Here we present the increased immunostimulatory effects of an α,ω-heterodimer composed of dendritic cell agonists. (Figure 1) These agonists are molecular signals used by the immune system to determine self from non-self often called Pathogen Associated Molecular Patterns (PAMPs).[1] Employing multiple agonists for effective immune stimulation is emerging as the key aspect in robust activation of the immune system in applications ranging from vaccine formulation[2] to cancer immunotherapy.[3] For example, in many attenuated whole virus vaccines, agonist “synergies” are used to generate durable immune responses and life-long immunity.[4] These agonist combinations activate a range of receptors found throughout dendritic cells (DCs).[5]
Figure 1.
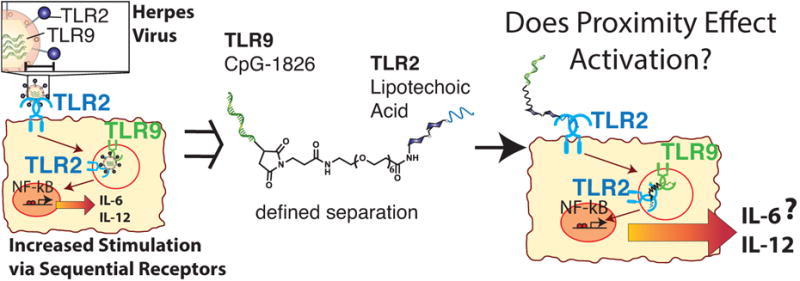
In native pathogens, such as the herpes virus, an inter-agonist spacing exists among different agonists. In this work we probe the immunostimulatory effects of two different agonists, LTA and CpG, conjugated at a discrete molecular distance.
The Toll-Like Receptor (TLR) family is the most highly studied DC receptor subset.[6] The molecular basis for the activation of TLRs relies on the formation of higher order complexes.[7] Inside a DC, the trans-membrane domains of these receptor complexes induce oligomerization of the signaling protein, myeloid differentiation primary response gene 88 (MYD88).[8] These MYD88 oligomers assemble with additional signaling proteins to activate an immune response.[9] We hypothesized that spatial confinement of heterodimers of receptor agonists would increase activation potentially via the confined association of multiple receptors by promoting MYD88 oligomerization and immune activation.
Activation of any one receptor on a DC leads to stimulation. Simultaneous activation of multiple TLRs results in increased stimulatory effects broadly called synergies that can direct the polarization of the immune response.[10] Stimulation of DCs by varied agonist combinations directs amplification of the immune response with a specific response (e.g. vaccine, cancer) requiring control over different combinations of pathways.[11] Recently, several approaches have combined multiple agonist signals including virus, nanoparticle, and dendrimer motifs.[12] Additionally, in other immune cell systems multivalent ligands enhance activation.[13] Increased synergistic activity appears to be correlated with proximity or multi-valency, but this observation has never been directly tested until this report. Our operating hypothesis is that multi-valent molecules might stimulate higher-order TLR structures. In this report, we tested this hypothesis and found that coupling TLR agonists results in increased stimulation, but we cannot yet conclude this is due exclusively to TLR ordering.
Well-defined and short inter-agonist spacings of imidazoquinoline homodimers result in modulation of immunostimulation of TLRs 7 and 8.[14] Similar to other biologic heterodimers,[15] we sought to explore the effect of inter-agonist proximity on the activation of multiple TLRs with different agonist combinations. We used an α,ω-heterotelechelic PEG linker to synthesize agonist-heterodimers consisting of immunostimulants (known to have a mild synergy) from two different signaling pathways with corresponding TLRs in two different cellular locations, on the cell-surface and in endosomal compartments.
We covalently conjugated lipoteichoic acid to a DNA fragment (Figure 2) using an α,ω-heterotelechelic PEG based linker. The DNA is the endosomal TLR 9 agonist, CpG ODN 1826 (CpG)[16] modified to contain a thiol and 6-fluorescein phosphoramidite (FAM) tag at the 3′ and 5′ ends respectively. CpG was attached to the cell-surface TLR 2/6 agonist, lipoteichoic acid (LTA), a class of glycosylated lipopolypeptides that contains a variable phosphate linked polymeric backbone.[17]
Figure 2.
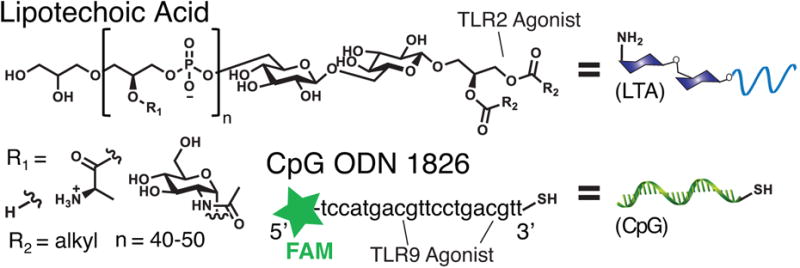
Lipoteichoic acid (LTA) is composed of alanine, GlcNAc, and hydroxyl moieties linked along a phosphate backbone, terminated with alkyl chains of varied structures. The CpG DNA, CpG ODN 1826 contained both a fluorescent FAM tag and thiol.
The two agonists were conjugated using a heterotelechelic PEG based linker (Figure 3) bearing NHS and maleimide end-groups. LTA was first coupled to the PEG linker before conjugation of the maleimide moiety to the thiol on CpG. Strict control of the solution pH was required during all stages of the bioconjugation process as LTA is susceptible to hydrolysis under acidic or basic conditions. LTA from Bacillus subtilis was conjugated to the PEG linker via coupling of the alanine side-chain in LTA to the NHS ester end-group of the PEG linker (experimental procedure in SI). The resulting conjugate was characterized by NMR (Figure S2), UV/Vis spectroscopy, SDS-PAGE, and FPLC (Figure S3). The LTA-PEG conjugate was quantified via the Absmax at 256 nm for use in the next step.
Figure 3.
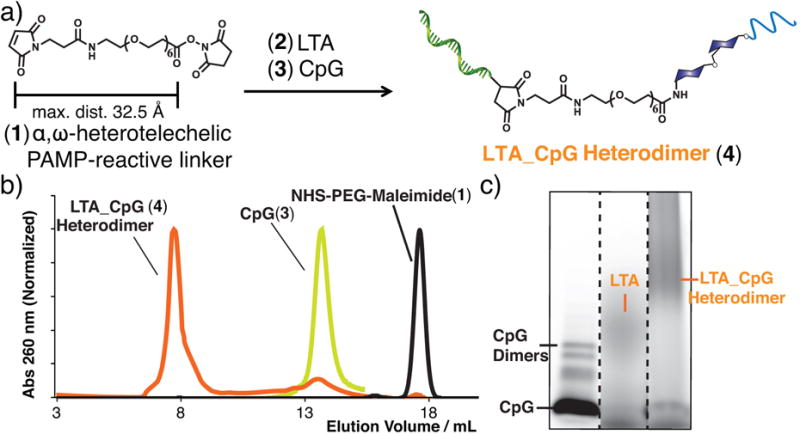
a) An α,ω-heterotelechelic PEG-based linker was conjugated first to a free amine on LTA followed by a thiol on CpG. c) The resulting crude reaction was purified by fast protein liquid chromatography. Characterization of the purified conjugate included d) SDS-PAGE (Lane 1: CpG, 2: FITC-labeled LTA, 3: purified LTA_CpG heterodimer).
LTA_PEG conjugate was further elaborated to synthesize the LTA_CpG heterodimer. The LTA_CpG heterodimer was purified via FPLC (Superdex G75, DPBS, 0.2 mL/min), and characterized by UV/Vis (Figure S4), SDS-PAGE (Figure S5) and dynamic light scattering (DLS, Figure S6). Stable particles were not observed by DLS, however the LTA_CpG heterodimer was found to agglomerate over time similar to LTA alone. The conjugate was quantified as the corresponding CpG by mass according to the FAM absorbance at 495 nm.
The LTA_CpG heterodimer was tested with two different cell lines, murine macrophage RAW-Blue cells and murine Bone Marrow Derived Dendritic Cells (BMDCs). RAW-Blue is a reporter cell line for NFκB activation, a transcription factor and general measure of immune cell stimulation. RAW-Blue cells secrete embryonic alkaline phosphatase (SEAP) upon activation of the NFκB pathway allowing quantification of cell stimulation. The RAW-Blue cell line was stimulated with LTA, CpG, an unconjugated mixture of LTA and CpG, or the LTA_CpG heterodimer (Figure S7). For comparison, lipopolysaccharide (LPS) was also tested. Concentrations were varied from 10-100 ng/mL with respect to CpG in the case of the LTA_CpG heterodimer and agonist mixture. The LTA_CpG heterodimer activated the RAW-Blue cell line to a greater extent than an unconjugated mixture of CpG and LTA. For concentrations >25 ng/mL the LTA_CpG heterodimer exhibited increased stimulation relative to LPS one of the most potent known immunostimulants (Figure 4a). The magnitude and polarization of the increased immune response was further examined in BMDCs to better understand the effect of the LTA_CpG heterodimer in the stimulation of primary antigen presenting cells. The LTA_CpG heterodimer provided greater up-regulation of cell-surface markers and release of cytokines associated with amplification of the immune response. Using antibody staining we monitored changes in cell-surface proteins on cells containing the BMDC phenotype CD11c+. T-cell adhesion (CD 40, 80, and 86) and antigen presentation (MHC-II) proteins were measured by flow cytometry. These proteins, always present on BMDCs, are up-regulated upon stimulation. Exposure to the LTA_CpG heterodimer increased the expression of each protein; this increase was most evident with CD 40 where surface expression was increased by over 40% for the LTA_CpG heterodimer relative to the unconjugated mixture (Figure 4b). The stimulation profile we observed is indicative of an increase in antigen cross-presentation and T-cell expansion based on increases in T-cell adhesion proteins and MHC-II. We therefore expect that the LTA_CpG heterodimers will perform as superior immunostimulants relative to either the agonist alone or an unconjugated mixture thereof.
Figure 4.
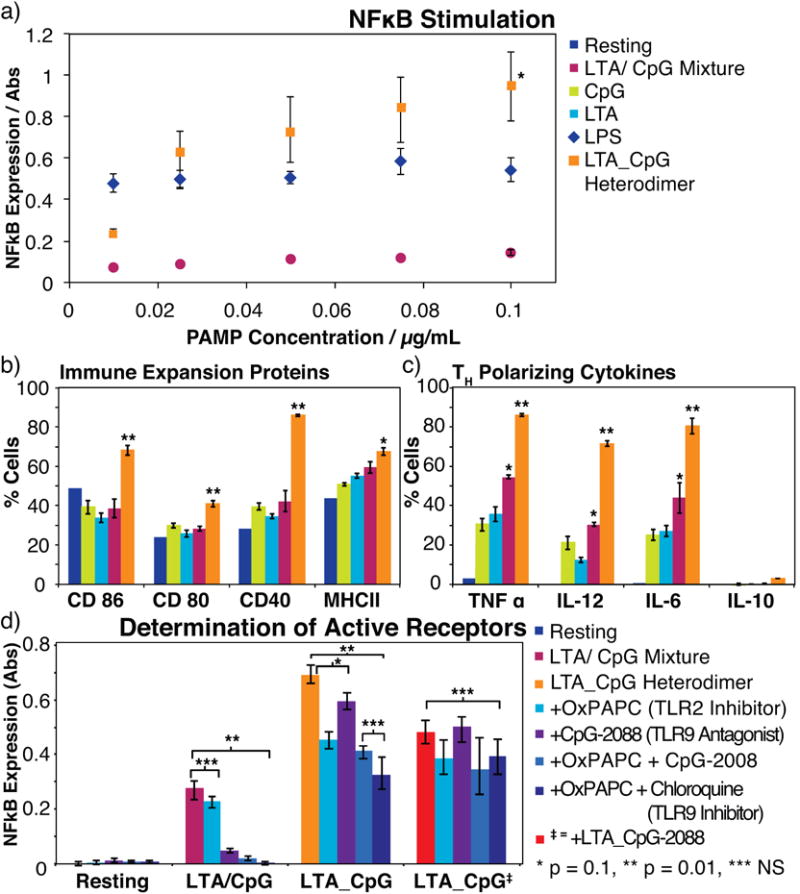
a) RAW-Blue cells were stimulated by addition of an unconjugated mixture of CpG/LTA, LPS or the LTA_CpG heterodimer. Activation was quantified using the Quanti-Blue assay of NFκB (*p < 0.005 for the heterodimer relative to LPS and < 0.001 for the heterodimer relative to the LTA/CpG mixture). b) Bone Marrow Derived Dendritic Cells (BMDCs) were incubated with the same agonist combinations at 100 ng/mL. Antibody staining and flow cytometry was used to determine changes in cell surfaces proteins involved with T-Cell adhesion and antigen presentation as well as c) polarizing cytokines. d) The effect of antagonists and inhibitors of TLR 2 and TLR 9 were also examined in RAW-Blue cells.
To determine the polarization of the increased stimulation, we measured expression of polarizing cytokines. Five cytokines were screened including TNF-α, ILs 6, 10, 12 (Figure 4c) and interferon-γ (Figure S8). The CpG, LTA, and mixture of CpG and LTA all induced production of cytokines associated with a TH1 (cytotoxic T-cell) type response (TNF-α, IL-6, IL-12). The LTA_CpG heterodimer induced over a 30% increase in production of each of these cytokines within this same TH1 profile and also induced production of IL-10 at low (3% of cells expressed IL-10) but significant (p < 0.001, for the heterodimer compared to the resting state) levels.
Mechanistic studies were performed with TLR2 and TLR9 antagonists (OxPAPC, TLR2 and ODN2088, TLR9), an endosomal activity inhibitor to block TLR9 (chloroquine), and LTA conjugated to ODN2088 (Figure 4d). First, we used OxPAPC (Invivogen, CA) to competitively inhibit the TLR2 pathway. The resulting decrease in stimulation confirmed CpG_LTA acts partially through TLR2. Addition of either ODN2088 or chloroquine further decreased stimulation confirming that the activity was dependent on both TLR2 and TLR9. Stimulation decreased upon addition of ODN2088 or chloroquine alone showing activation was partially dependent on the TLR9 pathway. The combination of ODN2088 and chloroquine produced an additive effect in decreasing stimulation. (SI) Cumulatively these results indicate that stimulation by LTA_CpG proceeds through traditional TLR2/6 and TLR9 pathways. To test if the increased activity of LTA_CpG was due to activation of both TLRs, LTA was conjugated to the antagonist CpG sequence ODN2088. This sequence competitively binds TLR9 thereby inhibiting activity. Stimulation for the antagonist construct was less than the LTA_CpG heterodimer but greater than a mixture of the two PAMPs. Taken together these results indicate that although the LTA_CpG heterodimer accessing the TLR2/6 and TLR9 pathways is partially responsible for the synergies, there may be a second mechanism at work as inhibitor and antagonist controls did not completely return baseline activity.
Our current operating hypothesis is that there is a molecular level synergy between TLR 2 and TLR 9 enhanced by tethering the TLR 2/6 agonist LTA to the TLR 9 agonist CpG. This is supported by synergies found for the herpes virus, which successively activates TLR 2 and then TLR 9 and by the synergistically decreased activity when inhibitors are added.[18] We are, as yet, unsure of how this synergy operates, but one possibility is that the LTA_CpG heterodimer might create an avidity effect for each TLR thereby promoting formation of the myddozome and increasing stimulation. Although we show that the LTA_CpG heterodimer is sensitive to both TLR2 and TLR9 antagonists we have not fully characterized the mechanism of increased stimulation so this remains only one possible explanation. We also observed the RAW-Blue cell line stimulated with CpG or the LTA_CpG heterodimer by confocal microscopy. Cell binding and entry was found to differ from CpG for the LTA_CpG heterodimer. The CpG localized more rapidly and the LTA_CpG provided greater, but more diffuse presence (Figure S9). This evidence in conjunction with the antagonist/inhibitor assay provides support for the presence of a molecular level synergy.
In summary we have synthesized a covalently coupled LTA_CpG heterodimer, and we have observed increases in immune stimulation with the LTA_CpG heterodimer relative to a mixture of LTA and CpG. This increase is conserved across two different cell types and among T-cell adhesion proteins, as well as polarizing cytokines. Our working hypothesis is that spatial patterning of agonists is enhancing formation of higher order, myddozome (MYD88 oligomer) structures, but an as yet unknown effect that operates outside of TLRs cannot be ruled out. Increases in cell adhesion and antigen presentation proteins were observed for the LTA_CpG heterodimer relative to a mixture of the two agonists. The corresponding cytokine profile is also larger in magnitude for the LTA_CpG heterodimer and is indicative of a TH1 response. These results indicate that controlling the spatial presentation of agonists to dendritic cell receptors dramatically alters the stimulation of dendritic cells. We plan to further probe this amplification in T-cell expansion assays. Further experiments will work to confirm a molecular-level effect by exploring different linker lengths and other combinations of agonists.
Supplementary Material
Acknowledgments
**The authors would like to thank the Cahalan, Tsai and Guan labs for reagents and instrument use. JKT acknowledges the NSF DGE-0808392 for a fellowship. We acknowledge UC-Irvine for startup funds and NIH New Innovator Award DP2 AI112194
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org
References
- 1.Akira S, Hemmi H. Immunol Lett. 2003;85:85–95. doi: 10.1016/s0165-2478(02)00228-6. [DOI] [PubMed] [Google Scholar]
- 2.Moyle PM, Toth I. ChemMedChem. 2013;8:360–376. doi: 10.1002/cmdc.201200487. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I, Coukos G, Dranoff G. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Querec T, Bennouna S, Alkan SK, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. J Exp Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Gordon S. Cell. 2002;111:927–930. doi: 10.1016/s0092-8674(02)01201-1. [DOI] [PubMed] [Google Scholar]; b) Banchereau J, Steinman RM. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]; c) Steinman RM. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 6.a) Akira S, Takeda K, Kaisho T. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]; b) Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, Wilson CB, Schroeder L, Aderem A. Proc Natl Acad Sci U S A. 2000;97:13766–13771. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Moresco EMY, LaVine D, Beutler B. Curr Biol. 2011;21:R488–R493. doi: 10.1016/j.cub.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 7.a) Botos I, Segal DM, Davies DR. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Yoon S, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, Wilson IA. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Science. 2013;339:1426–1429. doi: 10.1126/science.1229159. [DOI] [PubMed] [Google Scholar]; d) Chan M, Hayashi T, Kuy CS, Gray CS, Wu CCN, Corr M, Wrasidlo W, Cottam HB, Carson DA. Bioconjug Chem. 2009;20:1194–1200. doi: 10.1021/bc900054q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin SC, Lo YC, Wu H. Nature. 2010;465:885–U2. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Ferrao R, Li J, Bergamin E, Wu H. Sci Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gay NJ, Gangloff M, O'Neill LAJ. Trends Immunol. 2011;32:104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 10.a) Napolitani G, Rinaldi A, Bertoni F, Sallusto F, Lanzavecchia A. Nat Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Raman VS, Bhatia A, Picone A, Whittle J, Bailor HR, O'Donnell J, Pattabhi S, Guderian JA, Mohamath R, Duthie MS, et al. J Immunol. 2010;185:1701–1710. doi: 10.4049/jimmunol.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Chen K, Huang J, Liu Y, Gong W, Cui Y, Wang JM. J Neuroimmunol. 2009;213:69–77. doi: 10.1016/j.jneuroim.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Banchereau J, Palucka AK. Nat Rev Immunol. 2005;5:296–306. doi: 10.1038/nri1592. [DOI] [PubMed] [Google Scholar]
- 11.a) Kapsenberg ML. Nat Rev Immunol. 2003;3:984–993. doi: 10.1038/nri1246. [DOI] [PubMed] [Google Scholar]; b) Schuler-Thurner B, Schultz ES, Berger TG, Weinlich G, Ebner S, Woerl P, Bender A, Feuerstein B, Fritsch PO, Romani N, et al. J Exp Med. 2002;195:1279–1288. doi: 10.1084/jem.20012100. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Gupta R, Siber G. Vaccine. 1995;13:1263–1276. doi: 10.1016/0264-410x(95)00011-o. [DOI] [PubMed] [Google Scholar]; d) Shinohara ML, Cantor H. Nat Med. 2007;13:536–538. doi: 10.1038/nm0507-536. [DOI] [PubMed] [Google Scholar]; e) Kim CH, Rott L, Kunkel EJ, Genovese MC, Andrew DP, Wu LJ, Butcher EC. J Clin Invest. 2001;108:1331–1339. doi: 10.1172/JCI13543. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Ma JSY, Monu N, Shen DT, Mecklenbrauker I, Radoja N, Haydar TF, Leitges M, Frey AB, Vukmanovic S, Radoja S. J Immunol. 2007;178:7814–7821. doi: 10.4049/jimmunol.178.12.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Kovacs B, Maus MV, Riley JL, Derimanov GS, Koretzky GA, June CH, Finkel TH. Proc Natl Acad Sci U S A. 2002;99:15006–15011. doi: 10.1073/pnas.232058599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Reddy ST, van der Vlies AJ, Simeoni E, Angeli V, Randolph GJ, O'Neill CP, Lee LK, Swartz MA, Hubbell JA. Nat Biotechnol. 2007;25:1159–1164. doi: 10.1038/nbt1332. [DOI] [PubMed] [Google Scholar]; b) Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, et al. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Shukla NM, Salunke DB, Balakrishna R, Mutz CA, Malladi SS, David SA. PLoS ONE. 2012;7:e43612. doi: 10.1371/journal.pone.0043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Kiessling LL, Gestwicki JE, Strong LE. Angew Chem Int Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Puffer EB, Pontrello JK, Hollenbeck JJ, Kink JA, Kiessling LL. ACS Chem Biol. 2007;2:252–262. doi: 10.1021/cb600489g. [DOI] [PubMed] [Google Scholar]
- 14.Shukla NM, Mutz CA, Malladi SS, Warshakoon HJ, Balakrishna R, David SA. J Med Chem. 2012;55:1106–1116. doi: 10.1021/jm2010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Farrar MA, Alberol-Ila J, Perlmutter RM. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]; b) Fegan A, White B, Carlson JCT, Wagner CR. Chem Rev. 2010;110:3315–3336. doi: 10.1021/cr8002888. [DOI] [PubMed] [Google Scholar]; c) Belshaw PJ, Ho SN, Crabtree GR, Schreiber SL. Proc Natl Acad Sci U S A. 1996;93:4604–4607. doi: 10.1073/pnas.93.10.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lin H, Abida WM, Sauer RT, Cornish VW. J Am Chem Soc. 2000;122:4247–4248. [Google Scholar]; e) Miyamoto T, DeRose R, Suarez A, Ueno T, Chen M, Sun T, Wolfgang MJ, Mukherjee C, Meyers DJ, Inoue T. Nat Chem Biol. 2012;8:465–470. doi: 10.1038/nchembio.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu RS, Targoni OS, Krieg AM, Lehmann PV, Harding CV. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Wicken A, Knox K. Science. 1975;187:1161–1167. doi: 10.1126/science.46620. [DOI] [PubMed] [Google Scholar]; b) Ginsburg I. Lancet Infect Dis. 2002;2:171–179. doi: 10.1016/s1473-3099(02)00226-8. [DOI] [PubMed] [Google Scholar]; c) Fischer W, Mannsfeld T, Hagen G. Biochem Cell Biol. 1990;68:33–43. doi: 10.1139/o90-005. [DOI] [PubMed] [Google Scholar]; d) Behr T, Fischer W, Peterkatalinic J, Egge H. Eur J Biochem. 1992;207:1063–1075. doi: 10.1111/j.1432-1033.1992.tb17143.x. [DOI] [PubMed] [Google Scholar]
- 18.Sørensen LN, Reinert LS, Malmgaard L, Bartholdy C, Thomsen AR, Paludan SR. J Immunol Baltim Md 1950. 2008;181:8604–8612. doi: 10.4049/jimmunol.181.12.8604. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


