Abstract
Elimination of uniparental chromosomes occurs frequently in interspecific hybrid cells. For example, human chromosomes are always eliminated during clone formation when human cells are fused with mouse cells. However, the underlying mechanisms are still elusive. Here, we show that the elimination of human chromosomes in human–mouse hybrid cells is accompanied by continued cell division at the presence of DNA damage on human chromosomes. Deficiency in DNA damage repair on human chromosomes occurs after cell fusion. Furthermore, increasing the level of DNA damage on human chromosomes by irradiation accelerates human chromosome loss in hybrid cells. Our results indicate that the elimination of human chromosomes in human–mouse hybrid cells results from unrepaired DNA damage on human chromosomes. We therefore provide a novel mechanism underlying chromosome instability which may facilitate the understanding of carcinogenesis.
Keywords: chromosome instability, hybrid cells, chromosomal elimination, DNA damage repair, cell cycle regulation
Introduction
In many interspecific hybrid cells, the chromosomes from one parent are preferentially lost, while those from the other parent are retained. In somatic hybrid cells formed by artificial cell fusion between cells of different species, such as human–mouse hybrid cells1-4 and porcine–rodent hybrid cells,5-7 the extensive chromosomal elimination of one parental genome often occurs. In human–mouse hybrid cell clones, the retention of a few human chromosomes was used as an early tool for the determination of the physical location of genes or anonymous DNA fragments within the human genome.8-12 Moreover, in hybrid cells formed by sexual hybridizations, elimination of uniparental chromosomes occurs during embryonic development in interspecific fish13-17 and even in diverse taxa.18-23 Interspecific crosses are used to induce haploids in flowering plants by fertilizing eggs with sperm from a different species for the elimination of uniparental chromosomes.24,25
Several hypotheses have been proposed to explain the mechanisms underlying the elimination of uniparental chromosomes in the interspecific hybrid cells formed by sexual hybridization or artificial cell fusion. In hybrid cells of interspecific plants formed by sexual hybridization, asynchronous cell division,26 asynchronous nucleoprotein synthesis,27,28 genome elimination by nuclear extrusions,29,30 alien chromosomal degradation by host-specific nucleases,31 uniparental chromosomal nondisjunction at anaphase,18 and parent-specific centromere inactivation19-22,32 have been proposed to be responsible for uniparental chromosomal elimination. In interspecific fish formed by sexual hybridization, chromosomal lagging at anaphase was thought to be the mechanism of chromosomal elimination.17 In somatic hybrid cells induced by artificial cell fusion, premature centromeric separation33 or spatial separation of parental genomes34 have been reported for uniparental chromosomal elimination. However, these hypotheses were derived from observations on small numbers of fixed cells. The mechanisms underlying the elimination of uniparental chromosomes are still poorly understood.
Chromosome instability (CIN) involving gain or loss of whole chromosomes or chromosomal fragments is a hallmark of human cancers.35 CIN is thought to be an early stage of carcinogenesis and may be involved in tumor initiation.36 Despite its widespread prevalence, the mechanisms underlying CIN in cancers are elusive. Though CIN frequently occurs in cancers, the rate of chromosome changes is subtle. The observed chromosomal gains and losses in cancers could be the result of the genetically stable clones, which obtain an advantaged growth under certain selective pressures.35 However, human chromosome instability in human–mouse hybrid cells generated by artificial cell fusion is extensive and ongoing during clone formation. Here we study CIN in human–mouse hybrid cells formed by artificial cell fusion in a time course after fusion and provide direct evidence for a novel mechanism underlying CIN, which may facilitate the understanding of carcinogenesis.
Results
Progressive elimination of human chromosomes in human–mouse hybrid cells
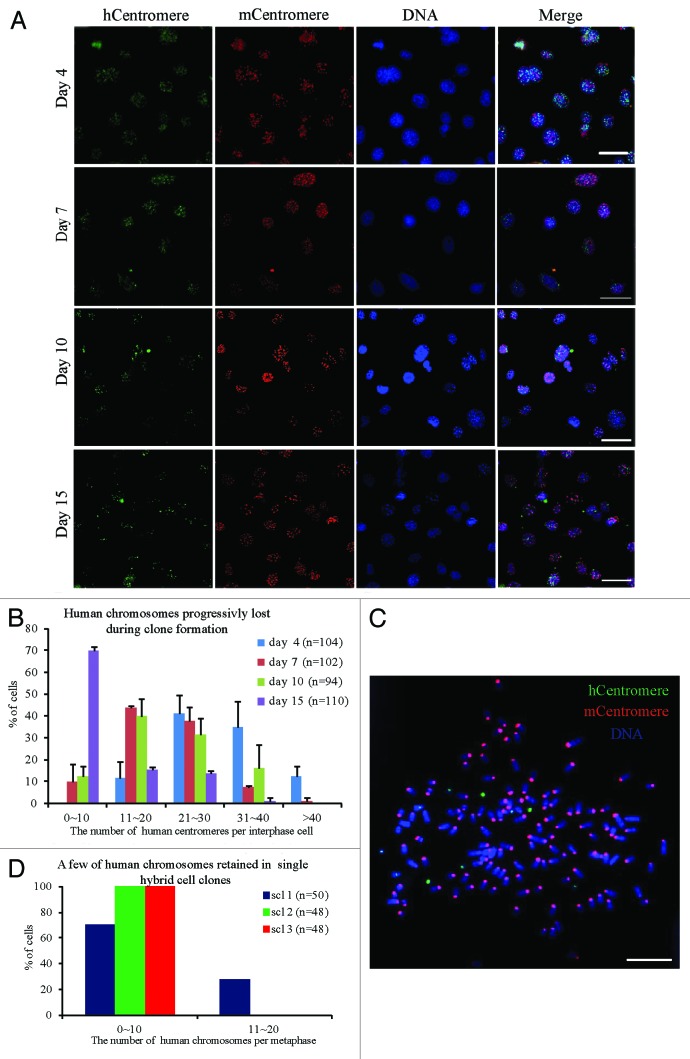
To produce human–mouse hybrid cells, mouse NIH/3T3 cells that stably expressed H2B-EGFP were fused with human HCT116 cells that stably expressed H2B-mCherry. To study the temporal progression of chromosome elimination, fluorescence in situ hybridization (FISH) was performed on hybrid cells using human and mouse pan-centromeric probes. After the binucleated hybrid cells were picked up on coverslips by micromanipulation, the chromosome composition of hybrid cells from 4 to 15 d after cell fusion was analyzed. Many human chromosomes were detected in the hybrid cells on day 4. On days 7, 10, and 15 continuous chromosome loss was observed along with culture (Fig. 1A and B). Binucleated hybrid cells were picked by micromanipulator to 96-well cell culture plates to establish single-cell clones. After about 1 month of culture, when the hybrid cells had divided many times, only a few human chromosomes were observed in the single-cell clones (Fig. 1C and D). Although multipolar divisions induced by extra centrosomes may cause rapid and extensive loss of chromosomes,37 we observed that 91.1% of the parental (P0) and 95.8% of progeny (P1) hybrid cells underwent bipolar mitosis (Fig. S1A and B). These observations suggest that human chromosomes in human–mouse hybrid cells were gradually and progressively eliminated during clone formation.
Figure 1. The chromosome composition of hybrid cells during clone formation. Human and mouse chromosomes were analyzed by fluorescence in situ hybridization (FISH) using human and mouse pan-centromeric probes. (A) Representative images and (B) statistical results of human chromosomes in interphase hybrid cells at different days (4, 7, 10, and 15 d) after cell fusion. The composition of human chromosomes was determined by metaphase spreads of the hybrid cells from 3 randomly selected hybrid single-cell clones. (C) Representative images and (D) statistical results. Red, mouse centromeres; green, human centromeres; blue, DNA; bars = 20 μm, n = the number of cells counted. Mean ± SD from 2 independent experiments.
Human chromosomes in hybrid cells exhibit structural aberrations
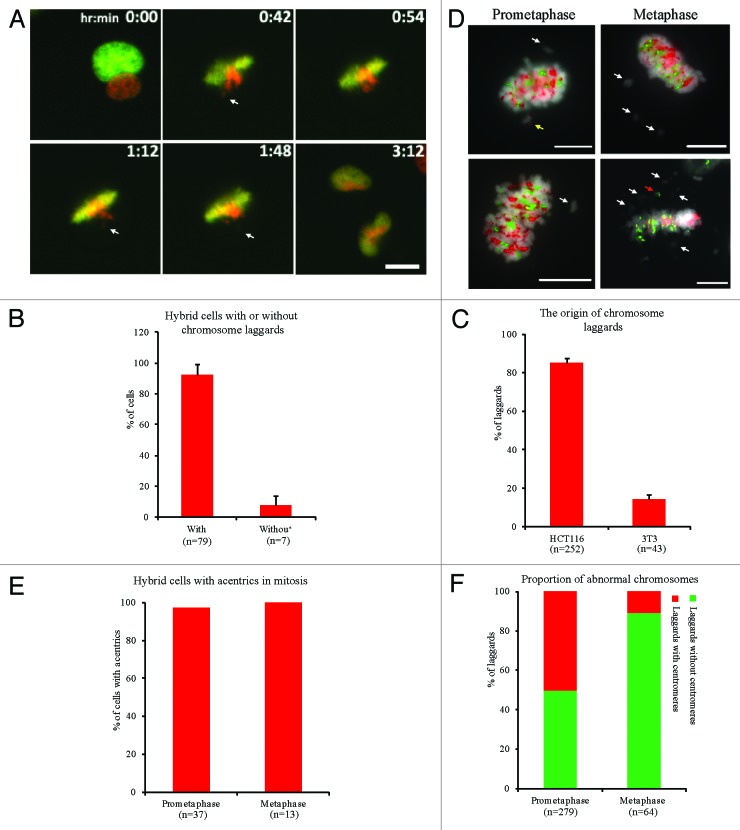
To investigate the cellular mechanism underlying human chromosomal elimination, the chromosomal behaviors of hybrid cells were observed by live cell imaging. The results showed that most hybrid cells (92.4%) exhibited laggards, 85.5% of which were caused by unaligned human chromosomes in metaphase (Fig. 2A–C; Video S1). Laggards can be divided into 2 types: whole chromosome and chromosomal fragment.38 To characterize the laggards, hybrid cells in premetaphase or metaphase were analyzed by FISH using human and mouse pan-centromeric probes. The results showed that most hybrid cells in premetaphase (97.3%) or metaphase (100%) contained acentrics. Many (49.8%) premetaphase laggards were acentrics. Furthermore, 89.1% of laggards in metaphase were acentrics (Fig. 2D–F), indicating that human chromosomes exhibited structural aberrations in metaphase. Moreover, many human chromosomes with structural aberrations and acentrics were also retained in the metaphase cells of 3 hybrid cell lines (Fig. S2). These results suggest that the elimination of human chromosomes is associated with chromosomal structural aberrations, and human chromosomes may be lost through chromosomal fragmentation.
Figure 2. Human chromosomes in hybrid cells exhibit chromosomal structural instability. Chromosome behaviors in hybrid binucleated cells during mitosis were detected by live cell imaging. (A) Representative images showing chromosome laggards from human genome (white arrow). Green, mouse nucleus; red, human nucleus. (B) Percentage of hybrid binucleated cells with chromosome laggards during first mitotic division. (C) Percentage of laggards from human or mouse genome. (D) Representative images of mitotic hybrid cells with chromosome laggards detected by FISH using human and mouse pan-centromeric probes. Red, mouse centromeres; green, human centromeres; gray, DNA; acentrics (white arrow), human or mouse chromosome laggards (red or yellow arrows, respectively). (E) Percentage of hybrid cells with acentrics. (F) Proportion of chromosome laggards with or without centromeres in prometaphase or metaphase. Bars = 20 μm. Mean ± SD from 3 independent experiments.
Defects in DNA damage repair of human chromosomes after cell fusion
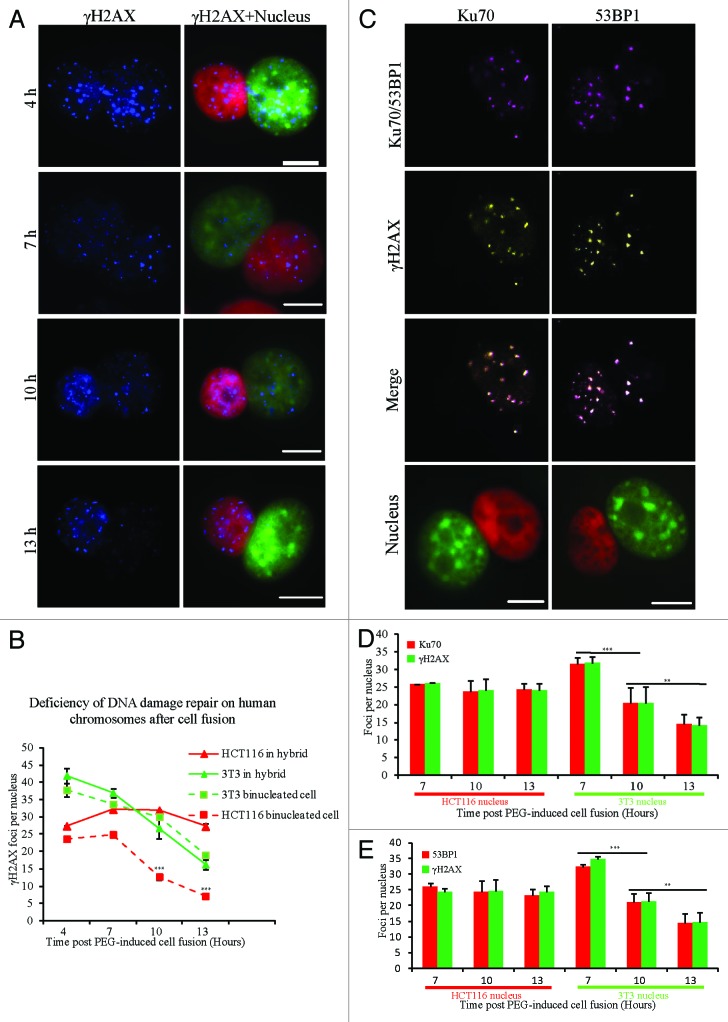
Defects in DNA damage repair can induce chromosomal structural aberrations, which is thought to be a reason for chromosome loss.39 To investigate DNA damage repair in hybrid cells, the dynamics of γH2AX foci as a DNA damage marker were examined at 4 different time points after cell fusion. The average number of γH2AX foci in mouse nuclei of binucleated hybrid cells was decreased from 41.9 at 4 h to 16.3 foci per nucleus at 13 h, a level also observed in the 2 types of homo-binucleated parental cells (37.8 to 19.0 foci per 3T3 nucleus or from 23.6 to 7.2 foci per HCT116 nucleus). However, there was no obvious reduction in the number of γH2AX foci in the human nuclei of binucleated hybrid cells at different time points, which are 27.4 at 4 h, 32.2 at 7 h, 31.9 at 10 h, and 27.3 foci per nucleus at 13 h (Fig. 3A and B). We then examined 53BP1 or Ku70 as surrogate markers40,41 for DNA damage in binucleated hybrid cells. 53BP1 and Ku70 foci colocalized with γH2AX foci in both nuclei of binucleated hybrid cells (Fig. 3C). Consistent with γH2AX staining (Fig. 3A and B), the average number of 53BP1 (25.9 at 7 h, 24.3 at 10 h, and 23.3 foci per nucleus at 13 h) or Ku70 foci (25.7 at 7 h, 23.8 at 10 h, and 24.3 foci per nucleus at 13 h) in human nuclei was not significantly changed at different time points, while it was significantly decreased in mouse nuclei (53BP1 foci from 32.2 at 7 h to 14.4 at 13 h, or Ku70 foci from 31.5 at 4 h to 14.4 at 13 h per nucleus) (Fig. 3D and E). These results indicate that a defect in DNA damage repair on human chromosomes occurs after cell fusion.
Figure 3. Dynamics of DNA damage foci in hybrid binucleated cells. (A) Representative images showing γH2AX staining at different time points (4, 7, 10, and 13 h) after PEG-induced cell fusion. Red, human genome; green, mouse genome; blue, γH2AX staining. (B) Statistical results for the average number of γH2AX foci per nucleus. Three different types of cells at each time point were analyzed in more than 20 cells. Asterisks denote statistically significant difference in the number of γH2AX foci in HCT116 nuclei between hybrids and binucleated cells. (C) Representative images of γH2AX and 53BP1 or γH2AX and Ku70 co-staining after cell fusion. Green, mouse genome; red, human genome; yellow, γH2AX; purple, 53BP1 or Ku70, respectively. (D and E) Statistical results for the average number of foci per nucleus. Asterisks denote statistically significant difference in the number of foci at different time points from more than 30 cells for each category. Bars = 20 μm. **P < 0.05, ***P < 0.001, 2-tailed t test. Mean ± SD from 2 independent experiments.
Binucleated hybrid cells with DNA damages could enter and complete mitosis
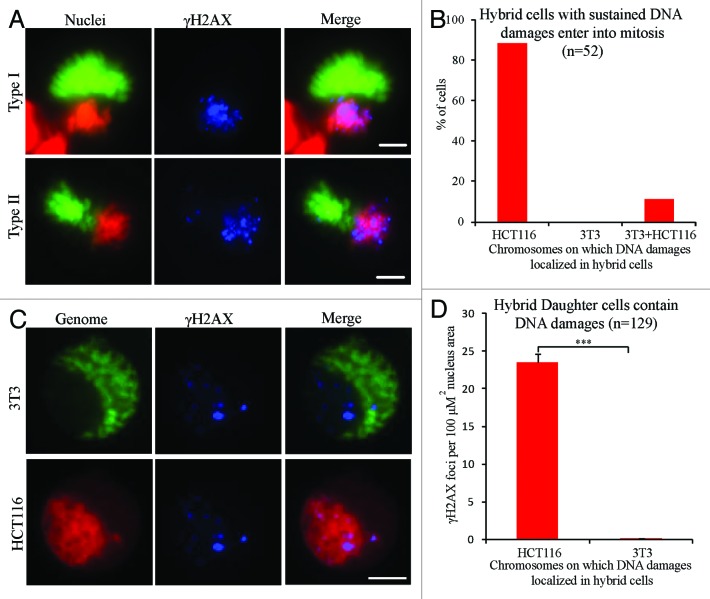
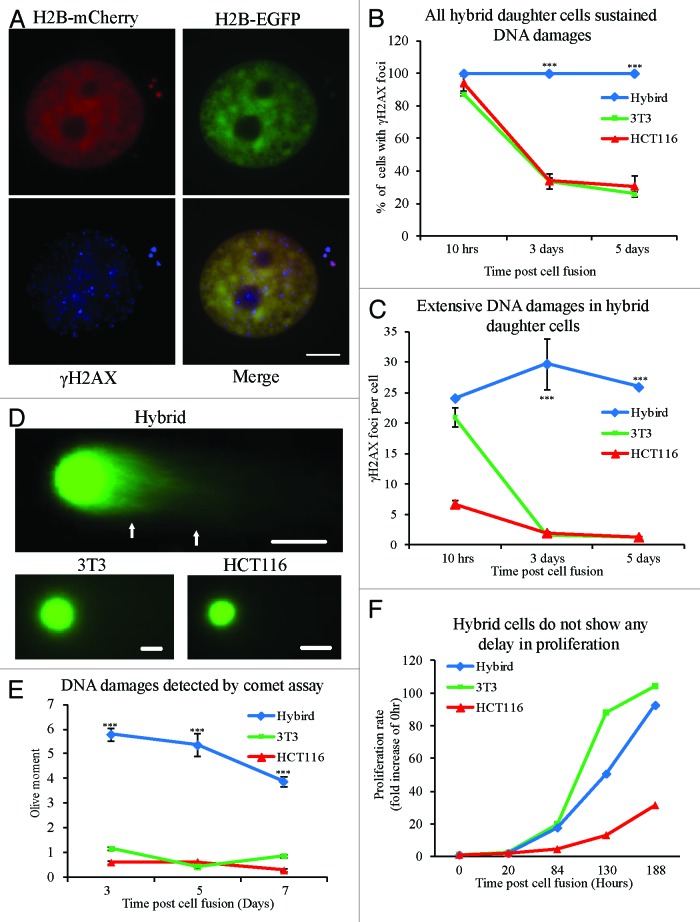
In mammalian normal cells, the cell cycle checkpoint works to ensure the efficiency and accurate rectification of DNA damage by delaying progression of the cell cycle until DNA damage is repaired.42,43 However, by live cell imaging, we observed that many hybrid cells (86/134) could enter mitosis, and all (86/86) those cells entering mitosis could complete division (data not shown). γH2AX staining showed that all binucleated hybrid cells exhibited many DNA damage sites on human chromosomes, while only a few sites were found on mouse chromosomes during mitosis (Fig. 4A and B). Furthermore, the hybrid daughter cells from first cell divisions exhibited a distinctive γH2AX labeling pattern. Many (average of 23.5 foci per unit nucleus area) γH2AX foci were found in the area of nuclei containing human genome, while only a few (average of 0.1 foci per unit nucleus area) were found in area of nuclei containing mouse genome (Fig. 4C and D). This phenotype of hybrid cells between NIH/3T3 and HCT116 (NIH/3T3 × HCT116) cells was also observed in 3 other types of hybrid cells, NIH/3T3 × RPE1, NIH/3T3 × DLD1, and mouse ovarian surface epithelial cells (Mosec) × DLD1 (Fig. S3A–B). These results implied that binucleated hybrid cells could enter and complete mitosis despite numerous unrepaired DNA damage on human chromosomes.
Figure 4. Hybrid binucleated cells with DNA damages enter and complete mitosis. (A) Representative images and (B) percentage of γH2AX-positive mitotic hybrid binucleated cells from 3T3 H2B-EGFP cells fused with HCT116 H2B–mCherry cells. Green, mouse genome; red, human genome; blue, γH2AX; Type I, γH2AX foci on HCT116 chromosomes only; Type II, γH2AX foci on both 3T3 and HCT116 chromosomes. (C) Representative images of hybrid daughter cells in interphase stained for γH2AX. (D) Statistical results. Bars = 20 μm. ***P < 0.001, 2-tailed t test. Mean ± SD, from 3 independent experiments.
Hybrid daughter cells sustain DNA damages and continually proliferate during cell proliferation
To determine whether hybrid daughter cells with unrepaired DNA damages could escape the DNA damage checkpoint in G1 phase to enter S phase, we labeled hybrid cells with EdU to mark DNA synthesis. After 2 h EdU addition, 13.5% of hybrid daughter cells were EdU-positive, not significantly different from NIH/3T3 (15%) and HCT116 (9%) cells (Fig. S4). To detect whether hybrid cells were able to repair DNA damage completely during cell proliferation, γH2AX staining and neutral comet assay were performed. We found that all of the hybrid daughter cells were γH2AX-positive (Fig. 5A and B), while the percentage of γH2AX-positive cells in NIH/3T3 and HCT116 cells was significantly decreased (Fig. 5B). The number of γH2AX foci per cell in hybrid cells was largely constant at 10 h, 3 d, and 10 d time points, while the number significantly decreased in NIH/3T3 cells and HCT116 cells (Fig. 5C). To obtain large numbers of fused cells, EGFP+mCherry+ hybrid cells and 2 parental cells were enriched by fluorescence-activated cell sorting (FACS) (Fig. S5). These cell populations were used to perform a neutral comet assay for DNA damage. These results showed that residual DNA damages in hybrid daughter cells were significantly higher than that in daughter cells from HCT116 or NIH/3T3 cells at all time points (Fig. 5D–E). Surprisingly, the proliferation of hybrid cells was not obviously disturbed as compared with NIH/3T3 and HCT116 cells (Fig. 5F). Altogether, these results implied that the hybrid daughter cells could proliferate with sustained DNA damages, which may be due to deficiency in DNA damage checkpoint.
Figure 5. Hybrid daughter cells sustain DNA damages and continually proliferate during clone formation. (A) Representative images of γH2AX staining in hybrid daughter cells. (B) Percentage of cells with γH2AX positive staining. (C) The average number of γH2AX foci per cell in 3 types of cells at each time point (10 h, 3 d, and 5 d) after PEG-induced cell fusion during clone formation. Statistical results in (B and C) from more than 60 cells at each time point in 3 different types of cells. At 1 d after PEG-induced cell fusion, mixed cells were sorted twice by flow cytometry to enrich hybrid cells or control cells respectively, then detection of DNA damage in 3 types of cells was performed by comet assay at different time points (3, 5, and 7 d) during clone formation. (D) Representative images and (E) statistical results from more than 100 cells at each time point in 3 different types of cells. (F) The proliferation rate of 3 types of cells during clone formation. The number of single-cell clones in 3 types of cells (n = 3 in HCT116, 7 in hybrid cells and 9 in 3T3). Bars = 20 μm. Asterisks denote statistically significant difference between hybrid cells and control cells. ***P < 0.001, 2-tailed t test (C and E), chi-square test (B). Mean ± SD, from 2 independent experiments.
Increasing DNA damages facilitates chromosomes loss in hybrid cells
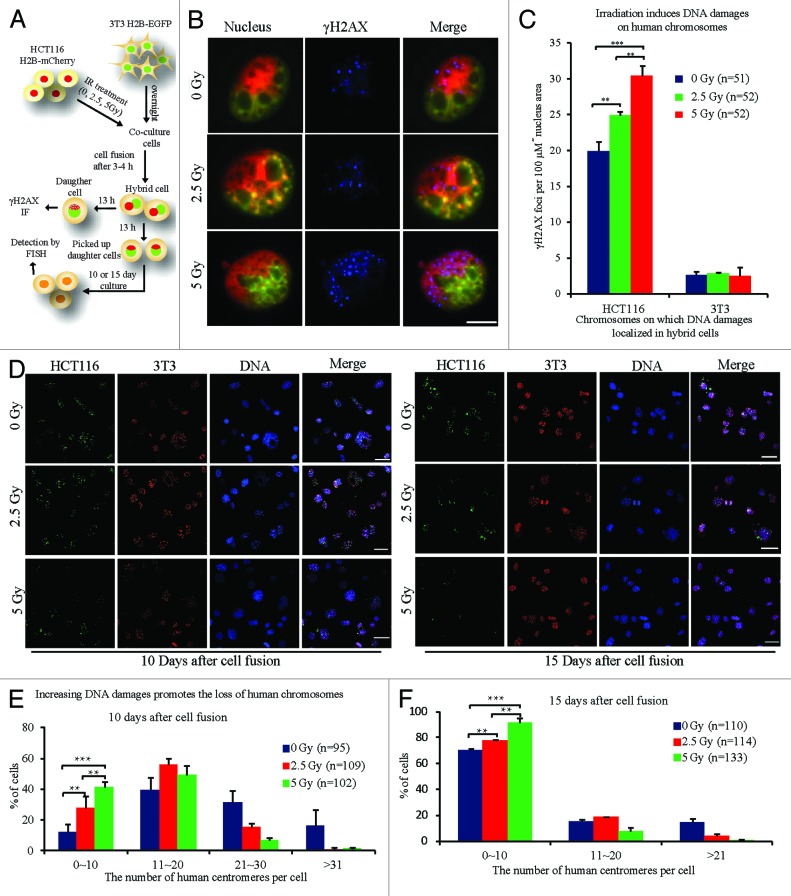
Based on the results above, we supposed that the elimination of human chromosomes was caused by unrepaired DNA damage during cell proliferation. To test if increasing DNA damage on human chromosomes would facilitate chromosome loss, HCT116 cells were irradiated at different doses to induce DNA damage on chromosomes before cell fusion. After 3 or 4 h of IR treatment, the number of γH2AX foci was detected on human chromosomes in HCT116 cells and positively correlated with IR dose (Fig. S7). HCT116 H2B–mCherry cells were then fused with NIH/3T3 H2B–EGFP cells. After about 13 h cell culture, the average number of DNA damage foci on human chromosomes of hybrid daughter cells was 19.9 per unit nucleus area, and significantly increased to 24.9 for 2.5 Gy treatment or to 30.4 for 5 Gy treatment (Fig. 6A-C). The hybrid daughter cells were picked up by micromanipulator on coverslips. At 10 and 15 d culture, the composition of human chromosomes in hybrid cells was analyzed by FISH. These results showed that the number of retained human chromosomes in hybrid cells treated with 2.5 Gy was significantly lower than that in normal hybrid cells and was even lower in cells receiving 5 Gy IR (Fig. 6D–F). Though increased DNA damages on human chromosomes slightly reduced the frequency of hybrid cells that completed the first cell division as well as the progression of hybrid cells during cell proliferation (Fig. S7), these results suggest that sustained DNA damages on human chromosomes were the cause of human chromosomal loss during clone formation.
Figure 6. Rapid loss of human chromosomes due to increased DNA damages. (A) Experimental scheme. 3T3 H2B-EGFP cells were seeded and cultured overnight first, and 3–4 h after different doses of ionizing radiation (IR) treatment to HCT116 H2B-mCherry cells, HCT116 H2B-mCherry cells fused with 3T3 H2B-EGFP cells. (B) Representative images and (C) statistical results of γH2AX staining in hybrid daughter cells at 13 h after cell fusion. Human or mouse, representing regions containing only human or mouse genome in nuclei. Bars = 20 μm. Besides, the chromosome composition of hybrid cells was analyzed by FISH using human and mouse pan-centromeric probes at 10 or 15 d after cell fusion. (D) Representative FISH images and (E and F) statistical results. Red, mouse centromeres; green, human centromeres; blue, DNA. Bars = 50 μm, n = the number of cells counted .**P < 0.05, ***P < 0.001, 2-tailed t test (C), Chi-square test (E and F). Mean ± SD, from 2 independent experiments.
Discussion
Our study revealed a novel mechanism underlying uniparental chromosomal elimination in interspecific somatic hybrid cells produced by artificial cell fusion. We showed that a progressive or gradual loss of human chromosomes occurs during the clone formation of human–mouse hybrid cells, and this elimination of human chromosomes is caused by chromosomal structural aberrations resulting from unrepaired DNA damages. We have also proved that the accumulation of unrepaired DNA damages on human chromosomes is caused by deficiency of DNA damage repair after cell fusion. However, the hybrid cells with unrepaired DNA damages proliferated, suggesting defects in DNA damage checkpoint in hybrid cells.
In the current study, for the first time, live cell imaging was used to track chromosomal behaviors of 2 species in hybrid cells. Although multipolar cell division was observed on fixed cells in previous report,33,44 the actual occurrence was rare determined by live cell image (Fig. S2). We also observed spatial separation of parental genomes in human–mouse somatic hybrid cells (Fig. 2A; Fig. S2), as previously reported.45,46 However, non-random chromosomal position is generally observed in other hybrid cells formed by sexual hybridization47-50 and mammalian cells,51-53 while no chromosomal loss occurs. Consistent with previous report,33 we found that there were many human laggards at metaphase, while only a few mouse chromosomes exhibited incomplete congression (Fig. 2C). Laggards were mainly acentrics in hybrid cells analyzed by FISH (Fig. 2F). Acentrics are unstable and are easily lost through cell divisions.54-56 Thus, the elimination of human chromosomes in human–mouse hybrid cells was through chromosome fragments. In hybrid cells of interspecific plants, the elimination of uniparental chromosomes is associated with chromosome fragments.18,57 In wheat and pearl millet crosses, the eliminated chromosomes from pearl millet entrapped in micronuclei at mitosis and interphase are heterochromatinized and fragmented.57 We also observed that most post-mitotically formed micronuclei containing human chromatins were found with DNA damages (data not shown). However, these potentially eliminated human chromosomes could be unrepaired breakage at interphase after cell fusion and be entrapped in micronuclei after mitosis (Figs. 3A–E, 4A and B). Besides a few of potentially eliminated human chromosomes entrapped in micronuclei, many of human chromosomes with unrepaired DNA damages were still in the main nuclei during the early stage of hybrid clone formation (Figs. 4C and D, 5A–C; Fig. S3). These results indicated that the fragmentation of human chromosomes occurs initially in main nuclei rather than in micronuclei. We cannot exclude the possibility that human chromosomes are eliminated by fragmentation occurring in micronuclei, a phenomenon which has been observed in the plant hybrid cells.57
Most chromosomal fragments and rearrangements may be caused by faults in DNA damage repair.58,59 In our results, deficiency in DNA damage repair on human chromosomes occurs after cell fusion (Fig. 4A and B). Furthermore, hybrid daughter cells sustain DNA damages during cell proliferation (Fig. 5). These results indicate that unrepaired DNA damages can contribute to the generation of human chromosomal fragments. Though DNA damage occurs frequently in normal cells, the DNA damage checkpoint monitors the efficient and accurate repair to maintain genomic stability.58,60 If the DNA damage remains unrepaired, the cells may enter cellular senescence or programmed cell death.42,61 However, our results from live cell imaging (Fig. 2A; Fig. S2) and γH2AX staining (Fig. 4) indicate that hybrid cells with sustained DNA damage can enter and complete mitosis, and the DNA damages were transmitted from binucleated hybrid cells to hybrid daughter cells. Additionally, the proliferation of hybrid daughter cells was not obviously disturbed during cell proliferation (Fig. 5). These results implied that the DNA damage checkpoint is faulty in hybrid cells. Furthermore, intraspecific hybrid cells increased sensitivity to hydrogen peroxide-induced DNA damage compared with parental cells.62 Perhaps cell fusion can produce hybrid cells with defects in DNA damage repair. In human–mouse hybrid cells, mouse genome may act as a “harbor” to help the human genome to escape from the DNA damage checkpoint.
DNA damage is repaired mainly by 2 pathways, homologous recombination repair (HR) and non-homologous end-joining (NHEJ).58,63 The NHEJ is the primary DNA repair pathway in somatic cells, especially in G1 phase.60 Though NHEJ has been suggested to be error-prone, its efficiency on DNA damage repair is high. In current research, we found that DNA damages still remained in hybrid daughter cells that proliferated and formed clones (Fig. 5), indicating that non-homologous end-joining (NHEJ) may be defective in hybrid cells. Ku70 and 53BP1 are key proteins involved in the NHEJ pathway.41,64 When NHEJ initiates, Ku70/Ku80 heterodimers are first clustered at DNA damage sites to recruit other components. 53BP1 promotes NHEJ and inhibits the end resection for HR.65 We found that Ku70 and 53BP1 could be recruited to DNA damage sites (Fig. 4), suggesting that NHEJ initiated and HR was inhibited in hybrid cells. Though NHEJ initiated, some unknown defects may disturb DNA repair through NHEJ. Furthermore, the efficiency of HR in DNA damage repair and the mechanism underlying its deficiency in hybrid cells are still unknown.
Another question is how the DNA damage was generated. Premature chromosome condensation (PCC), which is induced by the fusion between mitotic cells and interphase cells, may induce chromosome pulverization, an extensive DNA damage.66 However, we found that none (0/32) of the hybrid cells exhibited PCC, as tested by mitotic chromosome spreads, and all of them exhibited a normal mitotic pattern in our experiments (Fig. S8A). Asynchronous DNA replication between primary nucleus and micronuclei may result in extensive DNA damage and chromosomal fragmentation in micronuclei for incomplete DNA replication.67 However, 2 nuclei in most of the binucleated hybrid cells had synchronous DNA replication at different time points after cell fusion (Fig. S8B–D), indicating that the generation of most DNA damages is not related to asynchronous DNA replication. In hybrid cells of interspecific plants, DNA damages were generated from the breakage of chromosome bridges resulting from chromosomal nondisjunction caused by the retention of cohesins at anaphase.18 However, in human–mouse hybrid cells, many unrepaired DNA damages on human chromosomes in pro-metaphase may be transmitted from interphase after cell fusion (Fig. 3A). This difference may be due to the methods and species used for the generation of hybrid cells. The exact causes of DNA damages in hybrid cells are still unknown. These defects in the machinery of DNA damage repair are most likely to be the essence of sustained DNA damages. Most human cancers characterized by chromosome instability (CIN) involve gain or loss of whole chromosomes or chromosome fragments.35 Even though the relationship between CIN and carcinogenesis has been established for many decades; the mechanisms underlying CIN in cancers have been elusive. Though CIN occurs frequently in cancers, the rate of chromosome changes is subtle.35 However, human chromosome instability in human–mouse hybrid cells is drastic and progressive during clone formation.1,2 In this article, we studied CIN in human–mouse hybrid cells and provided direct evidence to support a novel underlying mechanism of CIN. This mechanism implies that the unrepaired DNA damage facilitates progressive chromosome elimination and is perhaps also a factor in CIN in carcinogenesis.
Materials and Methods
Cell culture and hybrid cells generation
HCT116, DLD1, NIH/3T3, and RPE1 cells were cultured in Dulbecco modified Eagle medium (Gibco 12800) supplemented with 10% fetal bovine serum (HyClone SV30087), 100 U/ml penicillin, and 100 U/ml streptomycin. Mouse ovarian surface epithelial cells (Mosec) were obtained and cultured as previously described.68 Cells stably expressing red fluorescent protein-tagged histone 2B (H2B-mCherry) or enhanced green fluorescent protein-tagged histone 2B (H2B-EGFP) were generated by retroviral transduction. Human and mouse cells were trypsinized for harvesting and mixed together according to parental cell ratio 1:1, then overlaid cells were seeded on coverslips or chambers. Cells were treated with polyethylene glycol (PEG-Sigma, MW 1000, prepared in serum-free DMEM with 10% DMSO, 45% w/v) for 1 min at room temperature, followed by 3 successive and 2 at 10-minute interval washes with DMEM medium at 37 °C. For irradiated hybrid cells, NIH/3T3 H2B-EGFP cells were seeded and cultured overnight first, and 3–4 h after different doses of ionizing radiation (IR) treatment to HCT116 H2B–mCherry cells, HCT116 H2B–mCherry cells fused with NIH/3T3 H2B-EGFP cells.
Fluorescence in situ hybridization (FISH)
The hybrid cells grown on coverslips were fixed by methanol, and FISH was performed according to our previous report,69 using human and mouse chromosome pan-centromeric probes (Cambio UK 1695-F-02, 1697-MF-02). Images were captured by a CCD camera (Retiga Exi FAST, Qimaging) using an Olympus BX-61 fluorescence microscope equipped with band pass filters. The images were analyzed using Image-Pro Plus 6.0 software (Media Cybernetic, Inc).
Live cell imaging
Cells were picked up by micromanipulator on gridded coverglass bottom dishes (MatTek Corporation) and then seeded into a poly-L-lysine-coated glass-bottom chamber. Images were captured immediately after cells attached to the chamber at 37 °C on the microscope stage. Images were automatically acquired with customized Nikon TE2000E inverted microscope equipped with a linearly encoded stage, a 20× Nikon Plan fluorescent objective, a Hamamatsu orca-ER CCD camera. The multiposition images of cells with autofocus microscope were implemented according to our previous report.70
Metaphase chromosome spreads preparation
Hybrid cells were grown in the presence of 0.05 μg/ml colcemid for 2 h, and harvested by trypsinization and hypotonic treatment with pre-warmed 0.075 M KCl for 5 min at 37 °C. Then cells were fixed in methanol:acetic (3:1). After 3 changes of the fixatives, cells were resuspended in fixative (about 1 ml) and added drop-wise onto cold slides. After that, cells in each clone were subjected to further analysis by FISH according to our previous report.69
Immunofluorescence assay
The antibodies, rabbit anti-53BP1 (Novus Biologicals, #100–305), mouse anti-γ-H2AX (Millipore, #05–636), rabbit anti-Ku70 (Abcam, #ab3114), goat anti-mouse, or goat anti-rabbit IgG conjugated to Alexa Fluor 647 (Invitrogen), or donkey anti-rabbit IgG or sheep anti-mouse conjugated to AMCA (Jakson ImmunoResearch) were diluted 1:100 for immunofluorescence staining assay. Cells grown on coverslips were fixed with 4% paraformaldehyde at room temperature for 10 min. Cells were permeabilized with PBS containing 0.25% Triton X-100 at room temperature for 5 min, and then were blocked by PBS containing 15% fetal bovine serum at room temperature for 1 h. The coverslips were incubated with primary antibodies at 4 °C overnight. After that, the primary antibodies were washed by PBS and secondary antibodies were added. After finishing immunofluorescence staining, images were captured by Olympus BX-61 fluorescence microscope.
EdU labeling assay
To investigate DNA replication in 2 nuclei of binucleated hybrid cells, 25 mM EdU was added into culture medium for 1 or 4 h at different time points after cell fusion. After that, the cells were washed 3 times with PBS and then fresh medium was added to culture. EdU-labeled assay was performed by Cell-Light EdU DNA Cell Proliferation Kit (RiboBio) according to the manufacturer’s instructions.
Fluorescence-activated cell sorting (FACS) and comet assay
NIH/3T3-expressing H2B–EGFP cells were fused with HCT116 H2B-mCherry stable-expressing cells using polyethylene glycol (PEG). To sort out the EGFP+mCherry+ hybrid cells and 2 parental cells, cells were trypsinized and resuspended in DMEM without serum. Cells were sorted out using a BD FACS Aria III (BD Biosciences). After several days culture, single-cell neutral gel electrophoresis was performed according to a previous report.71 DNA was stained by propidium iodide (PI), and slides were photographed digitally by using Retiga Exi FAST CCD camera. Tail moments were analyzed using Comet Score software (TriTek).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Qiaomei Hao for reagent management, and Professor M Wu, and our lab members for discussions. This work was supported by grants from National Natural Science Foundation of China (Grant 30900794), and Doctoral Fund of Ministry of Education of China (20123402130004).
Glossary
Abbreviations:
- FISH
fluorescence in situ hybridization
- CIN
chromosome instability
- NHEJ
non-homologous end-joining
- HR
homologous recombination repair
References
- 1.Matsuya Y, Green H, Basilico C. Properties and uses of human–mouse hybrid cell lines. Nature. 1968;220:1199–202. doi: 10.1038/2201199a0. [DOI] [PubMed] [Google Scholar]
- 2.Matsuya Y, Green H. Somatic cell hybrid between the established human line D98 (presumptive HeLa) and 3T3. Science. 1969;163:697–8. doi: 10.1126/science.163.3868.697. [DOI] [PubMed] [Google Scholar]
- 3.Migeon BR, Miller CS. Human-mouse somatic cell hybrids with single human chromosome (group E): link with thymidine kinase activity. Science. 1968;162:1005–6. doi: 10.1126/science.162.3857.1005. [DOI] [PubMed] [Google Scholar]
- 4.Nabholz M, Miggiano V, Bodmer W. Genetic analysis with human--mouse somatic cell hybrids. Nature. 1969;223:358–63. doi: 10.1038/223358a0. [DOI] [PubMed] [Google Scholar]
- 5.Nowak-Imialek M, Kues WA, Rudolph C, Schlegelberger B, Taylor U, Carnwath JW, Niemann H. Preferential loss of porcine chromosomes in reprogrammed interspecies cell hybrids. Cell Reprogram. 2010;12:55–65. doi: 10.1089/cell.2009.0045. [DOI] [PubMed] [Google Scholar]
- 6.Yerle M, Echard G, Robic A, Mairal A, Dubut-Fontana C, Riquet J, Pinton P, Milan D, Lahbib-Mansais Y, Gellin J. A somatic cell hybrid panel for pig regional gene mapping characterized by molecular cytogenetics. Cytogenet Cell Genet. 1996;73:194–202. doi: 10.1159/000134338. [DOI] [PubMed] [Google Scholar]
- 7.Zijlstra C, Bosma AA, de Haan NA. Comparative study of pig-rodent somatic cell hybrids. Anim Genet. 1994;25:319–27. doi: 10.1111/j.1365-2052.1994.tb00365.x. [DOI] [PubMed] [Google Scholar]
- 8.Wasmuth JJ. Overview of somatic cell hybrid mapping. Current protocols in human genetics / editorial board, Jonathan L Haines [et al] 2001; Chapter 3:Unit 3 1. [DOI] [PubMed] [Google Scholar]
- 9.Allikmets R, Kashuba VI, Huebner K, LaForgia S, Kisselev LL, Klein G, Dean M, Zabarovsky ER. Mapping of 22 Notl linking clones on human chromosome 3 by polymerase chain reaction and somatic cell hybrid panels. Chromosome Res. 1996;4:33–7. doi: 10.1007/BF02254942. [DOI] [PubMed] [Google Scholar]
- 10.Apte SS, Mattei MG, Olsen BR. Mapping of the human BAX gene to chromosome 19q13.3-q13.4 and isolation of a novel alternatively spliced transcript, BAX delta. Genomics. 1995;26:592–4. doi: 10.1016/0888-7543(95)80180-T. [DOI] [PubMed] [Google Scholar]
- 11.Hafezparast M, Cole CG, Kaur GP, Athwal RS, Jeggo PA. An extended panel of hamster-human hybrids for chromosome 2q. Somat Cell Mol Genet. 1994;20:541–8. doi: 10.1007/BF02255844. [DOI] [PubMed] [Google Scholar]
- 12.Kastury K, Taylor WE, Gutierrez M, Ramirez L, Coucke PJ, Van Hauwe P, Van Camp G, Bhasin S. Chromosomal mapping of two members of the human dynein gene family to chromosome regions 7p15 and 11q13 near the deafness loci DFNA 5 and DFNA 11. Genomics. 1997;44:362–4. doi: 10.1006/geno.1997.4903. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida MC. Uniparental chromosome elimination in the early embryogenesis of the inviable salmonid hybrids between masu salmon female and rainbow trout male. Chromosoma. 1997;106:44–52. doi: 10.1007/s004120050223. [DOI] [PubMed] [Google Scholar]
- 14.Iwamatsu T, Uwa H, Inden A, Hirata K. Experiments on Interspecific Hybridization between Oryzias-Latipes and Oryzias-Celebensis. Zoolog Sci. 1984;1:653–63. [Google Scholar]
- 15.Iwamatsu T, Watanabe T, Hori R, Lam TJ, Saxena OP. Experiments on Interspecific Hybridization between Oryzias-Melastigma and Oryzias Javanicus. Zoolog Sci. 1986;3:287–93. [Google Scholar]
- 16.Sakai C, Konno F, Nakano O, Iwai T, Yokota T, Lee J, Nishida-Umehara C, Kuroiwa A, Matsuda Y, Yamashita M. Chromosome elimination in the interspecific hybrid medaka between Oryzias latipes and O. hubbsi. Chromosome Res. 2007;15:697–709. doi: 10.1007/s10577-007-1155-9. [DOI] [PubMed] [Google Scholar]
- 17.Sakai C, Konno F, Nakano O, Iwai T, Yokota T, Lee J, Nishida-Umehara C, Kuroiwa A, Matsuda Y, Yamashita M. Chromosome elimination in the interspecific hybrid medaka between Oryzias latipes and O. hubbsi. Chromosome Res. 2007;15:697–709. doi: 10.1007/s10577-007-1155-9. [DOI] [PubMed] [Google Scholar]
- 18.Ishii T, Ueda T, Tanaka H, Tsujimoto H. Chromosome elimination by wide hybridization between Triticeae or oat plant and pearl millet: pearl millet chromosome dynamics in hybrid embryo cells. Chromosome Res. 2010;18:821–31. doi: 10.1007/s10577-010-9158-3. [DOI] [PubMed] [Google Scholar]
- 19.Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–81. doi: 10.1105/tpc.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim NS, Armstrong KC, Fedak G, Ho K, Park NI. A microsatellite sequence from the rice blast fungus (Magnaporthe grisea) distinguishes between the centromeres of Hordeum vulgare and H. bulbosum in hybrid plants. Genome. 2002;45:165–74. doi: 10.1139/g01-129. [DOI] [PubMed] [Google Scholar]
- 21.Mochida K, Tsujimoto H, Sasakuma T. Confocal analysis of chromosome behavior in wheat x maize zygotes. Genome. 2004;47:199–205. doi: 10.1139/g03-123. [DOI] [PubMed] [Google Scholar]
- 22.RA F. Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma. 1983;88:386–93. doi: 10.1007/BF00285861. [DOI] [Google Scholar]
- 23.Schwarzacher Robinson TFR, Smith JB, Bennett MD. Genotypic control of centromere positions of parental genomes in Hordeum X Secale hybrid metaphases. J Cell Sci. 1987;87:291–304. [Google Scholar]
- 24.Dunwell JM. Haploids in flowering plants: origins and exploitation. Plant Biotechnol J. 2010;8:377–424. doi: 10.1111/j.1467-7652.2009.00498.x. [DOI] [PubMed] [Google Scholar]
- 25.Ravi M, Chan SWL. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–8. doi: 10.1038/nature08842. [DOI] [PubMed] [Google Scholar]
- 26.Gupta SB. Duration of Mitotic Cycle and Regulation of DNA Replication in Nicotiana Plumbaginifolia and a Hybrid Derivative of N Tabacum Showing Chromosome Instability. Can J Genet Cytol. 1969;11:133. [Google Scholar]
- 27.Bennett MD, Finch RA, Barclay IR. Time Rate and Mechanism of Chromosome Elimination in Hordeum Hybrids. Chromosoma. 1976;54:175–200. doi: 10.1007/BF00292839. [DOI] [Google Scholar]
- 28.Laurie DABM. The timing of chromosome elimination in hexaploid wheat x maize crosses. Genome. 1989;32:953–61. doi: 10.1139/g89-537. [DOI] [Google Scholar]
- 29.Gernand D, Rutten T, Pickering R, Houben A. Elimination of chromosomes in Hordeum vulgare x H. bulbosum crosses at mitosis and interphase involves micronucleus formation and progressive heterochromatinization. Cytogenet Genome Res. 2006;114:169–74. doi: 10.1159/000093334. [DOI] [PubMed] [Google Scholar]
- 30.Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüss C, Kumlehn J, Matzk F, Houben A. Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell. 2005;17:2431–8. doi: 10.1105/tpc.105.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies DR. Chromosome elimination in inter specific hybrids. Heredity. 1974;32:267–70. doi: 10.1038/hdy.1974.30. [DOI] [Google Scholar]
- 32.Sanei M, Pickering R, Kumke K, Nasuda S, Houben A. Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc Natl Acad Sci U S A. 2011;108:E498–505. doi: 10.1073/pnas.1103190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelesco PA, Graves JA. Chromosome segregation from cell hybrids. IV. Movement and position of segregant set chromosomes in early-phase interspecific cell hybrids. J Cell Sci. 1988;89:49–56. doi: 10.1242/jcs.89.1.49. [DOI] [PubMed] [Google Scholar]
- 34.Vig BK, Athwal RS. Sequence of centromere separation: separation in a quasi-stable mouse-human somatic cell hybrid. Chromosoma. 1989;98:167–73. doi: 10.1007/BF00329680. [DOI] [PubMed] [Google Scholar]
- 35.Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR. Defining ‘chromosomal instability’. Trends Genet. 2008;24:64–9. doi: 10.1016/j.tig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 37.Vitale I, Senovilla L, Jemaà M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–84. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falck GC, Catalán J, Norppa H. Nature of anaphase laggards and micronuclei in female cytokinesis-blocked lymphocytes. Mutagenesis. 2002;17:111–7. doi: 10.1093/mutage/17.2.111. [DOI] [PubMed] [Google Scholar]
- 39.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–70. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 40.Asaithamby A, Chen DJ. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009;37:3912–23. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Postow L. Destroying the ring: Freeing DNA from Ku with ubiquitin. FEBS Lett. 2011;585:2876–82. doi: 10.1016/j.febslet.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12:440–50. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Giglia-Mari G, Zotter A, Vermeulen W. DNA damage response. Cold Spring Harb Perspect Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subrahmanyam NCKK. Selective chromosome elimination during haploid formation in barley following interspecific hybridization. Chromosoma. 1973;42:111–2. doi: 10.1007/BF00320934. [DOI] [Google Scholar]
- 45.Cieplinski W, Reardon P, Testa MA. Non-random human chromosome distribution in human-mouse myeloma somatic cell hybrids. Cytogenet Cell Genet. 1983;35:93–9. doi: 10.1159/000131848. [DOI] [PubMed] [Google Scholar]
- 46.Sengupta K, Camps J, Mathews P, Barenboim-Stapleton L, Nguyen QT, Difilippantonio MJ, Ried T. Position of human chromosomes is conserved in mouse nuclei indicating a species-independent mechanism for maintaining genome organization. Chromosoma. 2008;117:499–509. doi: 10.1007/s00412-008-0171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corredor E, Díez M, Shepherd K, Naranjo T. The positioning of rye homologous chromosomes added to wheat through the cell cycle in somatic cells untreated and treated with colchicine. Cytogenet Genome Res. 2005;109:112–9. doi: 10.1159/000082389. [DOI] [PubMed] [Google Scholar]
- 48.Gleba YY, Parokonny A, Kotov V, Negrutiu I, Momot V. Spatial separation of parental genomes in hybrids of somatic plant cells. Proc Natl Acad Sci U S A. 1987;84:3709–13. doi: 10.1073/pnas.84.11.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leitch AR, Schwarzacher T, Mosgoller W, Bennett MD, Heslopharrison JS. Parental Genomes Are Separated Throughout the Cell-Cycle in a Plant Hybrid. Chromosoma. 1991;101:206–13. doi: 10.1007/BF00365152. [DOI] [Google Scholar]
- 50.Mayer W, Fundele R, Haaf T. Spatial separation of parental genomes during mouse interspecific (Mus musculus x M. spretus) spermiogenesis. Chromosome Res. 2000;8:555–8. doi: 10.1023/A:1009227924235. [DOI] [PubMed] [Google Scholar]
- 51.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 52.Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–64. doi: 10.1016/S0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 53.Hübner MR, Spector DL. Chromatin dynamics. Annu Rev Biophys. 2010;39:471–89. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohashi H, Wakui K, Ogawa K, Okano T, Niikawa N, Fukushima Y. A stable acentric marker chromosome: possible existence of an intercalary ancient centromere at distal 8p. Am J Hum Genet. 1994;55:1202–8. [PMC free article] [PubMed] [Google Scholar]
- 55.Kanda T, Wahl GM. The dynamics of acentric chromosomes in cancer cells revealed by GFP-based chromosome labeling strategies. J Cell Biochem Suppl. 2000;(Suppl 35):107–14. doi: 10.1002/1097-4644(2000)79:35+<107::AID-JCB1133>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 56.Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23:2876–86. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gernand D, Rutten T, Varshney A, Rubtsova M, Prodanovic S, Brüss C, Kumlehn J, Matzk F, Houben A. Uniparental chromosome elimination at mitosis and interphase in wheat and pearl millet crosses involves micronucleus formation, progressive heterochromatinization, and DNA fragmentation. Plant Cell. 2005;17:2431–8. doi: 10.1105/tpc.105.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn K, Lowndes NF, Grenon M. Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell Mol Life Sci. 2012;69:1447–73. doi: 10.1007/s00018-011-0875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terradas M, Martín M, Tusell L, Genescà A. Genetic activities in micronuclei: is the DNA entrapped in micronuclei lost for the cell? Mutat Res. 2010;705:60–7. doi: 10.1016/j.mrrev.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 60.You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010;20:402–9. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grigsby RV, Fairbairn D, O’Neill KL. Differential DNA damage detected in hybridomas. Hybridoma. 1993;12:755–61. doi: 10.1089/hyb.1993.12.755. [DOI] [PubMed] [Google Scholar]
- 63.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–8. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noon AT, Goodarzi AA. 53BP1-mediated DNA double strand break repair: insert bad pun here. DNA Repair (Amst) 2011;10:1071–6. doi: 10.1016/j.dnarep.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 65.Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 2013;339:700–4. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson RT, Rao PN. Mammalian cell fusion: induction of premature chromosome condensation in interphase nuclei. Nature. 1970;226:717–22. doi: 10.1038/226717a0. [DOI] [PubMed] [Google Scholar]
- 67.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lv L, Zhang T, Yi Q, Huang Y, Wang Z, Hou H, Zhang H, Zheng W, Hao Q, Guo Z, et al. Tetraploid cells from cytokinesis failure induce aneuploidy and spontaneous transformation of mouse ovarian surface epithelial cells. Cell Cycle. 2012;11:2864–75. doi: 10.4161/cc.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi Q, Martin RH. Spontaneous frequencies of aneuploid and diploid sperm in 10 normal Chinese men: assessed by multicolor fluorescence in situ hybridization. Cytogenet Cell Genet. 2000;90:79–83. doi: 10.1159/000015668. [DOI] [PubMed] [Google Scholar]
- 70.Huang Y, Hou H, Yi Q, Zhang Y, Chen D, Jiang E, Xia Y, Fenech M, Shi Q. The fate of micronucleated cells post X-irradiation detected by live cell imaging. DNA Repair (Amst) 2011;10:629–38. doi: 10.1016/j.dnarep.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Bergqvist M, Brattström D, Stålberg M, Vaghef H, Brodin O, Hellman B. Evaluation of radiation-induced DNA damage and DNA repair in human lung cancer cell lines with different radiosensitivity using alkaline and neutral single cell gel electrophoresis. Cancer Lett. 1998;133:9–18. doi: 10.1016/S0304-3835(98)00178-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.