Abstract
Breast cancer is a disease of cell cycle, and the dysfunction of cell cycle checkpoints plays a vital role in the occurrence and development of breast cancer. We employed multi-gene fluorescence in situ hybridization (M-FISH) to investigate gene copy number aberrations (CNAs) of 4 genes (Rb1, CHEK2, c-Myc, CCND1) that are involved in the regulation of cell cycle, in order to analyze the impact of gene aberrations on prognosis in the young breast cancer patients. Gene copy number aberrations of these 4 genes were more frequently observed in young breast cancer patients when compared with the older group. Further, these CNAs were more frequently seen in Luminal B type, Her2 overexpression, and tiple-negative breast cancer (TNBC) type in young breast cancer patients. The variations of CCND1, Rb1, and CHEK2 were significantly correlated with poor survival in the young breast cancer patient group, while the amplification of c-Myc was not obviously correlated with poor survival in young breast cancer patients. Thus, gene copy number aberrations (CNAs) of cell cycle-regulated genes can serve as an important tool for prognosis in young breast cancer patients.
Keywords: breast cancer, Rb1, CCND1, c-Myc, CHEK2, survival analysis
Introduction
Breast cancer is the primary cause for female cancer death in the world. In 2008, breast cancer patients account for 23% of newly diagnosed cancer (about 1 380 000) and 13% of the total cancer deaths (about 458 400).1 Although the portion of the young patients in breast cancer is small, about 1.9%, the recurrence and metastasis risk is high, and prognosis is poor.1 Many factors are proposed to correlate with the adverse prognosis of the young breast cancer, such as tumor size at first diagnosis, high tumor stage and grade, lymph node infiltration, overexpression of HER2, expression deletion of estrogen and progesterone receptors, etc.2 After correction for stage, tumor characteristics, and treatment, age remains an independent risk factor for breast cancer death in women <35-years-of-age.3 There is no unified standard of age defining the young breast cancer research at present, but some researchers define it as ≤40 y.4 The latest researches show that in patients aged <35 y, the risk of death rose by 5% for every 1-year reduction in age, whereas there was no significant change in death risk with age in patients aged 35–50 y.5 Therefore, the breast cancer patients with age ≤35 were selected in our research.
Breast cancer is a complicated disease with multi-gene aberrations, and molecular classification can guide clinicians for better treatment and prognosis. Gene CNAs (copy number aberrations) are related to the breast cancer molecular classification and the gene instability, playing an important role in breast cancer occurrence and development. There are numerous CNAs, including those commonly known classic genes in breast cancer copy number mutation screening, which are closely correlated with the prognosis such as CCND1 and MYC gene amplification.6 The copy number profiling is very useful for clinical decision in terms of cancer therapy.6 In addition to studying CCND1 and c-Myc,6 we further selected CHEK2, and Rb1, which are involved in negative cell cycle regulation, for this research. The Rb1 protein, a G1/S gate keeper, can prevent G1 entry into S phase in order to avoid the duplication of damaged DNA,7,8 which ensures high fidelity of genetic material. CHEK2 protein can inhibit CDC25C phosphorylation to prevent cell proliferation. Meanwhile, CHEK2 protein can stabilize p53 protein, which, in turn, causes G1 phase arrest for further DNA repair.9,10 As an important oncogene, overexpression of c-Myc is frequently found in many tumors. c-Myc expression leads to DNA replication, and deregulation of c-Myc results in DNA damage in S phase.11 c-Myc contributes to tumorigenesis as a critical antagonist of the DNA damage repair mechanisms.12,13 It is important to note that c-Myc activates cell phase and facilitates the gene expression of cdc25A, CDK4, and Cyclin D1,14 etc. On the other hand, it inhibits gene transcription of p15, p21, and p27, which are CDK inhibitors and negative regulators of cell cycle progression, thereby facilitating cell proliferation. Cyclin D1 (CCND1) is one of the prime regulating proteins of G1/S-phase conversion, and CCND1 gene amplification in breast cancer correlates with poor prognosis.15,16
Here we employed the multi-gene fluorescence in situ hybridization (M-FISH) to analyze the copy number aberrations of these 4 genes, and to correlate the aberrations with prognosis in the young breast cancer patients.
Results
We analyzed the survival status of 196 young breast cancer patients (age ≤35) and 227 old patients (age ≥65). We showed that the 5-y disease free survival (DFS) of the young patients is poorer than that of the old (the young 5-DFS: 67.3%; the old 5-DFS: 78.9%), and that there is a significant difference between them (Fig. 1) (P = 0.049). Interestingly, there is a poor prognosis trend in the young group among 5-y overall survival (OS) when compared with the old group (the young 5-OS: 77.6%; the old 5-OS: 84.1%), but the statistical significance is not obvious (P = 0.074).
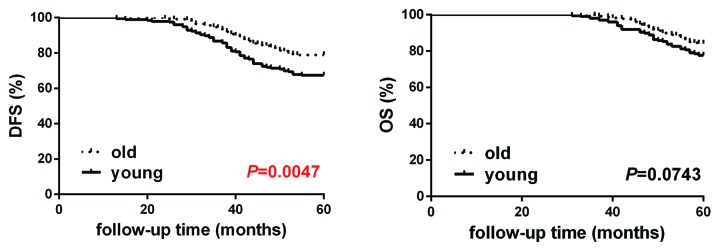
Figure 1. Kaplan–Meier curves of disease-free survival and overall survival in the young breast cancer group (n = 196) and the old breast cancer group (n = 227). DFS (disease-free survival) is defined as from the first operation to the first finding of local recurrence (including chest wall, local lymph node, contralateral breast) or distant metastasis. OS (overall survival) is defined as from first operation to the death caused by breast cancer or 5 y until the end of following-up.
After analyzing the characteristics of molecular classification of the 2 groups, we found that the percentage of Luminal A type in the young patient group is smaller than the old group with statistical significance (χ2 = 4.901, P = 0.027) (Table1). In contrast, the percentage of TNBC type is larger in the young patient group when compared with the old (χ2 = 5.250, P = 0.022). However, the rate differences of Luminal B type (χ2 = 0.207, P = 0.649) and HER2 overexpression type (χ2 = 0.002, P = 0.960) between the two groups are not significantly different (Table 1).
Table 1. Distribution of molecular subtypes in the young and old breast cancer patients.
| Young | Old | |||
|---|---|---|---|---|
| N | % | N | % | |
| Luminal A type | 48 | 24.5 | 78 | 34.4 |
| Luminal B type | 95 | 48.5 | 105 | 46.3 |
| Her-2 overexpression type | 17 | 8.7 | 20 | 8.8 |
| TNBC type | 36 | 18.4 | 24 | 10.6 |
| Total | 196 | 100 | 227 | 100 |
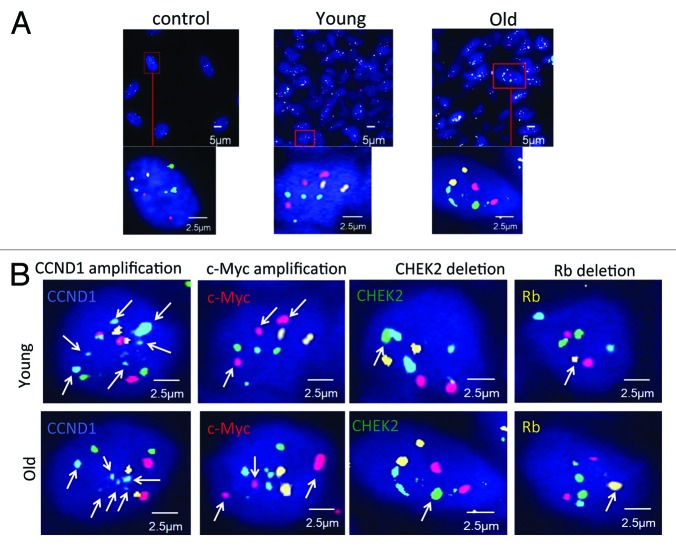
The M-FISH high-resolution technique is applied to examine the amplification and deletion status of the 4 cell cycle genes (Rb1, CHEK2, c-Myc, CCND1) in 423 cases (196 young breast cancer cases and 227 old cases), and all genes’ FISH testing are finished on one paraffin section. 411 gene amplifications and deletions events were found. The gene amplification and deletion events in the young and old breast cancer patients are summarized in Table 2. Representative M-FISH signals in control human diploid fibroblast-like (HDF) cell, young breast cancer patient, and old breast cancer patient were presented (Fig. 2A). The control group demonstrated FISH images of the 4-color probes in the HDF cells: Rb1 (orange), CHEK2 (green), c-Myc (red), CCND1 (blue), while representative young or old breast cancer patients had multiple copies or lost staining of one of these 4 probes. Different CNAs found in young and old breast tumor cells examined by M-FISH were demonstrated to indicate the accuracy of the technique. Arrowheads clearly indicated each alteration in representative young or old breast cancer patients (Fig. 2B).
Table 2. The gene amplification and deletion events in breast cancer patients.
| Young Group | Old Group | ||||
| Positive n | Negative n | Positive n | Negative n | ||
| Rb1 deletion* | 61 | 135 | 23 | 204 | 0.000 |
| CHEK2 deletion* | 91 | 105 | 44 | 183 | 0.000 |
| CCND1 amplification* | 46 | 150 | 52 | 175 | 0.891 |
| c-Myc amplification* | 56 | 140 | 30 | 197 | 0.000 |
*In all the gene mutation events, the following are excluded: 3 Rb1 amplification cases (1 old, 2 young), 1 CHEK2 amplification case (1 young), 2 CCND1 deletion case (1 old, 1 young), 2 c-Myc deletion cases (1 old, 1 young).
Figure 2. Gene status in breast tumor cells examined by M-FISH, (A) M-FISH signals in control human diploid fibroblast-like (HDF) cells, representative young breast cancer patient, and representative old breast cancer patient. The control group demonstrates FISH images of the 4-color probes in the HDF (human diploid fibroblast-like) cells. FISH images for representative image of a young patient and an old patient are presented. Orange signal, gene Rb1; green signal, gene CHEK2; red signal, gene C-myc; blue signal, gene CCND1 (B) Representative gene alteration found in young/old breast tumor cells examined by M-FISH. Representative FISH images for each type of alternation were indicated. Arrowheads indicated color probe of each alteration. Orange signal, gene Rb1; green signal, gene CHEK2; red signal, gene C-myc; blue signal, gene CCND1.
Among the 4 genes tested, there were 411 abnormal gene events out of 423 breast cancer cases. There are 259 gene variation events in the young patients (age ≤ 35), with the ratio of 63.0%, and 152 cases in the old (age ≥ 65), with the ratio of 37.0% (Table 2). In the 4 genes, mutation of Rb1, CHEK2, and c-Myc gene is frequently observed in young patients, and the difference is statistically significant. However, the CCND1 amplification can be observed in 2 groups (Table 2). Further correlating breast cancer subtypes with CNAs in young breast cancer patients, we found that the CNAs are frequently observed in Luminal B type (28%), HER2 overexpression (46%), and TNBC type (56%) (Table 3). In contrast, the CNAs in Luminal A type of young breast cancer is only 19%. Comparing breast cancer subtypes with CNAs in old and young breast cancer patients did not demonstrate that the percentage of CNAs in each subtype of the old group was higher than the young group. Also, CNAs in the Her-2 overexpression subtype were not significantly different among these two groups, which suggests that the genomic instability of this subtype is prevalent irrespective of the age difference. Together, this result demonstrates that CNAs are more prevalent in the young cancer group than the old group.
Table 3. Cancer subtypes with CNAs in breast cancer patients.
| CNA** (young group) | CNA** (old group) | P | |||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Luminal A type * | 37 (19%) | 154 (81%) | 26 (8%) | 284 (91%) | 0.000 |
| Luminal B type* | 105 (28%) | 271 (72%) | 61(15%) | 358(85%) | 0.000 |
| Her2 overexpression type | 31 (46%) | 37 (54%) | 27(34%) | 53 (66%) | 0.142 |
| TNBC type | 81 (56%) | 63 (44%) | 35(36%) | 61(64%) | 0.003 |
(1) **CNA: Copy number aberrations; (2) Luminal A type*: the followings are excluded: 1 Rb1 amplification event in the young breast cancer positive group, 1 Rb1 amplification event and 1 CCND1 deletion event in the old breast cancer positive group. TNBC type*: the followings are excluded: 1 Rb1 amplification event, 1 CHEK2 amplification event, 1 CCND1 and 1 C-myc deletion events in the young positive; 1 C-myc deletion event in the old positive group.
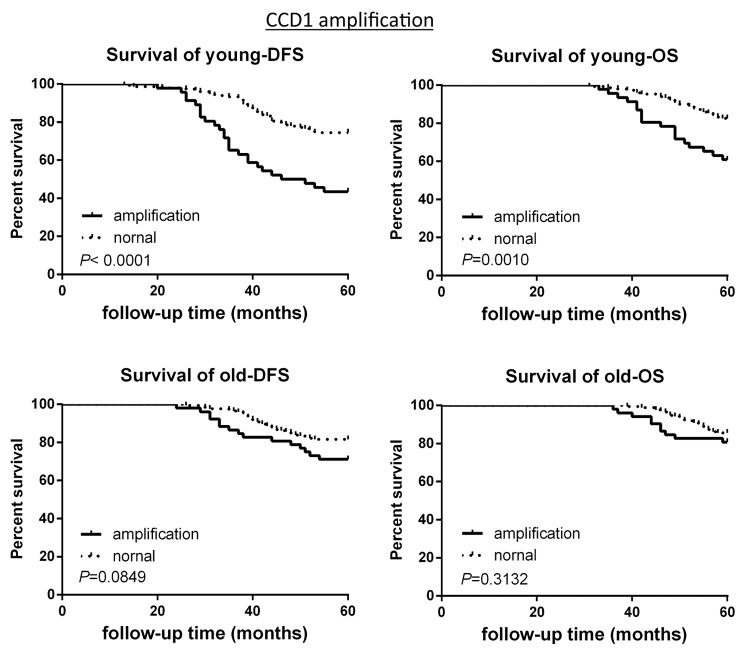
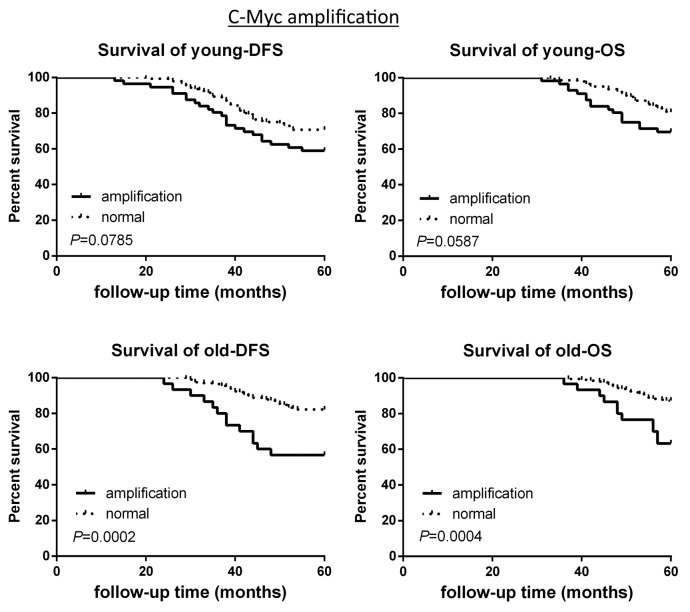
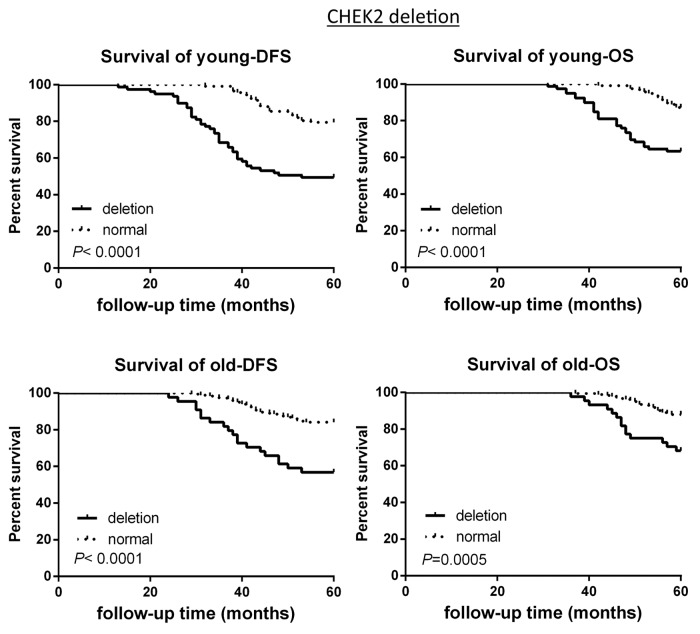
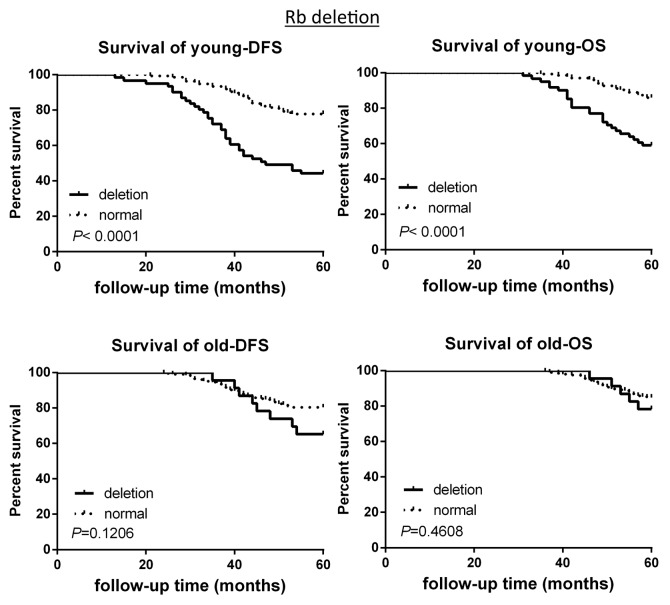
The survival analysis based on CNAs of the 2 groups was investigated. CCND1 gene amplification correlated with poor survival in the young patient group, as demonstrated by 5-y disease free survival rate (5-DFS) and the 5-y overall survival rate (5-OS) (Fig. 3). However, this observation is not statistically significant in the old breast cancer patient group (Fig. 3). Interestingly, c-Myc gene amplification did not correlate with poor survival in young patient group, while this is significantly correlated in older patient group (Fig. 4). CHEK2 deletion critically correlated with poor survival in both age groups (Fig. 5), while RB1 deletion correlated with poor survival only in young patient group, but not in old patient group (Fig. 6).
Figure 3. DFS and OS of young and old breast cancer patients with gene CCND1 amplification. Kaplan–Meier analysis of disease-free survival and overall survival in the young breast cancer group and the old breast cancer group with CCDN1 amplification.
Figure 4. DFS and OS of young and old breast cancer patients with gene c-Myc amplification. Kaplan–Meier analysis of disease-free survival and overall survival in the young breast cancer group and the old breast cancer group with c-Myc amplification.
Figure 5. DFS and OS of young and old breast cancer patients with gene CHK2 deletion. Kaplan–Meier analysis of disease-free survival and overall survival in the young breast cancer group and the old breast cancer group with CHK2 deletion.
Figure 6. DFS and OS of young and old breast cancer patients with gene Rb deletion. Kaplan–Meier analysis of disease-free survival and overall survival in the young breast cancer group and the old breast cancer group with Rb deletion.
Discussion
Recently, breast cancer morbidity has been rising annually, and the number of young patients has been increasing year by year. The prognosis of young breast cancer patients is worse than that of the old.17 However, the mechanism behind this phenomenon is not well-characterized. Here we showed that the 5-DFS of the young group is apparently worse than the old, but there is no obvious difference between the two in 5-OS, which is consistent with some reports of a Chinese young breast cancer study.18 The malignant characteristic of the primary tumor, easy local recurrence, and metastasis may contribute to the poor prognosis of the young breast cancer; however, there is no molecular characterization to provide a better understanding of why young breast cancer patients have a worse disease-free survival rate.
Here we showed that there are differences in the percentage of breast cancer subtypes between young and old groups. The percentage of Luminal A type in young patients is smaller than that in the old, but TNBC type in young patients is much larger than that in the old. This observation is consistent with a previous report by Cancello and others.19 It is shown by other researchers that there are frequent DNA CNAs in sporadic ER-negative, triple-negative breast cancer breast cancer.20 Here, we demonstrated that the CNAs are more prevalent in the young cancer group than the old group, which reflects the poor clinical outcome in the young breast cancer group. CNAs, including CCND1 gene amplification, Rb1 deletion, and CHEK2 deletion, were correlated with poor survival in the young breast cancer group, suggesting that the aberrations of these genes can be used as prognosis markers for young breast cancer. RB1 is frequently lost in TNBC21 and is involved in epithelial–mesenchymal transition.22 This also provides insights into the poor survival of the young breast cancer group. If CCND1 gene testing will be applied in the clinics in the future, targeting Cyclin D1 expression,23 such as using microRNA miR-338-3p,24,25 will be a good strategy for the young breast cancer patients with CCND1 gene amplification. The Rb1 gene deletions in breast ductal carcinoma in situ are closely related with breast cancer invasiveness and recurrence. In the multivariate analysis, the relation between Rb1 deletion and adverse prognosis has been documented.26 Here we showed that Rb1 gene deletion in the young breast cancer is more frequent than the old group, which may be one of the reasons to explain the poor prognosis of young breast cancer patients when compared with old group. The Chk2/hCds1 protein phosphorylates Cdc25C to inactivate Cdc25C activity, suggesting that it arrests cells in G2 in response to DNA damage. In addition, Chk2/hCds1 stabilizes the p53 tumor suppressor protein, leading to cell cycle arrest in response to DNA damage,9 while CHEK2 gene deletion will accelerate G1/S-phase progression. Some studies show that CHEK2 mutation is the independent risk factor of the breast cancer poor prognosis,27 which is consistent with our research results.
Importantly, CNAs are often seen in Luminal B type, HER2 overexpression, and TNBC type, but not in Luminal A type of young breast cancer. This observation suggests that we could use the CNAs to further stratify the young cancer patients for prognosis purposes. Further study of the young breast cancer, especially mechanistic analysis about these CNAs will be critical to explain the young breast cancer pathogenesis, to improve poor prognosis, and to explore new therapeutic strategies. The gene indexes described above can assist young breast cancer clinical pathologic diagnosis and will be used as the molecule testing index for prognosis evaluation, which can facilitate individual therapeutic plans.
Many studies showed that the young breast cancer has a close relationship with tumor pathologic type and molecular classification.4,28 Breast cancer is a multi-gene disease, and gene research is essential to understand further the mechanism. The commonly used FISH is to test HER-2 amplification status in breast cancer. Here, M-FISH in our study has examined 4 genes related to breast cancer cell cycle regulation, and we found that the gene mutation rates can correlate with poor prognosis in young breast cancer. It can be a tool for assisting accurate prognosis, facilitating the search of new therapeutic targets as well as the decision of individual therapeutic plans, and ultimately helps prolong survival of the young breast cancer patients.
Materials and Methods
Prime reagents and instruments
Plasmid Purification Kit: American (QIAGEN Corp.); plasmid DNA (BAC Clone), DNA Polymerase I, human cot-DNA, SSPE (American Invitrogen Corp); EcoR1, ssDNA, tRNA (American Sigma Corp); green dUTP (Abbott Molecular Corp); PromoFluor-415-aadUTP (blue), PromoFluor-555-aadUTP (orange), PromoFluor-590-aadUTP (red) (Germany PromoKine Corp); DAPI (Chroma Corp); ThermoBrite™ orthotopic hybridization instrument (NatureGene Corp); multi-color fluorescence microscope Axiom Imager Z2 type connected with high-resolution CCD (Charge-Coupled Device) camera (Zeiss Corp).
Patient samples
This research selected 221 young female breast cancer patients (age ≤35) and 347 old female breast cancer patients (age ≥65) with definite pathological diagnosis by Tianjin Medical University Cancer Institute and Hospital from January 2006 to December 2007. Six young and 15 old cases were excluded for incised biopsy in other hospitals or treated with new adjuvant chemotherapy. There are 19 young patients who failed to follow up, 57 old cases who did not have thermotherapy because of age or health factors, and 48 cases who failed to follow up (the deaths unrelated with breast cancer revisited in 5 y are regarded as failed to follow up). Therefore, there are 196 young cases and 227 old cases in this research. The excised tumor paraffin pathologic specimens and pathologic testing results are selected from the 2 groups. All the patients are documented from diagnosis and followed-up by telephone, letter, or at a follow-up clinic every quarter in the first 2 years and semi-annually after. The breast cancer death is regarded as the following-up end, and the cutting-off time is 5 y from first diagnosis.
In the research, a cutoff of a minimum of 1% of invasive tumor cells positive for ER/PgR for a specimen to be considered positive;29 the expression of Her-2 is tested by immunohistochemistry and FISH; Her-2 is regarded as positive when the immunohistochemistry result is +++ (> 10% cells with strong staining of complete membranes), and it needs further test by FISH when the result is ++ (> 10% cells with weak but complete membrane staining). The molecular classification bases on the breast cancer consensus by St. Gallen 2011:30 Luminal A (ER and/or PR positive/HER2 negative/Ki-67 low [< 14%]); Luminal B (ER and/or PR positive/HER2 negative/Ki-67 high; or ER and/or PR positive/any Ki-67/HER2 overexpressed or amplified); HER 2overexpression (HER2 overexpressed or amplified/ER and PR absent); “TNBC” (ER and PR absent/HER2 negative).
Disease-free survival (DFS) is defined as from the first operation to the first finding of local recurrence (including chest wall, local lymph node, contralateral breast), or distant metastasis. Overall survival (OS) is defined as from first operation to the death caused by breast cancer or 5 y until the end of following-up.
Probe making
The gene probes are designed by the UCSC gene browsing database (http://genome.ucsc.edu), and all the probes are bacterial artificial chromosomes (BACs) plasmid DNA. The plasmid purification kit was used to extract DNA, and after digestion by EcoR1, the DNA is precipitated. The purified and separated DNA is labeled by fluorescein with nick translation. There are 4 fluoresceins used in this research: PromoFluor-415-aadUTP (blue), PromoFluor-555-aadUTP (orange), PromoFluor-590-aadUTP (red), green dUTP (green). Four aimed genes were labeled with 4 fluoresceins separately, which are made into 4-color probes.
Specimen preparation and FISH testing
Paraffin specimens were dewaxed and hydrated in the 100%, 85%, and 70% gradient ethanol for 5 min separately. After conventional preprocessing, the specimens are digested in 0.25 mg/ml gastric enzyme and 0.01 M HCL solution at 37 °C for 10 min; they are refixed by 0.4% formaldehyde, dehydrated by 70%, 85%, 100% gradient ethanol for 5 min separately; dried in room temperature. The prepared specimens, hybridized with 4-color probe, were processed for 48 h at 47 °C. After hybridization, the specimens are washed by 47% and 55% saline sodium phosphate EDTA (SSPE) separately, dehydrated by gradient ethanol, and dried at room temperature. Then 8 μl 4,6-diamidino-2-pheny-lindole (DAPI) is added to counterstain the specimens, followed by fluorescence microscopy.
Image acquisition and analysis
The Zeiss Axioplan2 laser scanning confocal microscope (LSCM) was used to acquire image for analyzing. The microscope is connected with the high-resolution CCD camera and computer equipped with Axio Vision analyzing software. After hybridization, the specimens were observed and images acquired under Zeiss Axioplan2 LSCM. The view field was selected under DAPI image, and the location position is recorded. Photos were taken simultaneously for the DAPI, Green™, Orange™, Alexa594™, and CYTM5 staining using ×630 objective lens. Many view fields are selected to acquire image separately in each case to get at least 200 complete interphase nuclei with no overlapping to analyze. The exact copy number in nucleus of every gene is recorded. When the copy number in over 20% nucleus of each section exceeds the DNA ploidy number, it is regarded as gene amplification; and when the copy number in over 30% nucleus is under the ploidy number, it is regarded as gene deletion.
Statistic analyses
The SPSS17.0 statistic software and Graphpad 5.0 are applied to analyze the data. Comparisons of baseline characteristics between groups were made using chi-square tests for categorical variables. P values less than 0.05 were considered statistically significant. Kaplan–Meier survival analysis was applied to the prognosis survival analysis. The significance test was performed using the log-rank test.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This project was supported by Tianjin municipal Major Scientific and Technological Special Project for Significant Anticancer Development (No.: 12ZCDZSY15400).
This study was supported by International Cooperation, China Ministry of Science (No. 2010DFB30270). We thank Avira Som for editing.
References
- 1.Hartmann S, Reimer T, Gerber B. Management of early invasive breast cancer in very young women (<35 years) Clin Breast Cancer. 2011;11:196–203. doi: 10.1016/j.clbc.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 2.Anders CK, Acharya CR, Hsu DS, Broadwater G, Garman K, Foekens JA, Zhang Y, Wang Y, Marcom K, Marks JR, et al. Age-specific differences in oncogenic pathway deregulation seen in human breast tumors. PLoS One. 2008;3:e1373. doi: 10.1371/journal.pone.0001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredholm H, Eaker S, Frisell J, Holmberg L, Fredriksson I, Lindman H. Breast cancer in young women: poor survival despite intensive treatment. PLoS One. 2009;4:e7695. doi: 10.1371/journal.pone.0007695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narod SA. Breast cancer in young women. Nature reviews. Clin Oncol. 2012;9:460–70. doi: 10.1038/nrclinonc.2012.102. [DOI] [PubMed] [Google Scholar]
- 5.Han W, Kang SY, Korean Breast Cancer Society Relationship between age at diagnosis and outcome of premenopausal breast cancer: age less than 35 years is a reasonable cut-off for defining young age-onset breast cancer. Breast Cancer Res Treat. 2010;119:193–200. doi: 10.1007/s10549-009-0388-z. [DOI] [PubMed] [Google Scholar]
- 6.Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res. 2006;16:1465–79. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das SK, Hashimoto T, Shimizu K, Yoshida T, Sakai T, Sowa Y, Komoto A, Kanazawa K. Fucoxanthin induces cell cycle arrest at G0/G1 phase in human colon carcinoma cells through up-regulation of p21WAF1/Cip1. Biochim Biophys Acta. 2005;1726:328–35. doi: 10.1016/j.bbagen.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Lee MH, Yang HY. Regulators of G1 cyclin-dependent kinases and cancers. Cancer Metastasis Rev. 2003;22:435–49. doi: 10.1023/A:1023785332315. [DOI] [PubMed] [Google Scholar]
- 9.Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–88. [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MH, Lozano G. Regulation of the p53-MDM2 pathway by 14-3-3 sigma and other proteins. Semin Cancer Biol. 2006;16:225–34. doi: 10.1016/j.semcancer.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–51. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 12.Wen YY, Chou PC, Pham L, Su CH, Chen J, Hsieh YC, Xue Y-W, Qu C-J, Gully C, Parreno K, et al. DNA damage-mediated c-Myc degradation requires 14-3-3 sigma. Cancer Hallmarks. 2013;1:3–17. doi: 10.1166/ch.2013.1002. [DOI] [Google Scholar]
- 13.Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci U S A. 2003;100:9974–9. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MH, Yang HY. Negative regulators of cyclin-dependent kinases and their roles in cancers. Cell Mol Life Sci. 2001;58:1907–22. doi: 10.1007/PL00000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundgren K, Brown M, Pineda S, Cuzick J, Salter J, Zabaglo L, Howell A, Dowsett M, Landberg G, TransATAC investigators Effects of cyclin D1 gene amplification and protein expression on time to recurrence in postmenopausal breast cancer patients treated with anastrozole or tamoxifen: a TransATAC study. Breast Cancer Res. 2012;14:R57. doi: 10.1186/bcr3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown LA, Johnson K, Leung S, Bismar TA, Benítez J, Foulkes WD, Huntsman DG. Co-amplification of CCND1 and EMSY is associated with an adverse outcome in ER-positive tamoxifen-treated breast cancers. Breast Cancer Res Treat. 2010;121:347–54. doi: 10.1007/s10549-009-0479-x. [DOI] [PubMed] [Google Scholar]
- 17.Maggard MA, O’Connell JB, Lane KE, Liu JH, Etzioni DA, Ko CY. Do young breast cancer patients have worse outcomes? J Surg Res. 2003;113:109–13. doi: 10.1016/S0022-4804(03)00179-3. [DOI] [PubMed] [Google Scholar]
- 18.Tang J, Wu CC, Xie ZM, Luo RZ, Yang MT. Comparison of Clinical Features and Treatment Outcome of Breast Cancers in Young and Elderly Chinese Patients. Breast Care (Basel) 2011;6:435–40. doi: 10.1159/000332593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cancello G, Maisonneuve P, Rotmensz N, Viale G, Mastropasqua MG, Pruneri G, Veronesi P, Torrisi R, Montagna E, Luini A, et al. Prognosis and adjuvant treatment effects in selected breast cancer subtypes of very young women (<35 years) with operable breast cancer. Ann Oncol. 2010;21:1974–81. doi: 10.1093/annonc/mdq072. [DOI] [PubMed] [Google Scholar]
- 20.Vollebergh MA, Jonkers J, Linn SC. Genomic instability in breast and ovarian cancers: translation into clinical predictive biomarkers. Cell Mol Life Sci. 2012;69:223–45. doi: 10.1007/s00018-011-0809-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Z, Jones R, Liu JC, Deng T, Robinson T, Chung PE, Wang S, Herschkowitz JI, Egan SE, Perou CM, et al. RB1 and p53 at the crossroad of EMT and triple-negative breast cancer. Cell Cycle. 2011;10:1563–70. doi: 10.4161/cc.10.10.15703. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Zhou BP. Epithelial-Mesenchymal Transition—A Hallmark of Breast Cancer Metastasis. Cancer Hallmarks. 2013;1:38–49. doi: 10.1166/ch.2013.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MH, Yang HY. Molecular targets for cell cycle inhibition and cancer therapy. Expert Opinion on Therapeutic Patents. 2003;13:329–46. doi: 10.1517/13543776.13.3.329. [DOI] [Google Scholar]
- 24.Fu X, Tan D, Hou Z, Hu Z, Liu G, Ouyang Y, Liu F. The effect of miR-338-3p on HBx deletion-mutant (HBx-d382) mediated liver-cell proliferation through CyclinD1 regulation. PLoS One. 2012;7:e43204. doi: 10.1371/journal.pone.0043204. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Gangrade A, Calin GA. MicroRNAs and Cancer Hallmarks. Cancer Hallmarks. 2013;1:50–7. doi: 10.1166/ch.2013.1005. [DOI] [Google Scholar]
- 26.Knudsen ES, Pajak TF, Qeenan M, McClendon AK, Armon BD, Schwartz GF, Witkiewicz AK. Retinoblastoma and phosphate and tensin homolog tumor suppressors: impact on ductal carcinoma in situ progression. J Natl Cancer Inst. 2012;104:1825–36. doi: 10.1093/jnci/djs446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelova SG, Krasteva ME, Gospodinova ZI, Georgieva EI. CHEK2 gene alterations independently increase the risk of death from breast cancer in Bulgarian patients. Neoplasma. 2012;59:622–30. doi: 10.4149/neo_2012_079. [DOI] [PubMed] [Google Scholar]
- 28.Kothari AS, Fentiman IS. 11. Breast cancer in young women. Int J Clin Pract. 2002;56:184–7. [PubMed] [Google Scholar]
- 29.Hammond ME, Hayes DF, Wolff AC, Mangu PB, Temin S. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract. 2010;6:195–7. doi: 10.1200/JOP.777003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, Panel M, Panel members Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011;22:1736–47. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]