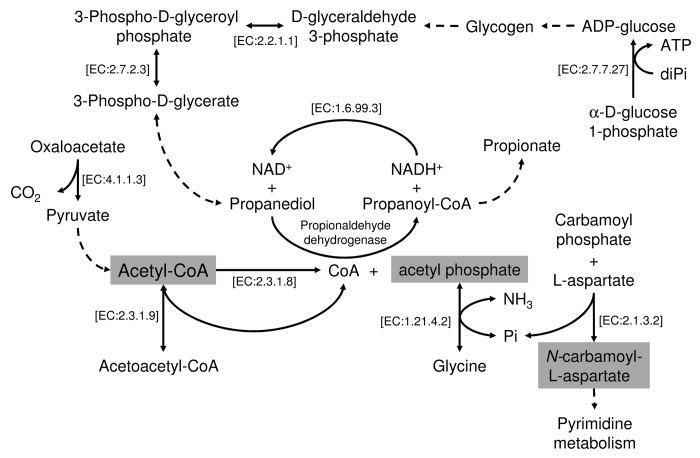
Our shotgun community protein expression data30 allowed us to characterize the time-span representation of active proteins and the functions assigned to them (Kyoto Encyclopedia of Genes and Genomes annotations, TGRFAM annotations, and Enzyme Commission numbers). Analysis of TGRFAM, KEGG, and EC proteome annotations identified 138 out of 394, 107 out of 304 and 58 out of 192 identifiers, respectively, that were differentially represented within the sampled populations; this constitutes approximately 30–35% of the total of the identifiers assigned to variable functions within the sampled populations. We ranked the variable functions according to the number of active/expressed proteins assigned to them; thus, we give more importance to those functions that have higher coverage. Table 2 shows the top-15 ranking of the most variable KEGG categories within the sampled populations. Nine of these categories correspond to pathways required for the production of acetyl phosphate and acetyl-CoA (Fig. 2). These are essential molecules involved in many cellular functions, including energy production, cellular respiration, gene expression, and proteolysis.50 Additionally, reactions possibly involved in pyrimidine (EC: 2.1.3.2) and propanediol (propionaldehyde dehydrogenase) metabolism (Fig. 2) were also variable in their presence or absence during and after the intervention. This suggests that the antibiotic-induced changes at the level of the active microbiota structure may have implications for the production of a limited set of key metabolic compounds involved in major cellular functions and whose abundance and expression levels vary over the duration of the therapy.
Table 2. Ranking of variable KEGG identifiers with high protein/enzyme coverage using metaproteomic data.
| KEGGID | Name | Rank | Score |
|---|---|---|---|
| K00975 | Gucose-1-phosphate adenylyltransferase [EC:2.7.7.27] | 1 | 40 |
| K02357 | Elongation factor Ts | 2 | 34 |
| K00609 | Aspartate carbamoyltransferase catalytic subunit [EC:2.1.3.2] | 3 | 30 |
| K00615 | Transketolase [EC:2.2.1.1] | 4 | 29 |
| K00625 | Phosphate acetyltransferase [EC:2.3.1.8] | 5 | 28 |
| K00626 | Acetyl-CoA C-acetyltransferase [EC:2.3.1.9] | 6 | 26 |
| K00927 | Phosphoglycerate kinase [EC:2.7.2.3] | 7 | 25 |
| K01572 | Oxaloacetate decarboxylase. beta subunit [EC:4.1.1.3] | 8 | 24 |
| K02029 | Polar amino acid transport system permease protein | 9 | 23 |
| K02117 | V-Type H+-transporting ATPase subunit A [EC:3.6.3.14] | 10 | 22 |
| K03798 | Cell division protease FtsH [EC:3.4.24.-] | 11 | 21 |
| K03885 | NADH dehydrogenase [EC:1.6.99.3] | 12 | 20 |
| K10549 | D-Allose transport system substrate-binding protein | 13 | 19 |
| K10670 | Glycine reductase [EC:1.21.4.2] | 14 | 18 |
| K13922 | Propionaldehyde dehydrogenase [EC:1.2.1.3] | 15 | 17 |
Score refers to the number of proteins/enzymes assigned to a particular KEGG ID as found in the metaproteomic dataset in all sampled populations.
Figure 2. Graphical representation of the pathways involved in the metabolism of acetyl phosphate and acetyl-CoA that have been shown to differ in their prevalence (based on the presence or absence of enzymes assigned to the pathways) in all six sampled communities. The enzymes (with EC numbers) implicated in each particular reaction are specifically indicated. Discontinuous lines indicate multiple reactions involved in the transformation.
Conclusions
Research efforts to elucidate the impact of antibiotic treatment on the GIT microbiota have mainly focused on monitoring community shifts and analyzing metagenomic data reflecting differences that result from shifts in community composition. However, little is known about the functional consequences of these shifts on the cross-talk between gut microbial metabolism and host response. Moreover, the role played by specific bacterial blooms in GIT composition and metabolism at defined stages remains to be established. In this study, we contribute to the understanding of the complex and dynamic interplay among antibiotics and the microbiota and mucosa of the GIT. We found that with the administration of β-lactam intravenous therapy consisting of ampicillin, sulbactam, and cefazolin, there is a reshaping of the distal GIT microbial community, in which only ~12.5% (or 14 out of 112) of the active bacterial lineages tentatively identified30 were significantly affected at defined stages. Additionally, phylum- and division-wide blooms were also identified that do not occur simultaneously. Interestingly, the observed blooms seem to play a presumptive role in mucin adhesion, synthesis, and degradation. It is therefore plausible that the composition of the intestinal epithelium is one of the main factors modulating bacterial community composition during β-lactam intravenous therapy. This is consistent with previous studies reporting that after metronidazole treatment, the thickness of the mucus layer in the gut of rats was increased approximately 2-fold.37 Further investigations are required to ascertain the role of β-lactam-induced bacterial blooms in the colonic mucosa. Finally, this study is also part of an initial effort to identify the groups of functions that are most affected within the time span of an antibiotic intervention. In this context, we demonstrated that functions leading to the production of key metabolic compounds, such as acetyl phosphate and acetyl-CoA, that are involved in major cellular functions were also among the most variable functions during β-lactam treatment.
The results presented here should be seen as those of an explorative study, as there are several limitations that prevent us from drawing definitive conclusions. First, we did not examine an additional cohort of subjects; consequently, it is unknown to what degree aging, weight, and alterations of diet components may affect the antibiotic-induced changes observed here. Second, we should stress the fact that only one type of antibiotic was studied, and therefore, the analysis of other antibiotics will be required in future investigations to properly establish the associations between bacterial blooms, biochemical shifts and antibiotic usage. Whatever the case, together with our recent article, the data presented herein suggests that future investigations should explore which antibiotics produce the least collateral effects in the gastrointestinal tract while maintaining efficacy in the treatment of bacterial infections. Indeed, future studies should investigate (i) how to reduce the antibiotic-induced blooms of bacteria that are likely involved in the degradation of intestinal/colonic epithelium components during antibiotic intervention, and (ii) the way antibiotics influence essential functional groups, such as biochemical activities and metabolites, that are required for maintaining human health. In both cases, it will be necessary to create a comprehensive data repository for microbial diversity (total and active) and functional data using computational and experimental strategies. Such strategies will help to generate knowledge and hypotheses that may lay a foundation for subsequent, systematic research and ultimately enable the design of personalized interventions.
Acknowledgments
The whole consortium was funded by the Spanish Ministry of Economy and Competitiveness and the Federal Ministry of Education and Research (BMBF) within the ERA NET PathoGenoMics2 program, grant number 0315441A. This work was further funded by grants BFU2008-04501-E, SAF2009-13032-C02–01, SAF2012-31187, and CSD2007-00005 from the Spanish Ministry of Economy and Competitiveness, Prometeo/2009/092 from Generalitat Valenciana (Spain), and AGL2006-11697/ALI. The authors gratefully acknowledge the financial support provided by the European Regional Development Fund (ERDF) and the European Union (FP7 project Systems medicine of chronic inflammatory bowel disease, Grant Agreement no. 305564). This work has been partially supported by the EVASYON study funded by the Spanish Ministry of Health and Consumption (Carlos III Institute of Health. FIS Grant PI 051579). We thank Rafael Bargiela for his excellent support in relation to the analysis of community proteome data sets and the production of Figure 1, David Rojo for the tentative identification of tyramine signatures in the HPLC-ESI-QTOF-MS chromatograms and María J Gosalbes for her excellent support in relation to the preparation of Table 1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest exist.
References
- 1.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tschöp MH, Hugenholtz P, Karp CL. Getting to the core of the gut microbiome. Nat Biotechnol. 2009;27:344–6. doi: 10.1038/nbt0409-344. [DOI] [PubMed] [Google Scholar]
- 3.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–9. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinross JM, Darzi AW, Nicholson JK. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3:14. doi: 10.1186/gm228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect. 2012;18(Suppl 4):2–4. doi: 10.1111/j.1469-0691.2012.03916.x. [DOI] [PubMed] [Google Scholar]
- 6.Feeney A, Sleator RD. The human gut microbiome: the ghost in the machine. Future Microbiol. 2012;7:1235–7. doi: 10.2217/fmb.12.105. [DOI] [PubMed] [Google Scholar]
- 7.Strader AD, Woods SC. Gastrointestinal hormones and food intake. Gastroenterology. 2005;128:175–91. doi: 10.1053/j.gastro.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly P. Nutrition, intestinal defence and the microbiome. Proc Nutr Soc. 2010;69:261–8. doi: 10.1017/S0029665110000108. [DOI] [PubMed] [Google Scholar]
- 10.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. MetaHIT Consortium A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–36. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brüls T, Weissenbach J. The human metagenome: our other genome? Hum Mol Genet. 2011;20(R2):R142–8. doi: 10.1093/hmg/ddr353. [DOI] [PubMed] [Google Scholar]
- 13.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12:562–8. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Mai V, Draganov PV. Recent advances and remaining gaps in our knowledge of associations between gut microbiota and human health. World J Gastroenterol. 2009;15:81–5. doi: 10.3748/wjg.15.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang JS, Im CR, Im SH. Immune disorders and its correlation with gut microbiome. Immune Netw. 2012;12:129–38. doi: 10.4110/in.2012.12.4.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nyangale EP, Mottram DS, Gibson GR. Gut microbial activity, implications for health and disease: the potential role of metabolite analysis. J Proteome Res. 2012;11:5573–85. doi: 10.1021/pr300637d. [DOI] [PubMed] [Google Scholar]
- 18.Shen D, Liu C, Xu R, Zhang F. Human gut microbiota: dysbiosis and manipulation. Front Cell Infect Microbiol. 2012;2:123. doi: 10.3389/fcimb.2012.00123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–75. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preidis GA, Versalovic J. Targeting the human microbiome with antibiotics, probiotics, and prebiotics: gastroenterology enters the metagenomics era. Gastroenterology. 2009;136:2015–31. doi: 10.1053/j.gastro.2009.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobsson HE, Jernberg C, Andersson AF, Sjölund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–23. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 24.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–61. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flint HJ. Obesity and the gut microbiota. J Clin Gastroenterol. 2011;45(Suppl):S128–32. doi: 10.1097/MCG.0b013e31821f44c4. [DOI] [PubMed] [Google Scholar]
- 26.Willing BP, Russell SL, Finlay BB. Shifting the balance: antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol. 2011;9:233–43. doi: 10.1038/nrmicro2536. [DOI] [PubMed] [Google Scholar]
- 27.Cotter PD, Stanton C, Ross RP, Hill C. The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov Med. 2012;13:193–9. [PubMed] [Google Scholar]
- 28.Flint HJ. The impact of nutrition on the human microbiome. Nutr Rev. 2012;70(Suppl 1):S10–3. doi: 10.1111/j.1753-4887.2012.00499.x. [DOI] [PubMed] [Google Scholar]
- 29.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62:1591–601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferrer M, Ruiz A, Lanza F, Haange SB, Oberbach A, Till H, Bargiela R, Campoy C, Segura MT, Richter M, et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol. 2013;15:211–26. doi: 10.1111/j.1462-2920.2012.02845.x. [DOI] [PubMed] [Google Scholar]
- 32.Montoliu I, Genick U, Ledda M, Collino S, Martin FP, le Coutre J, Rezzi S. Current status on genome-metabolome-wide associations: an opportunity in nutrition research. Genes Nutr. 2013;8:19–27. doi: 10.1007/s12263-012-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, Gao Z, Mahana D, Raju K, Teitler I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell SL, Gold MJ, Willing BP, Thorson L, McNagny KM, Finlay BB. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 2013;4:158–64. doi: 10.4161/gmic.23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Looft T, Allen HK. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012;3:463–7. doi: 10.4161/gmic.21288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antunes LC, Han J, Ferreira RB, Lolić P, Borchers CH, Finlay BB. Effect of antibiotic treatment on the intestinal metabolome. Antimicrob Agents Chemother. 2011;55:1494–503. doi: 10.1128/AAC.01664-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pélissier MA, Vasquez N, Balamurugan R, Pereira E, Dossou-Yovo F, Suau A, Pochart P, Magne F. Metronidazole effects on microbiota and mucus layer thickness in the rat gut. FEMS Microbiol Ecol. 2010;73:601–10. doi: 10.1111/j.1574-6941.2010.00916.x. [DOI] [PubMed] [Google Scholar]
- 38.Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M, Guigo R, Knight R, Guarner F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411–9. doi: 10.1101/gr.107987.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladero V, Linares DM, Del Rio B, Fernandez M, Martin MC, Alvarez MA. Draft genome sequence of the tyramine producer Enterococcus durans strain IPLA 655. Genome Announc. 2013;1:e00265–13. doi: 10.1128/genomeA.00265-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernández de Palencia P, Fernández M, Mohedano ML, Ladero V, Quevedo C, Alvarez MA, López P. Role of tyramine synthesis by food-borne Enterococcus durans in adaptation to the gastrointestinal tract environment. Appl Environ Microbiol. 2011;77:699–702. doi: 10.1128/AEM.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, Queipo-Ortuño MI. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, et al. Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 2013;11:61. doi: 10.1186/1741-7007-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raouf AH, Tsai HH, Parker N, Hoffman J, Walker RJ, Rhodes JM. Sulphation of colonic and rectal mucin in inflammatory bowel disease: reduced sulphation of rectal mucus in ulcerative colitis. Clin Sci (Lond) 1992;83:623–6. doi: 10.1042/cs0830623. [DOI] [PubMed] [Google Scholar]
- 44.Nakano V, Nascimento e Silva Ad, Merino VR, Wexler HM, Avila-Campos MJ. Antimicrobial resistance and prevalence of resistance genes in intestinal Bacteroidales strains. Clinics (Sao Paulo) 2011;66:543–7. doi: 10.1590/S1807-59322011000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Wu J, Li JV, Zhou NY, Tang H, Wang Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res. 2013;12:2987–99. doi: 10.1021/pr400263n. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol. 2006;56:1599–605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 47.Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep. 2013;14:319–27. doi: 10.1038/embor.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hernández E, Bargiela R, Diez MS, Friedrichs A, Pérez-Cobas AE, Gosalbes MJ, Knecht H, Martínez-Martínez M, Seifert J, von Bergen M, et al. Functional consequences of microbial shifts in the human gastrointestinal tract linked to antibiotic treatment and obesity. Gut Microbes. 2013;4:306–15. doi: 10.4161/gmic.25321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates Gut inflammation in Salmonella typhimurium-infected gnotobiotic Mice. PLoS One. 2013;8:e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizrahi I, Biran D, Ron EZ. Requirement for the acetyl phosphate pathway in Escherichia coli ATP-dependent proteolysis. Mol Microbiol. 2006;62:201–11. doi: 10.1111/j.1365-2958.2006.05360.x. [DOI] [PubMed] [Google Scholar]