Abstract
All-trans retinoic acid (ATRA) is a widely used differentiation drug that can effectively induce osteogenic differentiation of osteosarcoma cells, but the underlying mechanism remains elusive, which limits the clinical application for ATRA in osteosarcoma patients. In this study, we identified E2F1 as a novel regulator involved in ATRA-induced osteogenic differentiation of osteosarcoma cells. We observed that osteosarcoma cells are coupled with individual differences in the expression levels of E2F1 in patients, and E2F1 impairs ATRA-induced differentiation of osteosarcoma cells. Moreover, remarkable anti-proliferative and differentiation-inducing effects of ATRA treatment are only observed in E2F1 low to negative expressed primary osteosarcoma cultures. These results strongly suggested that E2F1 may serve as a potent indicator for the effectiveness of ATRA treatment in osteosarcoma. Interestingly, E2F1 is found to downregulate retinoic acid receptor α (RARα), a key factor determines the effectiveness of ATRA. E2F1 specifically binds to RARα and promotes its ubiquitination-mediated degradation; as a consequence, RARα-mediated differentiation is inhibited in osteosarcoma. Therefore, our studies present E2F1 as a potent biomarker, as well as a therapeutic target for ATRA-based differentiation therapeutics, and raise the hope of using differentiation-based approaches for osteosarcoma patients.
Keywords: ATRA, E2F1, RARα, all-trans retinoic acid, osteogenic differentiation, osteosarcoma
Introduction
Osteosarcoma is the most frequent primary bone sarcoma,1-4 which is generally regarded as a differentiation disease that is caused by genetic and epigenetic disruptions of terminal differentiation of osteoblasts.5-7 Despite modern treatment protocols that combine chemotherapy, surgery, and sometimes radiotherapy, the 5-year survival rate for patients diagnosed with osteosarcoma remains at 60%~70% since the 1970s.8 As surgical techniques and implants have evolved, chemotherapeutic agents used today seem to be wholly similar to those used 40 y ago. Moreover, adverse effects associated with chemotherapies, chemoresistance, recurrence, and pulmonary metastasis make clinical management of osteosarcoma face numerous challenges.3 Hence, novel therapies based on noncytotoxic induction of cell differentiation responsive pathways could represent a significant advance.
Numerous experiments and clinical trials have shown that all-trans retinoic acid (ATRA) and its derivatives are potent agents used for cancer therapy by inducing differentiation of acute promyelocytic leukemia (APL) and various other tumor types, including neuroblastoma, breast cancer, and melanoma.9-13 We, along with others, also have found that ATRA induces osteoblastic differentiation of osteosarcoma cells both in vivo and in vitro.14,15 Moreover, ATRA has been found to induce osteoblastic differentiation of mouse and rat mesenchymal stem cells and pre-osteoblasts.16,17 All these studies suggest that ATRA can restore normal osteogenesis, and ATRA-based differentiation therapeutics might be feasible for osteosarcoma. However, no clinical applications for ATRA in patients with osteosarcoma have been reported to date. Considering various responses to ATRA therapeutics have been observed in APL patients in clinical, it is particularly important to characterize dominant regulators that determine the differentiation response to ATRA, which may provide the criteria of patient selection for future differentiation therapy in osteosarcoma patients.
Osteoblasts are derived from multipotent mesenchymal stem cells (MSCs). Notably, besides osteocytes, MSCs can also give rise to several other lineages, including myocytes, megakaryocytes, and adipocytes.18-20 What interests us is that the differentiation of myocyte, megakaryocyte, and adipocyte are known to be regulated by the same factor, E2F1, a member of the E2F family of transcription factors that displays properties of both a proto-oncogene and a tumor suppressor.21-24 In the development of myocytes and megakaryocytes, E2F1 suppression is required for the initiation of differentiation.22,23 Paradoxically, E2F1 is also reported to play an active role in adipogenesis.24 In this case, the role of E2F1 in differentiation may be cell type- and tissue-specific. Since osteosarcoma is also reported to originate from MSCs,7,25 and ATRA is able to induce osteogenic differentiation of osteosarcoma, we are thus encouraged to address if E2F1 similarly plays a critical role in ATRA-induced osteogenic differentiation of osteosarcoma.
In the present study, we dissected the expression of E2F1 in osteosarcoma patient samples and provided evidences that E2F1 impairs all-trans RA (ATRA)-induced osteoblastic differentiation through interacting with retinoic acid receptor α (RARα): E2F1 physically associates with RARα and promotes the ubiquitination-dependent degradation of RARα, thus, RARα-mediated differentiation is inhibited. Moreover, our studies in primary osteosarcoma cultures demonstrated that the E2F1 expression level serves as a potential indicator of the effectiveness of ATRA treatment for osteosarcoma. These observations are meaningful for further understanding the mechanism of ATRA-induced osteogenic differentiation, and provide future direction to develop efficacious differentiation therapies for osteosarcoma.
Results
Osteosarcoma cells are coupled with individual differences in the expression levels of E2F1 in vivo
In this study, we first evaluated the expression levels of E2F1 in primary osteosarcoma tissues from patients by immunohistochemistry assay. A total of 37 separate osteosarcoma tissue blocks were analyzed, and the intensity and percentage of staining were determined. The immunohistochemical staining patterns of E2F1 were evaluated by an experienced pathologist and scored as: (1) negative (“−”, no positive staining or up to 1% of scattered positive cells); (2) low (“+”, heterogeneous staining, where an area corresponding to at least 20% of the section showed 2~10% positive cells); (3) medium (“++”, heterogeneous, with at least 20% of the section showing 10~50% positive cells); (4) high positivity (“+++”, variable to almost homogeneous staining, with at least 20% of the section showing 51~90% positive cells). As illustrated in Figure 1A, 4 reprehensive cases with different E2F1 expression levels were shown. Furthermore, the distributional patterns of E2F1 in all patient samples were also analyzed: only 5 of 37 osteosarcoma cases (13.5%) were determined to demonstrate negative expression of E2F1 in tumor tissues, whereas the remaining 32 samples (86.5%) showed mild to intense E2F1 immunoreactivities (Fig. 1B). Consequently, the results demonstrated that the expression level of E2F1 is coupled with individual differences in human osteosarcoma tissues.
Figure 1.
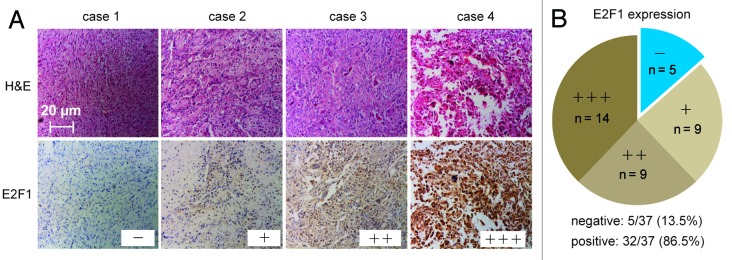
Osteosarcoma cells are coupled with individual differences in the expression levels of E2F1 in vivo. (A) Four reprehensive cases of immunohistochemical analysis of E2F1 expression levels in human osteosarcoma tissues. Four cases were subjected to immunohistochemical staining using an anti-E2F1, and cryosections were stained with H&E. (B) The expression levels of E2F1 in 37 detected osteosarcoma tumor tissues were graded and summarized in pie charts. (A and B) “-”, negative expression; ‘+”, low expression; “++”, medium expression; “+++”, high positive expression.
E2F1 impairs ATRA-induced osteogenic differentiation in osteosarcoma cells
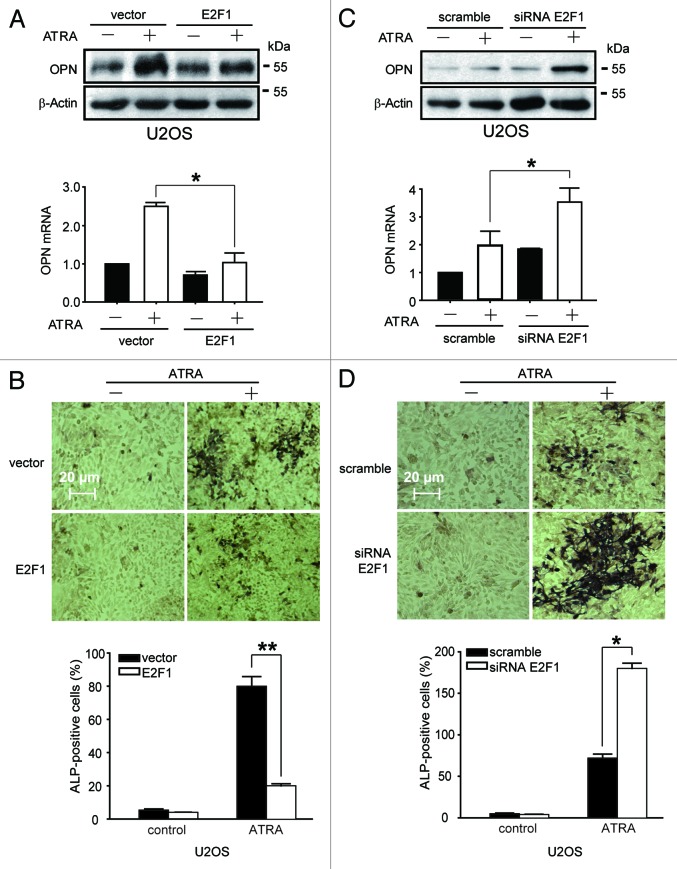
We previously reported that ATRA could effectively induce osteogenic differentiation of U2OS cells;14 thus, the role of E2F1 was determined by using this established cell model in this study. By overexpressing or silencing E2F1, the differentiation efficiency of ATRA was evaluated. Western blotting results showed that transfection with pSG5L-HA-E2F1 plasimd led to significant elevation of E2F1 protein (Fig. S1A). Since osteoblasts have the properties to synthesize osteopontin (OPN) and stimulate high alkaline phosphatase (ALP) activity,26,27 western blotting and real-time PCR analysis were performed to evaluate the production of OPN, and BCIP/NBT staining was further applied to detect ALP activity. Our results showed that ATRA induced apparent upregulation of OPN expression and ALP activity in U2OS cells as we previously reported,14 while there were no active osteoblasts present in E2F1-overexpressed U2OS cells even after ATRA treatment (Fig. 2A and B). Consistently, silencing of endogenous E2F1 expression by specific siRNA (Fig. S1B) remarkably potentiated cytodifferentiating activity of ATRA in U2OS cells, as indicated by OPN as well as ALP activity (Fig. 2C and D). Therefore, these results revealed that E2F1 impairs ATRA-induced osteogenic differentiation in osteosarcoma cells.
Figure 2.
E2F1 impairs ATRA-induced osteogenic differentiation of U2OS cells. (A and B) Overexpression of E2F1 inhibits ATRA-induced osteogenic differentiation. The protein and mRNA levels of OPN were determined (A), and the activities of ALP were evaluated (B). (C and D) Silencing of E2F1 promotes ATRA-induced osteogenic differentiation. The levels of OPN were determined (C), and the activities of ALP were evaluated (D). (A–D) U2OS cells transfected with E2F1 plasmid (A and B) or E2F1 siRNA (C and D) along with corresponding control were treated with 1 μM ATRA for 7 d. *, P < 0.05.
ATRA-induced osteogenic differentiation is only effective in E2F1 low expressed primary osteosarcoma cells
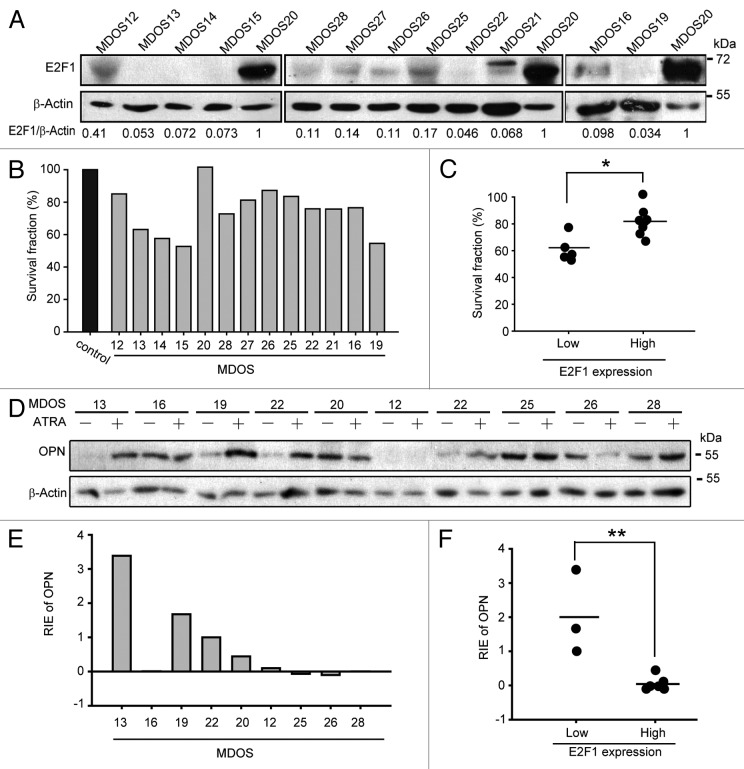
As E2F1 plays negative regulatory roles in ATRA-induced osteogenic differentiation, we further intended to validate this probable correlation between E2F1 and ATRA-induced differentiation in primary osteosarcoma cells. The basal protein expression levels of E2F1 in 13 primary osteosarcoma cells cultured from fresh tissue sections from biopsies of osteosarcoma patients were examined by western blotting, as indicated in Figure 3A. In addition, the effects of ATRA on the anti-proliferation were detected by trypan blue exclusion. After ATRA treatment for 7 d, the proliferation of several osteosarcoma cultures, including MDOS12, MDOS13, MDOS14, MDOS15, MDOS19, were significantly inhibited (Fig. 3B). We adopted the value of relative E2F1 expression of MDOS16 as a cutoff point to divide the cultures into low and high E2F1 expression groups. Among all cultures, those with low E2F1 expression were considerably more sensitive to the growth-inhibitory effect of ATRA when comparing to those with high E2F1 expression with a P value < 0.5 (Fig. 3C). To further evaluate the relationship between E2F1 expression and the sensitivity to cyto-differentiating activity of ATRA, the protein levels of OPN were also detected after ATRA treatment in 9 selected primary osteosarcoma cultures (Fig. 3D). The relative increase of OPN protein level in each culture were summarized in Figure 3E, and the results showed that primary osteosarcoma cultures with low E2F1 expressions displayed remarkably more relative increased OPN protein level, while almost no increase in OPN expression were observed in cultures with high E2F1 expressions (Fig. 3F). Collectively, these results strongly suggested that E2F1 may inhibit the effectiveness of ATRA during differentiation therapy of osteosarcoma in vivo, and ATRA-induced osteogenic differentiation is only effective in E2F1 low expressed primary osteosarcoma cells.
Figure 3.
Inverse correlation between E2F1 expression levels and sensitivity of primary osteosarcoma cells to ATRA-inducd cyto-differentiating. (A) The protein levels of E2F1 in 13 primary osteosarcoma cultures. Expression levels of E2F1 were determined and normalized to β-actin, and then the relative E2F1 expression was set based on the value of sample MDOS20. (B) The anti-proliferation effect of ATRA in 13 primary osteosarcoma cultures. Primary osteosarcoma cultures were treated with 5 μM ATRA for 7 d followed by trypan blue exclusion. (C) The relationship between E2F1 expression levels and anti-proliferation effects of ATRA in 13 primary osteosarcoma cultures. According to the relative E2F1 expression value of MDOS16, cultures were divided into 2 groups: E2F1 expression low (MDOS13, 14, 15, 19, 21, 22) and high (MDOS12, 16, 20, 25, 26, 27, 28). Then cell viabilities were analyzed. *, P < 0.05. (D and E) The protein levels of OPN in the absence or presence of ATRA treatment in primary osteosarcoma cultures. Primary osteosarcoma cultures were treated with 5 μM ATRA for 7 d, and the levels of OPN were determined and normalized to β-actin; then the relative increased OPN expression was set based on the value of sample MDOS16. RIE, relative increased expression. (F) The correlation between relative increased OPN expression and basal E2F1 expression levels in primary osteosarcoma cultures. As presented in (C), 2 groups were set first, then the relative increased OPN expression for each culture were analyzed as presented. **, P < 0.01.
RARα expression is inversely related to the level of E2F1 in osteosarcoma tissues and cells
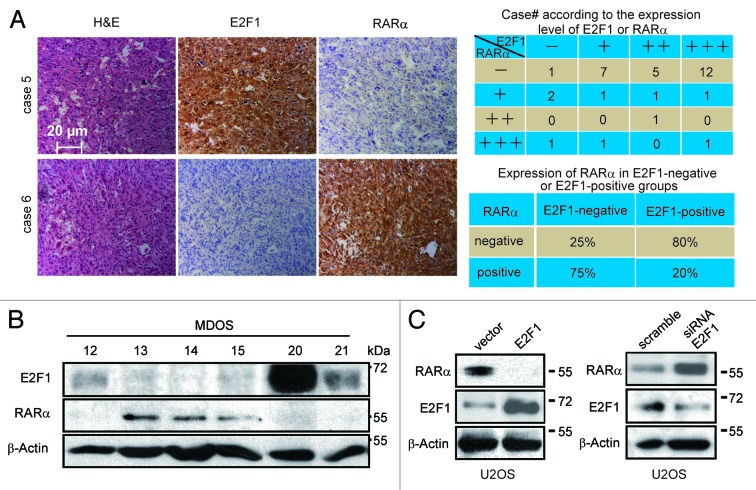
Our previous studies reported that retinoic acid receptors play a critical role in ATRA-induced osteogenic differentiation;14 therefore, we next asked how E2F1 finely regulates ATRA-induced differentiation, and whether RARα is involved. The RARα expression in tissue sections from biopsies of patients was assessed by immunohistochemistry assay first. Our subsequent finding showed a low frequency of RARα in osteosarcoma tissues, with a negative expression rate of 71.4% (25/35) (Fig. S2A). Since the expression patterns of E2F1 and RARα vary significantly in osteosarcoma tissues, we sought to explore whether there were some probable correlations between these 2 proteins by comparing the expression of E2F1 against RARα. Thirty-four osteosarcoma cases were included in this analysis, as demonstrated in Figure 4A. Out of 4 cases with negative E2F1 expression, 3 cases were positive for RARα expression (75%). Among the remaining 30 cases with moderate to intense E2F1 expression, 24 cases showed negative RARα expression (80%). Therefore, a significant inverse correlation was existed between those 2 proteins in patient tumor samples. Consistent with the immunohistochemistry results, the same phenomenon was also found among a panel of primary human osteosarcoma cultures (Fig. 4B).
Figure 4.
RARα expression is inversely related to the levels of E2F1 in osteosarcoma tissues and cells. (A) Correlation analysis of E2F1 with RARα in osteosarcoma tumor tissues. The immunoreactivities of E2F1 and RARα in osteosarcoma tumor tissues were illustrated (left) and the analyzed results as presented (right). (B) Inverse correlation between E2F1 and RARα expression levels in primary osteosarcoma cultures. The protein levels of E2F1 and RARα in primary osteosarcoma cultures were determined. (C) E2F1 inversely regulates RARα. U2OS cells were transfected with E2F1 plasmid (left) or E2F1 siRNA (right) along with corresponding control groups, and the protein levels of RARα and E2F1 were detected.
To determine whether the inverse correlation between E2F1 and RARα reflects an inhibition effect of E2F1 on the protein levels of RARα, we evaluated the involvement of E2F1 in controlling the stability of RARα. As shown in Figure 4C, overexpression of E2F1 caused a remarkable decrease in the amount of RARα in U2OS cells. Consistent with this result, in transient transfection studies, we found that coexpression of E2F1 dramatically reduced the levels of ectopic RARα expression in COS7 cells (Fig. S2B). Likewise, silencing E2F1 increased the basal level of RARα in U2OS cells (Fig. 4D). Given that (1) E2F1 is highly expressed in osteosarcoma cells and tissues, and E2F1 impairs the differentiation effect of ATRA in osteosarcoma; (2) E2F1 is accompanied by negative RARα expression in the majority of osteosarcoma tumors; (3) studies indicated that RARα is the receptor for ATRA and RARα may be inhibited by E2F1, we proposed that RARα may be critical downstream of E2F1 to regulate ATRA-induced differentiation in osteosarcoma.
E2F1 directly interacts with RARα and inhibits RARα-mediated osteogenic differentiation
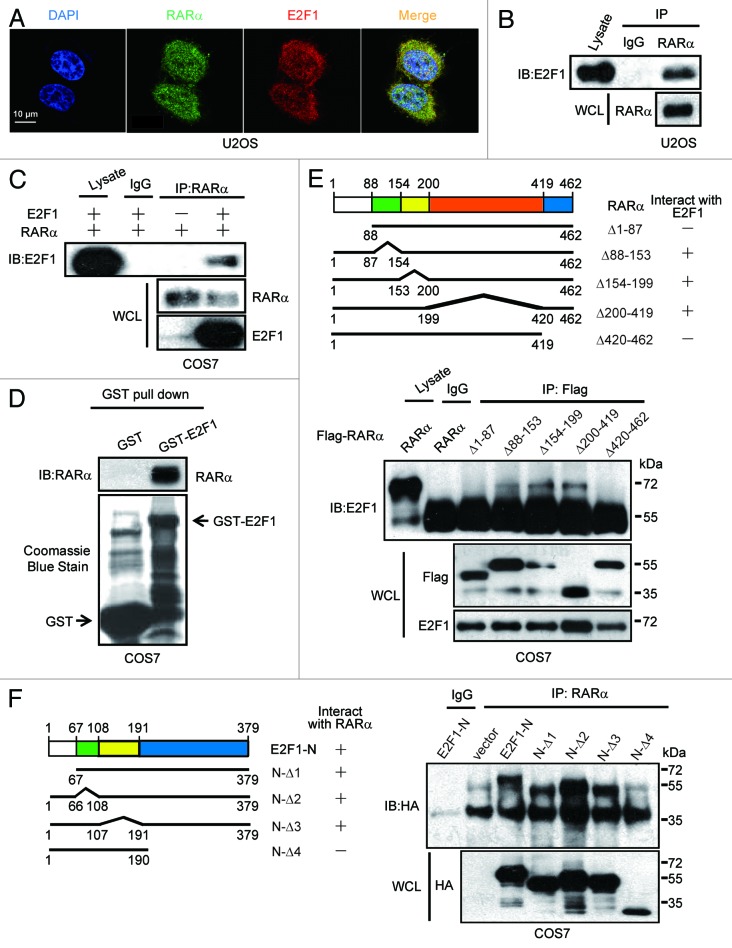
To gain insight into the mechanisms underlying the inverse relationship between E2F1 and RARα, we first examined whether E2F1 regulates RARα through direct interaction. Immunofluorescence results demonstrated that endogenous E2F1 was partially colocalized with endogenous RARα in U2OS cells, suggesting the interaction between these 2 proteins may exist (Fig. 5A). Furthermore, this interaction was further validated by immunoprecipitation experiments. U2OS cell extracts were subjected to immunoprecipitation with anti-RARα or anti-IgG as control. As shown in Figure 5B, E2F1 was precipitated by anti-RARα but not by the control, indicating that endogenous E2F1 may interact with endogenous RARα. Likewise, ectopically expressed E2F1 and RARα were coprecipitated with each other in COS7 cells, which was overexpressed with both proteins, whereas little or no coprecipitation occurred in cells that overexpressed RARα only, suggesting that the coprecipitation was not due to cross-reactivity of antibodies, and exogenous E2F1 indeed interacts with RARα (Fig. 5C). In order to more accurately assess whether this interaction is ligand-independent, GST pull-down assay was applied. Results shown in Figure 5D demonstrated that RARα was precipitated with GST-E2F1 but not with GST, indicating that E2F1 and RARα form a complex in vitro. In total, the current data supports our hypothesis that E2F1 and RARα interact specifically.
Figure 5.
Interaction between E2F1 and RARα. (A) Colocalization of endogenous E2F1 and RARα. The subcellular localization of E2F1 and RARα in U2OS cells was assessed by immunofluorescence. (B) Interaction between endogenous E2F1 and RARα. U2OS cell lysates were immunoprecipitated with anti-RARα followed by immunoblotting with anti-E2F1. WCL, whole-cell lysates. (C) Interaction between ectopical expressions of E2F1 and RARα. COS7 cells were transfected with RARα and E2F1; 48 h later, cell lysates were coprecipitation with anti-RARα and immunoblotting with anti-E2F1 or anti-RARα. (D) Interaction between E2F1 and RARα in vitro. GST or GST-E2F1 fusion proteins were used in a pull-down assay with in vitro translated. (E) The N terminus and C terminus of RARα are both required for E2F1 binding. COS7 cells transfected with HA-E2F1 along with different deletion mutants of Flag-RARα were immunoprecipitated with anti-Flag and immunoblotted with anti-E2F1 or anti-Flag (lower). Different deletion mutants of RARα were presented (upper). (F) N terminus of E2F1 involved in the interaction with RARα. RARα and E2F1-N mutants were transfected into COS7 cells followed by immunoprecipitated with anti-RARα and immunoblotted with anti-HA (right). Different E2F1-N deletion mutant constructs were illustrated (left).
To define the region of RARα that is required for E2F1 binding, COS7 cells were transfected with E2F1 together with 5 deletion constructs of RARα fused to Flag-tag. Cellular extracts were then immunoprecipitated with anti-Flag antibodies. Results showed that deletion of either the N terminus (1–87) or C terminus (420–462) of RARα abolished its interaction with E2F1, suggesting that both the N terminus and C terminus of RARα are necessary for E2F1 binding (Fig. 5E). Since E2F1-N (1–379) was previously demonstrated to be required for association with RARα in our experiment (data not shown), different E2F1-N (1–379) constructs were co-transfected with RARα into COS7 cells, and the region of E2F1 that is required for RARα binding was defined. As shown in Figure 5F, all E2F1-N (1–379) mutants, except E2F1-N-Δ4, interacted with RARα. Consistent with this interpretation, overexpression of E2F1-N-Δ4 was observed as not able to decrease protein expression levels of RARα as well as inhibit ATRA-induced osteogenic differentiation (Fig. S3A–C). Thus, these results obviously suggest that E2F1 interacts with RARα directly, and this interaction is responsible for E2F1-impaired RARα expression as well as osteoblastic differentiation.
E2F1 promotes ubiquitination-mediated degradation of RARα
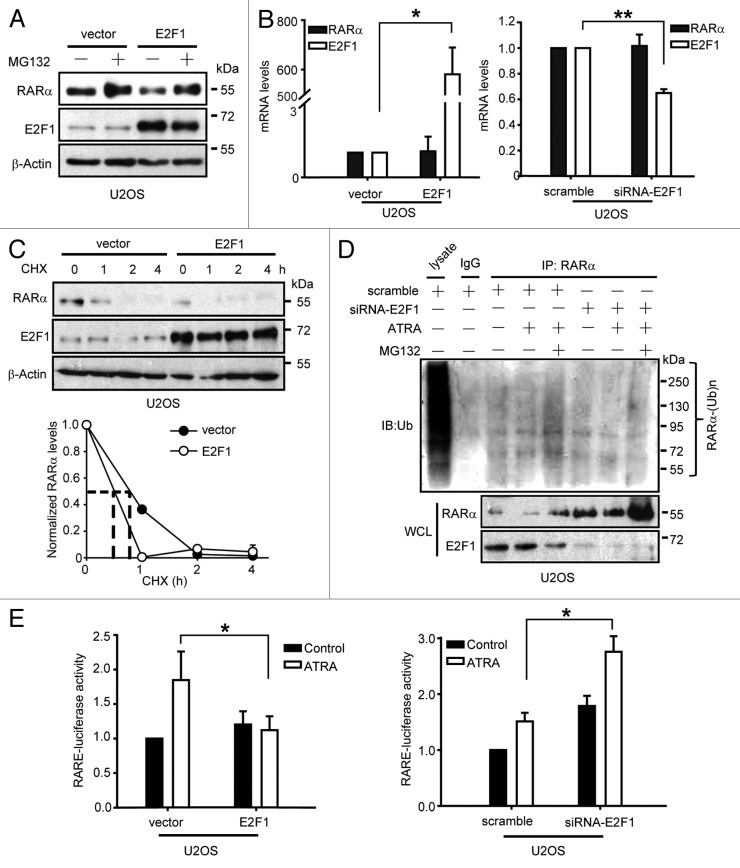
Finally, we sought to determine the probable mechanism responsible for E2F1-impaired RARα expression. We first observed that MG132, a known proteasome inhibitor, was able to prevent downregulation of RARα caused by E2F1 (Fig. 6A). This probably indicated that E2F1 regulates the basal turnover of RARα via control of its degradation by proteasome. Likewise, along with overexpressing or silencing E2F1, the mRNA levels of RARα remain stable (Fig. 6B), further suggesting that a transcription-independent mechanism is involved in negative regulation of RARα by E2F1. To further investigate whether downregulation of RARα by E2F1 is due to protein degradation, the half-life of endogenous RARα was measured. U2OS cells transfected with pSG5L-HA or pSG5L-HA-E2F1 plasmid were treated with protein synthesis inhibitor CHX, and the protein levels of endogenous RARα were examined. Results showed that the protein levels of RARα decreased much more rapidly in E2F1-overexpressed U2OS cells than control cells (Fig. 6C). This data further raised the possibility that E2F1 may function as an inhibitor of RARα by promoting its degradation. Since previous studies have demonstrated that the degradation of RARα is mainly mediated through the ubiquitination–proteasome pathway, ATRA-induced ubiquitination of RARα was therefore examined.28,29 Our results demonstrated that deletion of E2F1 by siRNA significantly reduced RARα ubiquitination (Fig. 6D). Therefore, E2F1 can be identified as a critical regulator involved in RARα ubiquitination. To assess the functional consequences of E2F1-inhibited RARα, E2F1 plasmid or specific E2F1 siRNA was transfected along with retinoid-responsive reporter, RARE-luciferase, into U2OS cells, then the activation of RARα was evaluated. Results showed that exogenous E2F1 dramatically inhibited ATRA-induced RARE-luciferase activity, whereas E2F1 siRNA upregulated it (Fig. 6E). This result was also supported by the analysis of 3 selected retinoid-dependent genes (Fig. S4A and B). Taken together, our results clearly indicated that E2F1 functions as a negative regulator of RARα by interacting with it and promoting its ubiquitination–proteasome-mediated degradation.
Figure 6.
E2F1 destabilizes RARα by promoting ubiquitination-mediated degradation of RARα. (A) MG132 blocks RARα degradation induced by ectopic expression of E2F1. U2OS cells were treated with or without 20 μM MG132 for 12 h; then the protein levels of RARα and E2F1 were determined. (B) E2F1 does not influence mRNA levels of RARα. The mRNA levels of RARα and E2F1 in U2OS cells transfected with E2F1 plasmid (left) or E2F1 siRNA (right) were determined. Expression levels were normalized to GAPDH. (C) Overexpression of E2F1 decreases half-time of RARα. U2OS cells were treated with 10 μg/ml cycloheximide (CHX), and then the protein levels of RARα and E2F1 were determined. The relative levels of RARα were normalized to β-actin as indicated (low). (D) Knocking down of E2F1 inhibits ubiquitination of RARα. U2OS cells were transfected with scramble or E2F1 siRNA followed by treatment with ATRA and MG132 for 12 h; then cell lysates were immunoprecipitated with anti-RARα and immunoblotted with anti-ubiquitin. (E) E2F1 inversely regulates RARα promoter activity. U2OS cells were transfected with RARE and Renilla reporter genes plus vector and E2F1 plasmids (left) or scramble and E2F1 siRNA (right) followed by treatment with 1 μM ATRA for 6 h. Firefly luciferase activities were measured and normalized to Renilla. (A and C) U2OS cells were transfected with vector and E2F1 plasmids. *, P < 0.05; **, P < 0.01.
Discussion
Induction of terminal differentiation may present a promising alternative to conventional chemotherapy for osteosarcoma, since ATRA can induce osteoblastic differentiation of osteosarcoma and restore normal osteogenesis.15,30 However, no clinical application for ATRA in osteosarcoma patients has been reported to date. Hence, increasing attention has been focused on the effectiveness of ATRA treatment in patients as well as the discovery of a proper biomarker for selecting respondent osteosarcoma patients for differentiation therapy. In our study, for the first time, we provided evidences for the involvement of E2F1 in controlling osteogenic differentiation. We found that enforced expression of E2F1 in U2OS cells significantly attenuates ATRA-stimulated osteoblastic differentiation, while downregulation of E2F1 potentiates the differentiation-inducing activity of ATRA (Fig. 2). Moreover, by using many primary osteosarcoma cultures, ATRA treatment is observed only effective in E2F1 low expressed cells (Fig. 3). In addition, E2F1 is found to specifically bind to RARα and promotes its ubiquitination-mediated degradation, leading to inhibition of differentiation in osteosarcoma (Figs. 4–6). Therefore, these data suggest that ATRA-based differentiation therapy is indeed effective in primary osteosarcoma cells, only if these cells are E2F1 low expressed. Our study provides evidence to support E2F1 serve as a potent biomarker of osteosarcoma patient selection for differentiation therapy and may open new opportunities to use ATRA as a more promising approach for osteosarcoma patients in future.
Including osteosarcoma, various solid cancers are already being treated with retinoid-based therapies, and several are undergoing clinical evaluation.31-33 For example, Kaposi sarcoma and head-and-neck cancer have already been treated with ATRA-based therapies.34,35 Retinoids are clearly a promising class of compounds that will greatly enlarge our arsenal in the fight against cancer. However, there is no report of biomarker for retinoid-based differentiation therapy. Here we found that E2F1 may serve as a potent indicator for the effectiveness of ATRA treatment in chemotherapy of osteosarcoma, because the response rates of ATRA treatment are dependent on the levels of E2F1 (Fig. 3): under high expression levels of E2F1, only 2 of 7 primary cultures (29%) are responsive to ATRA-inhibited proliferation, and 2 of 6 samples (33%) are induced to express OPN; under mild to negative levels of E2F1, up to 100% of the primary cells are sensitive to ATRA-inhibited proliferation (6 of 6 samples) and ATRA-induced OPN upregulation (3 of 3 samples). Besides our study, E2F1 is also reported to serve as a putative biomarker that could gauge the anti-melanoma activity of p53–MDM2 inhibitors.36,37 Actually, in the past few years, pre-clinical experiments mainly using E2F1 itself as a biomarker or anti-cancer therapeutic target have been initiated. For example, E2F1 is found to be applied as molecular biomarkers for Parkinson disease (PD).38 Additionally, Gorgoulis et al. revealed that E2F1 overexpression is associated with a poor prognosis in NSCLC patients.39 E2F1 transcript levels are also reported as a strong determinant of favorable breast cancer outcome.40 Unfortunately, in our study, only 5 of 37 osteosarcoma cases (13.5%) are determined to demonstrate negative expression of E2F1 (Fig. 1), and this result also provided some explanations for clinical resistance to ATRA of osteosarcoma, since upregulation of E2F1 is very common in osteosarcoma. In this case, prior measurement of E2F1 expression should be performed as a potential index for selecting osteosarcoma patients for ATRA treatment. Further studies should be performed to confirm the expression levels of E2F1 with more osteosarcoma patients, and prospective trials of ATRA treatment for osteosarcoma should also be performed to determine the cutoff point of E2F1 expression in each tumor tissue for ATRA treatment to be an effective clinical chemotherapy.
RARα, a retinoic acid receptor, plays an important role in ATRA-induced osteosarcoma differentiation, and insufficient RARα is not beneficial for differentiation progress.14,41 Interestingly, we found that E2F1 directly interacts with RARα and promotes ubiquitination–proteasome-mediated degradation of RARα (Fig. 5 and 6). Hence, for the critical role of RARα in ATRA-induced differentiation, the modulation of RARα by E2F1 is bound to affect ATRA-induced differentiation in osteosarcoma. There have been only a few studies indicating the role of E2F1 in regulation of protein stability instead of expression. It was demonstrated that besides increases the mRNA level of p73, deregulated expression of E2F1 promotes proteolytic degradation of p73 in a proteasome-independent manner, suggesting that E2F1 has a dual role in the regulation of p73.42 In addition, E2F1 was reported to increase protein degradation of MDM2 via directly activating expression of p14/ARF tumor suppressor, thereby elevating p53 levels and leading to apoptosis.43 Further studies showed that the expression of an E2F1 mutant, E2F1 (180–437), which lacks the DNA binding domain, results in proteolytic degradation of MDM2, MDMX, and MDMX-S proteins, raising a possibility of promoting cell death independent of E2F1-driven transcription.44 Although the specific mechanism by which E2F1 promoting ubiquitination of RARα is unknown, our findings indicated for the first time that RARα can be inversely regulated by E2F1, which adds to our understanding of novel mechanisms underlying RARα degradation.
In summary, our data demonstrated that E2F1 impairs ATRA-induced osteogenic differentiation by promoting ubiquitination–proteasome pathway-mediated degradation of RARα via physical interaction. Reducing the expressions of E2F1 by means of biological treatments may therefore have a great potential for potentiating differentiation-inducing activity of ATRA. Furthermore, the correlation between the expression levels of E2F1 and effectiveness of ATRA treatment in primary osteosarcoma cultures raises the possibility that E2F1 may serve as a predictor for the sensitivity of osteosarcoma to ATRA treatment. This knowledge may help us develop efficacious differentiation therapies for osteosarcoma to a large extent.
Materials and Methods
Cell culture and chemicals
Human osteosarcoma U2OS cells and COS7 cells were from the Shanghai Institute of Biochemistry and Cell Biology, maintained in 1640 and DMEM medium, respectively. All primary osteosarcoma cultures were from fresh tissue sections from biopsies of osteosarcoma patients and maintained in DMEM/F12 medium. All cells were routinely authenticated.45 All media were supplemented with 10% fetal bovine serum (Hyclone) plus 1% penicillin/streptomycin. All cells were incubated at 37 °C in a 5% CO2 atmosphere.
All-trans retinoic acid (ATRA) and cycloheximide (CHX) were obtained from Sigma-Aldrich. MG132 was purchased from EMD Biosciences, Inc.
Immunohistochemistry and immunofluorescence
Human osteosarcoma tissues were paraffin embedded. After deparaffinization, the slides were blocked with 3% hydrogen peroxide and preincubated in 20% normal goat serum, then probed with anti-E2F1 or anti-RARα followed by biotinylated secondary antibodies and HRP-conjugated avidin. E2F1 and RARα were visualized with 3, 3′-diaminobenzidine. Immunofluorescence was conducted as previously described.46
Transient transfection and siRNA transfection
pSG5L-HA-E2F1 plasmid was purchased from Addgene, and pccl-RARα plasmid was a kind gift from Dr Lingtao Wu (University of Southern California, Keck School of Medicine). Scramble and E2F1 siRNA were synthesized by Shanghai GenePharma Co, Ltd. The sense strand of E2F1 siRNA was as follows: UGGACCACCU GAUGAAUAU.47 Briefly, cells were transfected with indicated plasmids using Lipofectamine 2000 (Invitrogen) or siRNA with oligofectamine (Invitrogen). The deletion mutants were constructed by GenScript.
Real-time PCR
Total RNA was prepared using Trizol reagent (Bio Basic Inc). Real-time PCR was performed as previously described.29 Primers used for PCR were as follows: OPN, forward: GGATCCCTCA CTACCATGAG; reverse: AAGCTTGACC TCAGAAGATG CACT48; E2F1, forward: CCGCCATCCA GGAAAAGG; reverse: GCCCTCAAGG ACGTTGGT49; RARα, forward: ACCCCCTCTA CCCCGCATCT ACAAG; reverse: CATGCCCACT TCAAAGCACT TCTGC29; GAPDH, forward: GTCATCCATG ACAACTTTGG; reverse: GAGCTTGACA AAGTGGTCGT.50 GAPDH was used as an internal standard.
Alkaline phosphatase assay
Alkaline phosphatase activity was assessed by colorimetric assay using BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime Institute of Biotechnology) according to the manufacturer’s instructions. Quantitative analysis of ALP-positive cells was made, and vector or scramble group was used as an internal standard.
Cell proliferation assay
Primary osteosarcoma cultures were treated with ATRA for 7 d, and then total cell number and viability were determined by trypan blue exclusion with manual counting in Burker chambers.
Immunoprecipitation and western blotting
To detect E2F1-RARα conjugation, immunoprecipitation, and western blotting were conducted as previously described.29 Antibodies against E2F1 (KH20), RARα (C-20), ubiquitin (P4D1), OPN (LFMb-14), β-Actin (C-11), and protein A/G plus agarose were from Santa Cruz Biotechnology Inc. HA-tag and Flag-tag were purchased from GeneScript Corporation. Secondary antibodies were purchased from Jackson ImmunoResearch Laboratories, Inc.
GST pull-down assay
pGEX-KG-GST and pGEX-KG-GST-E2F1 were expressed in E. coli BL21 (DE3) and induced with 0.4 mM isopropyl-thio-β-D-galactopyranoside. After overnight incubation, the bacterial lysates containing GST or GST-E2F1 were incubated with Glutathione Sepharose 4B (GE) beads at 4 °C for 6 h. Then COS7 cell lysates overexpressing RARα protein were added to the beads and incubated for 3 h followed by proteins eluted from the beads and analyzed by western blotting.
Luciferase reporter assay
U2OS cells (transfected with the indicated plasmids or siRNAs) were cotransfected with a Renilla luciferase plasmid and pGL3-RARE-luciferase plasmid. After ATRA treatment, luciferase activities were measured using Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activities were normalized by Renilla luciferase activities.
Statistical analysis
ANOVA or Student unpaired, 2-tailed t test was used when appropriate.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
This work was supported by grants from National Natural Science Foundation of China (No.81273534 and No.81202558), Program for New Century Excellent Talents in University, Zhejiang Province Program for the Cultivation of High-level Innovative Health Talents, Zhejiang Provincial Program for Qianjiang Talents (2013R10025), the Fundamental Research Funds for the Central Universities.
Glossary
Abbreviations:
- RARα
retinoic acid receptor α
- RARE
retinoic acid response element
- ATRA
all-trans retinoic acid
- CHX
cycloheximide
- ALP
alkaline phosphatase
References
- 1.Dorfman HD, Czerniak B. Bone cancers. Cancer. 1995;75(Suppl):203–10. doi: 10.1002/1097-0142(19950101)75:1+<203::AID-CNCR2820751308>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 2.Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: a review. Sarcoma. 2011;2011:959248. doi: 10.1155/2011/959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG, Meyers PA, Lafleur B, McDonough WS, Henry MM, et al. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005;65:1748–54. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 4.Sabile AA, Arlt MJ, Muff R, Bode B, Langsam B, Bertz J, Jentzsch T, Puskas GJ, Born W, Fuchs B. Cyr61 expression in osteosarcoma indicates poor prognosis and promotes intratibial growth and lung metastasis in mice. J Bone Miner Res. 2012;27:58–67. doi: 10.1002/jbmr.535. [DOI] [PubMed] [Google Scholar]
- 5.Thomas DM, Johnson SA, Sims NA, Trivett MK, Slavin JL, Rubin BP, Waring P, McArthur GA, Walkley CR, Holloway AJ, et al. Terminal osteoblast differentiation, mediated by runx2 and p27KIP1, is disrupted in osteosarcoma. J Cell Biol. 2004;167:925–34. doi: 10.1083/jcb.200409187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haydon RC, Luu HH, He TC. Osteosarcoma and osteoblastic differentiation: a new perspective on oncogenesis. Clin Orthop Relat Res. 2007;454:237–46. doi: 10.1097/BLO.0b013e31802b683c. [DOI] [PubMed] [Google Scholar]
- 7.Mohseny AB, Szuhai K, Romeo S, Buddingh EP, Briaire-de Bruijn I, de Jong D, van Pel M, Cleton-Jansen AM, Hogendoorn PC. Osteosarcoma originates from mesenchymal stem cells in consequence of aneuploidization and genomic loss of Cdkn2. J Pathol. 2009;219:294–305. doi: 10.1002/path.2603. [DOI] [PubMed] [Google Scholar]
- 8.Dai X, Ma W, He X, Jha RK. Review of therapeutic strategies for osteosarcoma, chondrosarcoma, and Ewing’s sarcoma. Med Sci Monit. 2011;17:RA177–90. doi: 10.12659/MSM.881893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degos L, Dombret H, Chomienne C, Daniel MT, Micléa JM, Chastang C, Castaigne S, Fenaux P. All-trans-retinoic acid as a differentiating agent in the treatment of acute promyelocytic leukemia. Blood. 1995;85:2643–53. [PubMed] [Google Scholar]
- 10.Reynolds CP, Matthay KK, Villablanca JG, Maurer BJ. Retinoid therapy of high-risk neuroblastoma. Cancer Lett. 2003;197:185–92. doi: 10.1016/S0304-3835(03)00108-3. [DOI] [PubMed] [Google Scholar]
- 11.Zanardi S, Serrano D, Argusti A, Barile M, Puntoni M, Decensi A. Clinical trials with retinoids for breast cancer chemoprevention. Endocr Relat Cancer. 2006;13:51–68. doi: 10.1677/erc.1.00938. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Satyamoorthy K, Herlyn M, Rosdahl I. All-trans retinoic acid (atRA) differentially induces apoptosis in matched primary and metastatic melanoma cells -- a speculation on damage effect of atRA via mitochondrial dysfunction and cell cycle redistribution. Carcinogenesis. 2003;24:185–91. doi: 10.1093/carcin/24.2.185. [DOI] [PubMed] [Google Scholar]
- 13.Garattini E, Paroni G, Terao M. Retinoids and breast cancer: new clues to increase their activity and selectivity. Breast Cancer Res. 2012;14:111. doi: 10.1186/bcr3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo P, Yang X, Ying M, Chaudhry P, Wang A, Shimada H, May WA, Adams GB, Mock D, Triche TJ, et al. Retinoid-suppressed phosphorylation of RARalpha mediates the differentiation pathway of osteosarcoma cells. Oncogene. 2010;29:2772–83. doi: 10.1038/onc.2010.50. [DOI] [PubMed] [Google Scholar]
- 15.Haydon RC, Zhou L, Feng T, Breyer B, Cheng H, Jiang W, Ishikawa A, Peabody T, Montag A, Simon MA, et al. Nuclear receptor agonists as potential differentiation therapy agents for human osteosarcoma. Clin Cancer Res. 2002;8:1288–94. [PMC free article] [PubMed] [Google Scholar]
- 16.Hisada K, Hata K, Ichida F, Matsubara T, Orimo H, Nakano T, Yatani H, Nishimura R, Yoneda T. Retinoic acid regulates commitment of undifferentiated mesenchymal stem cells into osteoblasts and adipocytes. J Bone Miner Metab. 2013;31:53–63. doi: 10.1007/s00774-012-0385-x. [DOI] [PubMed] [Google Scholar]
- 17.Dingwall M, Marchildon F, Gunanayagam A, Louis CS, Wiper-Bergeron N. Retinoic acid-induced Smad3 expression is required for the induction of osteoblastogenesis of mesenchymal stem cells. Differentiation. 2011;82:57–65. doi: 10.1016/j.diff.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–56. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda K, Fujita J. Mesenchymal, but not hematopoietic, stem cells can be mobilized and differentiate into cardiomyocytes after myocardial infarction in mice. Kidney Int. 2005;68:1940–3. doi: 10.1111/j.1523-1755.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 20.Heino TJ, Hentunen TA. Differentiation of osteoblasts and osteocytes from mesenchymal stem cells. Curr Stem Cell Res Ther. 2008;3:131–45. doi: 10.2174/157488808784223032. [DOI] [PubMed] [Google Scholar]
- 21.Bell LA, Ryan KM. Life and death decisions by E2F-1. Cell Death Differ. 2004;11:137–42. doi: 10.1038/sj.cdd.4401324. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Helin K, Jin P, Nadal-Ginard B. Inhibition of in vitro myogenic differentiation by cellular transcription factor E2F1. Cell Growth Differ. 1995;6:1299–306. [PubMed] [Google Scholar]
- 23.Guy CT, Zhou W, Kaufman S, Robinson MO. E2F-1 blocks terminal differentiation and causes proliferation in transgenic megakaryocytes. Mol Cell Biol. 1996;16:685–93. doi: 10.1128/mcb.16.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J. E2Fs regulate adipocyte differentiation. Dev Cell. 2002;3:39–49. doi: 10.1016/S1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 25.Dani N, Olivero M, Mareschi K, van Duist MM, Miretti S, Cuvertino S, Patané S, Calogero R, Ferracini R, Scotlandi K, et al. The MET oncogene transforms human primary bone-derived cells into osteosarcomas by targeting committed osteo-progenitors. J Bone Miner Res. 2012;27:1322–34. doi: 10.1002/jbmr.1578. [DOI] [PubMed] [Google Scholar]
- 26.Sodek J, Chen J, Nagata T, Kasugai S, Todescan R, Jr., Li IW, Kim RH. Regulation of osteopontin expression in osteoblasts. Ann N Y Acad Sci. 1995;760:223–41. doi: 10.1111/j.1749-6632.1995.tb44633.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu PP, Leung KS, Kumta SM, Lee KM, Fung KP. Bone-specific alkaline phosphatase in plasma as tumour marker for osteosarcoma. Oncology. 1996;53:275–80. doi: 10.1159/000227573. [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Gianni M, Kopf E, Honoré N, Chelbi-Alix M, Koken M, Quignon F, Rochette-Egly C, de Thé H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci U S A. 1999;96:14807–12. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang Y, Zhou X, Lin M, Ying M, Luo P, Zhu D, Lou J, Yang B, He Q. Inhibition of all-trans-retinoic acid-induced proteasome activation potentiates the differentiating effect of retinoid in acute myeloid leukemia cells. Mol Carcinog. 2011;50:24–35. doi: 10.1002/mc.20687. [DOI] [PubMed] [Google Scholar]
- 30.Laue K, Jänicke M, Plaster N, Sonntag C, Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development. 2008;135:3775–87. doi: 10.1242/dev.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–93. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- 32.Kelly WK, Osman I, Reuter VE, Curley T, Heston WD, Nanus DM, Scher HI. The development of biologic end points in patients treated with differentiation agents: an experience of retinoids in prostate cancer. Clin Cancer Res. 2000;6:838–46. [PubMed] [Google Scholar]
- 33.Connolly RM, Nguyen NK, Sukumar S. Molecular pathways: current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin Cancer Res. 2013;19:1651–9. doi: 10.1158/1078-0432.CCR-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrari N, Morini M, Pfeffer U, Minghelli S, Noonan DM, Albini A. Inhibition of Kaposi’s sarcoma in vivo by fenretinide. Clin Cancer Res. 2003;9:6020–9. [PubMed] [Google Scholar]
- 35.Smith MA, Parkinson DR, Cheson BD, Friedman MA. Retinoids in cancer therapy. J Clin Oncol. 1992;10:839–64. doi: 10.1200/JCO.1992.10.5.839. [DOI] [PubMed] [Google Scholar]
- 36.Verhaegen M, Checinska A, Riblett MB, Wang S, Soengas MS. E2F1-dependent oncogenic addiction of melanoma cells to MDM2. Oncogene. 2012;31:828–41. doi: 10.1038/onc.2011.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pützer BM. E2F1 death pathways as targets for cancer therapy. J Cell Mol Med. 2007;11:239–51. doi: 10.1111/j.1582-4934.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diao H, Li X, Hu S, Liu Y. Gene expression profiling combined with bioinformatics analysis identify biomarkers for Parkinson disease. PLoS One. 2012;7:e52319. doi: 10.1371/journal.pone.0052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorgoulis VG, Zacharatos P, Mariatos G, Kotsinas A, Bouda M, Kletsas D, Asimacopoulos PJ, Agnantis N, Kittas C, Papavassiliou AG. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J Pathol. 2002;198:142–56. doi: 10.1002/path.1121. [DOI] [PubMed] [Google Scholar]
- 40.Vuaroqueaux V, Urban P, Labuhn M, Delorenzi M, Wirapati P, Benz CC, Flury R, Dieterich H, Spyratos F, Eppenberger U, et al. Low E2F1 transcript levels are a strong determinant of favorable breast cancer outcome. Breast Cancer Res. 2007;9:R33. doi: 10.1186/bcr1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gianni M, Peviani M, Bruck N, Rambaldi A, Borleri G, Terao M, Kurosaki M, Paroni G, Rochette-Egly C, Garattini E. p38αMAPK interacts with and inhibits RARα: suppression of the kinase enhances the therapeutic activity of retinoids in acute myeloid leukemia cells. Leukemia. 2012;26:1850–61. doi: 10.1038/leu.2012.50. [DOI] [PubMed] [Google Scholar]
- 42.Ozaki T, Okoshi R, Ono S, Kubo N, Nakagawara A. Deregulated expression of E2F1 promotes proteolytic degradation of tumor suppressor p73 and inhibits its transcriptional activity. Biochem Biophys Res Commun. 2009;387:143–8. doi: 10.1016/j.bbrc.2009.06.141. [DOI] [PubMed] [Google Scholar]
- 43.Itoshima T, Fujiwara T, Waku T, Shao J, Kataoka M, Yarbrough WG, Liu TJ, Roth JA, Tanaka N, Kodama M. Induction of apoptosis in human esophageal cancer cells by sequential transfer of the wild-type p53 and E2F-1 genes: involvement of p53 accumulation via ARF-mediated MDM2 down-regulation. Clin Cancer Res. 2000;6:2851–9. [PubMed] [Google Scholar]
- 44.Strachan GD, Rallapalli R, Pucci B, Lafond TP, Hall DJ. A transcriptionally inactive E2F-1 targets the MDM family of proteins for proteolytic degradation. J Biol Chem. 2001;276:45677–85. doi: 10.1074/jbc.M103765200. [DOI] [PubMed] [Google Scholar]
- 45.Ying M, Zhou X, Zhong L, Lin N, Jing H, Luo P, Yang X, Song H, Yang B, He Q. Bortezomib sensitizes human acute myeloid leukemia cells to all-trans-retinoic acid-induced differentiation by modifying the RARα/STAT1 axis. Mol Cancer Ther. 2013;12:195–206. doi: 10.1158/1535-7163.MCT-12-0433. [DOI] [PubMed] [Google Scholar]
- 46.Cao J, Xu D, Wang D, Wu R, Zhang L, Zhu H, He Q, Yang B. ROS-driven Akt dephosphorylation at Ser-473 is involved in 4-HPR-mediated apoptosis in NB4 cells. Free Radic Biol Med. 2009;47:536–47. doi: 10.1016/j.freeradbiomed.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 47.Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21:2112–23. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- 48.Lee JL, Wang MJ, Sudhir PR, Chen GD, Chi CW, Chen JY. Osteopontin promotes integrin activation through outside-in and inside-out mechanisms: OPN-CD44V interaction enhances survival in gastrointestinal cancer cells. Cancer Res. 2007;67:2089–97. doi: 10.1158/0008-5472.CAN-06-3625. [DOI] [PubMed] [Google Scholar]
- 49.Paik JC, Wang B, Liu K, Lue JK, Lin WC. Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J Biol Chem. 2010;285:6348–63. doi: 10.1074/jbc.M109.072074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadowaki S, Chikumi H, Yamamoto H, Yoneda K, Yamasaki A, Sato K, Shimizu E. Down-regulation of inducible nitric oxide synthase by lysophosphatidic acid in human respiratory epithelial cells. Mol Cell Biochem. 2004;262:51–9. doi: 10.1023/B:MCBI.0000038215.89821.7f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.