Abstract
Deubiquitination has emerged as an important mechanism of p53 regulation. A number of deubiquitinating enzymes (DUBs) from the ubiquitin-specific protease family have been shown to regulate the p53-MDM2-MDMX networks. We recently reported that Otub1, a DUB from the OTU-domain containing protease family, is a novel p53 regulator. Interestingly, Otub1 abrogates p53 ubiquitination and stabilizes and activates p53 in cells independently of its deubiquitinating enzyme activity. Instead, it does so by inhibiting the MDM2 cognate ubiquitin-conjugating enzyme (E2) UbcH5. Otub1 also regulates other biological signaling through this non-canonical mechanism, suppression of E2, including the inhibition of DNA-damage-induced chromatin ubiquitination. Thus, Otub1 evolves as a unique DUB that mainly suppresses E2 to regulate substrates. Here we review the current progress made towards the understanding of the complex regulation of the p53 tumor suppressor pathway by DUBs, the biological function of Otub1 including its positive regulation of p53, and the mechanistic insights into how Otub1 suppresses E2.
Keywords: p53, MDM2, Ubiquitination, Deubiquitinating enzymes, Otub1, Cell cycle, Apoptosis
Core tip: p53 is tightly regulated by dynamic ubiquitination and deubiquitination. A number of deubiquitinating enzymes (DUBs) have been shown to regulate p53 stability and activity by either directly deubiquitinating p53 or indirectly deubiquitinating its regulators. We recently discovered that Otub1, an OTU family DUB, stabilizes and activates p53 via distinct and non-canonical mechanism wherein it suppresses the MDM2 cognate ubiquitin-conjugating enzymes UbcH5. Here we review the current progress made towards the understanding of the Otub1 functions as a potent E2 inhibitor and the underlying mechanisms.
MDM2 AND MDMX: KEEPING P53 UNDER CONTROL
The p53 tumor suppressor plays a central role in maintaining the genomic stability and preventing the organism from cancer[1-3]. Loss of p53 function, either through direct mutations in the p53 gene or indirectly through alterations in the p53 regulatory networks, is associated with most, if not all, human cancers[4,5]. Germline mutations of p53 result in the cancer-prone Li-Fraumeni syndrome in human[6] and deletion of the p53 gene leads to spontaneous tumors in mice[7,8]. p53 is a stress-induced transcription factor that activates or represses the expression of many target genes, thereby executing its anti-proliferative activity by inducing cell cycle arrest, apoptosis, or senescence[1,2,9-11]. Under normal circumstances, p53 is tightly controlled at low levels mainly by its negative regulator MDM2[12-14]. As a RING-finger-containing ubiquitin ligase (E3)[15,16] MDM2 mediates p53 ubiquitination and degradation through the proteasomal system[17,18]. MDM2 also directly suppresses p53 transactivation activity by binding and concealing the N-terminal transactivation domain of p53[19-21]. The centrality of the MDM2-mediated p53 suppression has been demonstrated by mouse genetic studies showing that deletion of the mdm2 gene caused embryonic lethal phenotype, which is completely rescued by concomitant deletion of p53[22,23]. This essential function of MDM2 requires its E3 activity, as mice with homozygous knock-in of the E3 inactivation mutant, MDM2C464A, are also embryonic lethal, which can be rescued by deleting p53 as well[24]. Consistently, MDM2 is overexpressed in a number of human cancers, most of which contain wild-type p53[25-29].
The MDM2 homolog MDMX has emerged as an equally important p53 regulator as MDM2[30]. MDMX shares high homology with MDM2 in their C-terminal RING-finger domain and the N-terminal p53-binding domain. Like MDM2, MDMX binds to the N-terminal transactivation domain of p53 and suppresses its activity. However, MDMX does not have appreciable ubiquitin ligase activity towards p53[31,32], yet it assists MDM2 to suppress p53 function. MDMX directly binds to MDM2 via their RING domains[33-35] and renders MDM2 sufficiently stable to ubiquitinate and degrade p53[33,36-38]. Also, MDMX suppresses p53 function by specifically promoting p53-induced MDM2 transcription following DNA damage[39]. MDM2, in turn, ubiquitinates and degrades MDMX in response to DNA damage[40-42]. Thus, the mutual regulation between MDM2 and MDMX ensures a proper cellular level and activity of p53. Supporting the indispensible role of MDMX towards p53, deleting the p53 gene also rescues the lethal phenotype of knocking out the mdmx gene in mice[43-45]. Like MDM2, MDMX is also overexpressed or amplified in several types of human cancers that harbor wild-type p53[46-49]. Recent studies have provided further molecular insights into the non-redundant and indispensible role for MDMX in MDM2-mediated p53 degradation. First, like MDM2, the RING domain of MDMX and resulting MDM2-MDMX heterodimerization are required for the regulation of MDM2, as deletion of the RING-finger domain of MDMX or knock-in of the MDM2-binding defective MDMX mutant (C462A) resulted in embryonic lethal phenotype, which was completely rescued by deletion of p53[50,51]. Second, The extreme C-terminal short sequences outside of the RING domain of both MDM2 and MDMX contribute to the MDM2 E3 activity, owing to their role in the formation of MDM2-MDMX heterodimer and perhaps the E3 holoenzyme mediating p53 polyubiquitination[37,38,52]. Third, a recent in vitro study has shown that while MDM2 alone is sufficient to mediate multi-monoubiquitination of p53, the MDM2-MDMX complex is required for p53 polyubiquitination[53]. Thus, the stoichiometry of the p53-MDM2-MDMX complex is critical for the determination of whether targeting p53 for polyubiquitination or monoubiquitination.
The p53-MDM2-MDMX axis is among the most highly regulated pathways. Enormous molecules regulate the interplay among the three proteins in response to diverse stressors, leading to p53 stabilization and consequent activation. These include various post-translational modifications of all three proteins. Ubiquitination plays a key role in controlling the protein stability and activity of all three proteins. Under stress conditions, p53 ubiquitination mediated by MDM2/MDMX is crippled as a result of either dissociation of MDM2/MDMX from p53 or suppression of MDM2/MDMX activity towards p53. For example, DNA damage-mediated phosphorylation of both p53 and MDM2 disrupts their interaction, resulting in p53 stabilization[54-57]. DNA damage also triggers phosphorylation and degradation of MDMX, alleviating its suppressive effect on p53[58-63]. Oncogenic stress induces p53 via suppression of MDM2 by ARF[64-68], whereas ribosomal stress induces p53 via suppression of MDM2 by a number of ribosomal proteins[69-85]. Again, ARF also promotes MDM2-mediated MDMX degradation[40] and ribosomal stress-induced p53 activation requires MDM2-mediated MDMX degradation[86]. Thus, barricading the inhibition of p53 imposed by MDM2 and MDMX is centrally important for p53 activation in response to most, if not all, stressors. Indeed, both MDM2 and MDMX bind to p53 at its target gene promoters and suppress its transactivation activity[87-89]. Thus, p53 activation is thought to involve the release of such repression, called anti-repression under stress conditions, through diverse posttranslational modifications[90]. In addition, p53 is also ubiquitinated by a number of other ubiquitin ligases such as ARF-BP1[91], PIRH2[92], COP1[93], etc.[94,95]. For example, p53, under certain cellular levels, is thought no longer regulated by the MDM2/MDMX complex. Instead, the basal level of p53 is mainly regulated by ARF-BP1. Deletion of ARF-BP1 completely activates p53 in the presence of MDM2[91]. Adding to the complexity of the ubiquitination regulation of the p53 pathway, deubiquitination regulation has recently emerged as an equally important mechanism for p53 control.
REGULATION OF THE P53-MDM2-MDMX PATHWAY BY DEUBIQUITINATING ENZYMES
Like other posttranslational modifications, ubiquitination of p53, MDM2 and MDMX can be reversed through a process called deubiquitination, which is catalyzed by a different class of enzymes called deubiquitinating enzymes (DUBs). The human genome encodes approximately 95 predicted DUBs that are classified into 5 families: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor associated proteases (OTUs), Machado-Joseph disease (or Josephin domain) proteins (MJDs), and JAB1/MPN/MOV34 proteins (JAMMs). Except that the JAMMs are zinc metalloproteases, all other DUBs are cysteine proteases[96,97].
Recently, several DUBs from the USP family have been shown to regulate the p53-MDM2-MDMX loop (Figure 1). USP7, also called herpesvirus associated USP (HAUSP), is the first DUB reported to be a bona fide p53 deubiquitinase[98-100]. Overexpression of USP7 stabilizes and activates p53[99]. Intriguingly, MDM2 seems to be a better substrate of USP7 compared to p53 under physiological circumstances, as substantial knockdown of USP7 results in destabilization of MDM2 and activation of p53[98,101]. Further, USP7 also deubiquitinates MDMX in cells and in vitro and depletion of USP7 results in destabilization of the otherwise stable MDMX[100]. DNA damage triggers ATM-dependent phosphorylation of MDMX, which disrupts its binding to USP7 and leads to the consequent increase of ubiquitination and degradation of MDMX[100], whereas the interaction between p53 and USP7 is increased following DNA damage. Thus USP7 scrutinizes the homeostatic levels of p53, MDM2, and MDMX under both normal and stress conditions. The second p53 DUB, USP10, has also been shown to play a critical role in p53 activation following DNA damage[102]. Unlike USP7, USP10 is a cytoplasmic DUB and specifically deubiquitinates p53, but not MDM2 and MDMX[102], reversing MDM2-mediated ubiquitination, nuclear export, and cytoplasmic degradation of p53. Following DNA damage, ATM phosphorylates USP10 at Thr42 and Ser337, resulting in not only the stabilization of USP10, but also the translocation of a fraction of USP10 into the nucleus to deubiquitinate and activate p53. Consistent with its function in regulating p53, USP10 expression is down-regulated in high percentage of clear cell carcinomas[102]. Recently, USP42 was reported to be another DUB that positively regulates p53 stability and activity. Interestingly, USP42 deubiquitinates p53 only during the early stages of stress response, without significant effect on p53 regulation under unstressed conditions. Despite of this, it has been shown that USP42 is required for rapid p53 activation and cell cycle arrest in response to mild or transient DNA damage stress[103]. In addition, Liu et al[104] has shown that USP29 positively regulates p53 stability and function following oxidative stress. This is achieved by the increased transcription of USP29 induced by oxidative stress, which in turn cleaves polyubiquitinated p53, leading to p53-dependent apoptosis in cells.
Figure 1.
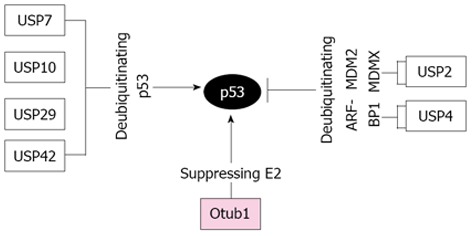
Diagram of the regulation of the p53 pathway by deubiquitinating enzymes. Arrows indicate activation and bars indicate inhibition. USP7, USP10, USP29, and USP42 deubiquitinate and activate p53, whereas USP2 destabilizes p53 by deubiquitinating MDM2 and MDMX and USP4 destabilizes p53 by deubiquitinating and stabilizing ARF-BP1. Otub1 stabilizes and activates p53 via non-canonical suppression of the MDM2 cognate E2 UbcH5, thereby inhibiting MDM2-mediated p53 ubiquitination and degradation. USP: Ubiquitin-specific protease
In contrast to above USPs positively regulating p53, USP2a and USP4 were reported to destabilize p53 and suppress p53 function, albeit via targeting different p53 E3s. USP2a destabilizes p53 by deubiquitinating and stabilizing both MDM2[105] and MDMX[106], whereas USP4 destabilizes p53 by deubiquitinating and stabilizing ARF-BP1[107]. Consistently, USP2a is overexpressed in a subset of prostate cancers[108,109], whereas USP4 is overexpressed in a broad range of human cancers[107]. Thus, USP2a and USP4 are likely oncogenic DUBs.
Together, these studies demonstrate that deubiquitination plays a crucial role in finely tuning the normal homeostasis of the p53-MDM2-MDMX loop as well as its response to stress. They also imply that different DUBs could regulate the p53 pathway via different mechanisms within different cellular compartments following different stress. However, whether p53 is regulated by DUBs other than USP family members is previously unknown. We recently identified that the OTU domain-containing ubiquitin aldehyde-binding proteins 1 (Otubain 1, Otub1 thereafter), an OTU family DUB, controls p53 stability and activity via a novel non-canonical mechanism[110].
OTUB1: A UNIQUE MEMBER OF OTU DUB FAMILY
Otub1 was identified along with its close homolog Otub2 by affinity purification using the DUB-specific inhibitor, Ub aldehyde[111]. Subsequent studies, including our own, revealed that Otub1 possesses in vitro deubiquitinating enzyme activity preferentially towards K48-linked polyubiquitin chains[110,112,113]. Like other cysteine proteases, Otub1 contains a catalytic triad consisting of Cys (C) 91, His (H) 265, and Asp (D) 268[112]. However, crystal structure studies demonstrated that Otub1 possesses unique structure features wherein H265 is located distantly from the catalytic C91 and D268 and the access of C91 to ubiquitin is blocked by Glu (E) 214 residue, forming a conformation incompatible with catalysis by typical cysteine proteases[112], implying that the activity of Otub1 may be highly regulated in cells and its activation may be subjected to conformational change (See below). Otub1 is ubiquitously expressed in tested human tissues. A longer isoform called Otub1 ARF (alternative reading frame)-1, resulting from alternative splicing and start codon, is predominantly expressed in peripheral blood mononuclear cells, lymph nodes, spleen, and the tonsils[114]. The function of Otub1 ARF-1 is thought to antagonize the function of Otub1 in cells[114].
Functionally, Otub1 has been implicated in the regulation of immune response, estrogen signaling, DNA damage response, as well as pathogen biology. Soares et al[114] first reported that Otub1 regulates CD4+ T cell clonal anergy by enhancing degradation of the ubiquitin ligase called GRAIL (gene related to anergy in lymphocytes) and promoting interleukin 2 production following antigenic stimulation, whereas the Otub1 ARF-1 has an opposite effect. Interestingly, the effect of Otub1 does not depend on its catalytic activity. As a matter of fact, the role of Otub1 in degrading GRAIL is opposite to its predicted role as a DUB[114]. A possible explanation is that Otub1 forms a ternary complex with GRAIL and USP8, another USP family DUB, thereby suppressing the deubiquitination of GRAIL by USP8. In this case, Otub1 may act as an ubiquitin editing protease[114]. Li et al[115] reported that Otub1 (and Otub2) mediate virus-induced deubiquitination of TNF receptor-associated factor 3 (TRAF3) and TRAF6, two ubiquitin ligases required for virus-induced Interferon regulatory factor 3 (IRF3) and NF-kB activation, leading to the inhibition of viral-induced production of INFβ. However, whether this effect requires the DUB enzymatic activity of Otub1 is not clear[115]. Further, Otub1 has recently been shown to enhance TGFβ signaling by inhibiting ubiquitination and degradation of SMAD2/3[116]. Otub1 also plays a role in pathogen invasion of the host cells. The Yersinia-encoded virulence factor YpkA interacts with and phosphorylates Otub1[117] and recruits the small GTPase RhoA, leading to the stabilization of the active RhoA[118]. Consequently, overexpression of wild-type, but not the C91S mutant, Otub1 increased the susceptibility of host cells to the Yersinia evasion[118]. Otub1 has been shown to deubiquitinate and stabilize ERα in chromatin[119], albeit this stabilization results in the inhibition of ERα-mediated transcription. Adding to the complexity, the catalytic mutant Otub1, C91S in which the catalytic C91 is mutated to S, did not abolish Otub1-mediated suppression of ERα activity[119]. Otub1 has been shown to inhibit DNA-damage-induced chromatin ubiquitination, which is also independent of its DUB activity. Instead, Otub1 suppresses RNF168-dependent chromatin polyubiquitination by binding to and inhibiting the RNF168 cognate E2 enzyme UBC13[120]. Recently, Otub1 has been shown to regulate apoptosis by deubiquitinating the cellular inhibitor of apoptosis (c-IAP1)[121].
Together, Otub1 has been implicated in multiple biological processes. In most cases, the effects of Otub1 do not require its DUB activity, such as the regulation of DNA damage-induced chromatin ubiquitination[120], T-cell anergy[114], ERα[119], and SMAD2/3[116], implying a unique model of ubiquitination regulation by a DUB: suppression of the ubiquitin-conjugating enzyme (E2) (see below). Because of this and the fact that it is expressed in most tissues, Otub1 may have a broad function in cells.
OTUB1 IS A NOVEL POSITIVE P53 REGULATOR
We recently found that Otub1 positively regulates the stability and activity of p53[110]. Overexpression of Otub1, but not its close homolog Otub2, markedly stabilizes and activates p53 and induces p53-dependent apoptosis and cell growth inhibition. Interestingly, Otub1 regulation of p53 does not require its catalytic activity, as mutating C91 to either A or S did not abolish the activity of Otub1 to block MDM2-mediated p53 ubiquitination and degradation, to stabilize and activate p53, and to induce p53-dependent cell growth inhibition[110]. Mechanistically, Otub1 suppresses MDM2-mediated p53 ubiquitination by binding to and inhibiting the MDM2 cognate E2 enzyme UbcH5s[110]. This is consistent with the non-canonical role for Otub1 in suppressing DNA damage-induced chromatin ubiquitination by inhibiting UBC13[120]. Therefore, our study further supports that the suppression of substrate ubiquitination through inhibiting cognate E2s by Otub1 represents a unique noncanonical mode of DUB regulation compared to classical cysteine proteases and this may be a general mechanism for Otub1 to regulate the substrate protein ubiquitination and stability.
Consistent with the noncanonical mode of regulation, mutating C91 to either A or S did not abolish the activity of Otub1 to bind to and suppress UbcH5[110]. However, a point mutation of Asp 88 to Ala (Otub1D88A) abolished the function of Otub1 to suppress p53 ubiquitination and degradation and this mutant interacts with p53 stronger than wild-type Otub1, indicating this mutation might create a dominant-negative effect. D88 is located closely to the donor ubiquitin-binding surface and thus its mutation would affect the binding of Otub1 to donor ubiquitin conjugated to UbcH5. Although D88 is not located directly in the E2 binding surface, our experimental data revealed that this mutation clearly disrupted the Otub1-E2 interaction in cells[110]. This might be due to the overall structure change after D88 mutation. Supporting this conformational change is that D88A mutant also results in the loss of Otub1’s DUB activity.
Our functional studies of the endogenous Otub1 suggest that Otub1 plays an important role in p53 stabilization and activation following DNA damage induced by diverse agents. This is consistent, but not completely, with the observation that Otub1 suppresses DNA damage-induced chromatin ubiquitination, thereby suppressing DNA repair pathway[120]. One explanation is that upon DNA damage, Otub1 might target UbcH5-MDM2 to stabilize p53, while it may dissociate from the RNF168-Ubc13 complex, allowing RNF168 to catalyze K63-linked chromatin ubiquitination and subsequent DNA repair response. Whether DNA damage-induced posttranslational modification plays a role in this functional switch remains unclear. However, phosphorylation of Otub1 has been observed at several residues such as T134. Further, it has been shown that the phosphorylation mimicking Otub1 mutant T134E, but not T134A, failed to rescue the DNA damage response in Otub1-depleted cells[122]. Thus it is interesting to examine the signaling pathways involved in the phosphorylation of Otub1 and how this phosphorylation plays a role in regulating Otub1 function in response to DNA damage stress.
MECHANISTIC INSIGHTS INTO THE NON-CANONICAL SUPPRESSION OF E2 BY OTUB1
Recent biochemical and structural studies have shed a light on how Otub1 suppresses E2s[122-124]. It has been shown that Otub1 preferentially binds to ubiquitin-charged E2[120,122]. Otub1 contains two ubiquitin-binding motifs: a distal site that binds to free ubiquitin and a proximal site that binds to donor ubiquitin conjugated to the active site of an E2 (e.g., Ubc13 or UbcH5). The structure of two ubiquitin binding to Otub1 is reminiscent of that of K48-linked di-ubiquitin[122]. Interestingly, the binding of a free ubiquitin to the distal site allosterically causes the conformational change of Otub1, allowing the formation of a N-terminal ubiquitin-binding helix where the E2-charged donor ubiquitin then binds[122,124]. Consequently, this binding limits the donor ubiquitin interaction with the backside of another E2 and the attack on the thioester bond by an acceptor ubiquitin, a step important for ubiquitin transfer[122,124]. On the other hand, Otub1 also makes contacts with E2 and the Otub1-binding surface in E2 (UbcH5 and Ubc13) overlaps with the E3-binding surface. Thus this Otub1-E2 interaction may also attenuate the E2-E3 engagement[122,124]. Collectively, Otub1 is a potential inhibitor of the E2 enzymes. Further supporting this notion, Otub1 has recently been shown to be a major DUB that interacts with the D and E classes of E2 as well as UbcE2N[125]. Thus disruption of the Otub1-E2 interaction or donor ubiquitin-Otub1 interaction would theoretically abolish Otub1’s activity to suppress E2. This could distinguish Otub1’s E2 suppressing activity from its DUB enzyme activity. Indeed, several mutants involved in the E2-contacting surface of the Otub1, such as F133A, T134R, F138A, have been shown to lack the E2-suppressing activity but retain the DUB activity[122,124]. Therefore, it is interesting to examine whether these mutants could fail to stabilize and activate p53 in cells. On another note, we recently found that Otub1 can be monoubiquitinated by UbcH5 and this monoubiquitination in turn plays a critical role in the Otub1’s E2 suppressing activity. We further found that UbcH5 preferentially binds to monoubiquitinated Otub1, through the ubiquitin interaction with the backside ubiquitin-interacting surface of E2[126]. This binding could potentially disrupt the formation of self-assembled ubiquitin-charged UbcH5 (UbcH5-Ub) conjugates that is critical for ubiquitin transfer, polyubiquitin chain formation and efficient polyubiquitination of substrates[127,128], suggesting another novel mechanism of Otub1 suppression of E2.
CONCLUSION
Recent studies have convincingly demonstrated Otub1 as a unique DUB that executes diverse biology functions by non-canonically suppressing E2 enzymes. Therefore it is expected that Otub1 may play broad functions in cells. One question would be how these broad functions coordinate with each other in cells. We also do not know how Otub1’s activity is regulated in cells. Interestingly, a recent observation showed that Otub1 DUB activity can be regulated by UbcH5, which stimulates the binding of the Lys48-linked polyubiquitin substrate by stabilizing the folding of the N-terminal ubiquitin-binding helix of Otub1, thereby promoting its deubiquitinating enzyme activity[129]. It is interesting to know how these mutually regulatory functions are controlled in cells. It is also important to test how Otub1’s activity and levels are regulated in cells under physiologic and stress conditions. As Otub1 is a potent activator of p53[110] and plays a role in DNA damage repair[120], Otub1 may act as a tumor suppressor. Thus it is important to determine whether Otub1 is deregulated in human cancers. Gene targeting in mice could provide further information regarding the function of Otub1 and whether Otub1 indeed possesses tumor suppression function in vivo. Further characterization of mechanistic insights into the Otub1 suppression of E2 could also be useful for developing strategies that target the E2 enzymes for cancer therapy, e.g., small molecule compounds that resemble Otub1 interaction with E2.
Together, p53 is ubiquitinated by MDM2/MDMX and several other E3s whereas it is deubiquitinated by a number of DUBs, including USP7, USP10, USP29 and USP42. One obvious question is how these multiple DUBs are coordinated to ensure the tight, precise, and dynamic control of p53 stability and activity. Different DUBs may regulate the p53 pathway in response to different cellular stress (e.g., USP29 deubiquitinates p53 in response to oxidative stress[104] whereas USP10 deubiquitinates p53 following DNA damage[102]). Different DUBs may also regulate p53 in different cellular compartments (e.g., USP7 regulates p53 in the nucleus whereas Otub1 regulates p53 in the cytoplasm[110] and USP10 relocates from the cytoplasm to the nucleus to regulate p53 in response to DNA damage[102]). It is interesting to examine whether different DUBs may cooperate with each other to synergistically regulate p53 stability and activity in future studies.
Nevertheless, efforts have been made towards targeting the ubiquitin-proteasome system (UPS) for reactivating p53 in cancer therapy. For example, compounds have been developed to target the p53-MDM2 interaction such as Nutlin-3s[130], the p53-MDMX interaction such as WK298[131], or both such as RO-2443[132]. Targeting DUBs has promising potential as well. For example, the cyano-indenopyrazine derivatives small molecule compounds HBX 41108, HBX 19818, and HBX 28258[133] and P22077[134] were discovered as USP7 inhibitors. For further details about targeting the UPS for cancer therapy, please refer our recent review[135]. Future directions will aim to discover more potent and specific DUB inhibitors that can be used for cancer treatment.
ACKNOWLEDGEMENTS
Views and opinions of, and endorsements by, the author(s) do not reflect those of the United States Army or the Department of Defense.
Footnotes
Supported by NIH/NCI, No. R00 CA127134 and No. R01 CA160474; and a Department of Defense, No. W81XWH-10-1-1029, to Dai MS; and A Grant from Medical Research Foundation (MRF) of Oregon, to Sun XX
P- Reviewers: Chui YL, Jia J, Wang Y, Yang JH S- Editor: Wen LL L- Editor: A E- Editor: Lu YJ
References
- 1.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 4.Hollstein M, Rice K, Greenblatt MS, Soussi T, Fuchs R, Sørlie T, Hovig E, Smith-Sørensen B, Montesano R, Harris CC. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 5.Soussi T, Dehouche K, Béroud C. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum Mutat. 2000;15:105–113. doi: 10.1002/(SICI)1098-1004(200001)15:1<105::AID-HUMU19>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 6.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2:475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 8.Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9:831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 9.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 11.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 12.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 14.Picksley SM, Lane DP. The p53-mdm2 autoregulatory feedback loop: a paradigm for the regulation of growth control by p53? Bioessays. 1993;15:689–690. doi: 10.1002/bies.950151008. [DOI] [PubMed] [Google Scholar]
- 15.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–8951. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 16.26 Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 21.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 22.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 23.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 24.Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindström MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Bueso-Ramos CE, Yang Y, deLeon E, McCown P, Stass SA, Albitar M. The human MDM-2 oncogene is overexpressed in leukemias. Blood. 1993;82:2617–2623. [PubMed] [Google Scholar]
- 26.Cordon-Cardo C, Latres E, Drobnjak M, Oliva MR, Pollack D, Woodruff JM, Marechal V, Chen J, Brennan MF, Levine AJ. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 27.Dworakowska D, Jassem E, Jassem J, Peters B, Dziadziuszko R, Zylicz M, Jakóbkiewicz-Banecka J, Kobierska-Gulida G, Szymanowska A, Skokowski J, et al. MDM2 gene amplification: a new independent factor of adverse prognosis in non-small cell lung cancer (NSCLC) Lung Cancer. 2004;43:285–295. doi: 10.1016/j.lungcan.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–3459. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deb SP. Cell cycle regulatory functions of the human oncoprotein MDM2. Mol Cancer Res. 2003;1:1009–1016. [PubMed] [Google Scholar]
- 30.Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. MDMX: a novel p53-binding protein with some functional properties of MDM2. EMBO J. 1996;15:5349–5357. [PMC free article] [PubMed] [Google Scholar]
- 31.Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP, Saville MK, Jochemsen AG. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2001;2:1029–1034. doi: 10.1093/embo-reports/kve227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson MW, Berberich SJ. MdmX protects p53 from Mdm2-mediated degradation. Mol Cell Biol. 2000;20:1001–1007. doi: 10.1128/mcb.20.3.1001-1007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J, Kawai H, Nie L, Kitao H, Wiederschain D, Jochemsen AG, Parant J, Lozano G, Yuan ZM. Mutual dependence of MDM2 and MDMX in their functional inactivation of p53. J Biol Chem. 2002;277:19251–19254. doi: 10.1074/jbc.C200150200. [DOI] [PubMed] [Google Scholar]
- 34.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 35.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett. 1999;447:5–9. doi: 10.1016/s0014-5793(99)00254-9. [DOI] [PubMed] [Google Scholar]
- 36.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, Prives C. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biderman L, Poyurovsky MV, Assia Y, Manley JL, Prives C. MdmX is required for p53 interaction with and full induction of the Mdm2 promoter after cellular stress. Mol Cell Biol. 2012;32:1214–1225. doi: 10.1128/MCB.06150-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan Y, Chen J. MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol. 2003;23:5113–5121. doi: 10.1128/MCB.23.15.5113-5121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Graaf P, Little NA, Ramos YF, Meulmeester E, Letteboer SJ, Jochemsen AG. Hdmx protein stability is regulated by the ubiquitin ligase activity of Mdm2. J Biol Chem. 2003;278:38315–38324. doi: 10.1074/jbc.M213034200. [DOI] [PubMed] [Google Scholar]
- 42.Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23:4939–4947. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22:5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 45.Finch RA, Donoviel DB, Potter D, Shi M, Fan A, Freed DD, Wang CY, Zambrowicz BP, Ramirez-Solis R, Sands AT, et al. mdmx is a negative regulator of p53 activity in vivo. Cancer Res. 2002;62:3221–3225. [PubMed] [Google Scholar]
- 46.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, et al. Amplification of Mdmx (or Mdm4) directly contributes to tumor formation by inhibiting p53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramos YF, Stad R, Attema J, Peltenburg LT, van der Eb AJ, Jochemsen AG. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type p53. Cancer Res. 2001;61:1839–1842. [PubMed] [Google Scholar]
- 48.Riemenschneider MJ, Büschges R, Wolter M, Reifenberger J, Boström J, Kraus JA, Schlegel U, Reifenberger G. Amplification and overexpression of the MDM4 (MDMX) gene from 1q32 in a subset of malignant gliomas without TP53 mutation or MDM2 amplification. Cancer Res. 1999;59:6091–6096. [PubMed] [Google Scholar]
- 49.Riemenschneider MJ, Knobbe CB, Reifenberger G. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer. 2003;104:752–757. doi: 10.1002/ijc.11023. [DOI] [PubMed] [Google Scholar]
- 50.Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S, et al. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci USA. 2011;108:12001–12006. doi: 10.1073/pnas.1102309108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pant V, Xiong S, Iwakuma T, Quintás-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA. 2011;108:11995–12000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 53.Wang X, Wang J, Jiang X. MdmX protein is essential for Mdm2 protein-mediated p53 polyubiquitination. J Biol Chem. 2011;286:23725–23734. doi: 10.1074/jbc.M110.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 55.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 56.Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24:3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H. 14-3-3gamma binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J. 2006;25:1207–1218. doi: 10.1038/sj.emboj.7601010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LeBron C, Chen L, Gilkes DM, Chen J. Regulation of MDMX nuclear import and degradation by Chk2 and 14-3-3. EMBO J. 2006;25:1196–1206. doi: 10.1038/sj.emboj.7601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine JC, et al. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol. 2005;25:9608–9620. doi: 10.1128/MCB.25.21.9608-9620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereg Y, Lam S, Teunisse A, Biton S, Meulmeester E, Mittelman L, Buscemi G, Okamoto K, Taya Y, Shiloh Y, et al. Differential roles of ATM- and Chk2-mediated phosphorylations of Hdmx in response to DNA damage. Mol Cell Biol. 2006;26:6819–6831. doi: 10.1128/MCB.00562-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 65.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Honda R, Yasuda H. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol. 2001;3:445–452. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- 68.Midgley CA, Desterro JM, Saville MK, Howard S, Sparks A, Hay RT, Lane DP. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- 69.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23:2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26:5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 71.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 72.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24:7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 75.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32:180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun XX, DeVine T, Challagundla KB, Dai MS. Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J Biol Chem. 2011;286:22730–22741. doi: 10.1074/jbc.M111.223651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xiong X, Zhao Y, He H, Sun Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30:1798–1811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Wang W, Wang H, Wang MH, Xu W, Zhang R. Identification of ribosomal protein S25 (RPS25)-MDM2-p53 regulatory feedback loop. Oncogene. 2013;32:2782–2791. doi: 10.1038/onc.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, Jin C, Chen H, Zhang L, Yang X, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010;38:6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35:316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bai D, Zhang J, Xiao W, Zheng X. Regulation of the HDM2-p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic Acids Res. 2014;42:1799–1811. doi: 10.1093/nar/gkt971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cui D, Li L, Lou H, Sun H, Ngai SM, Shao G, Tang J. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2014;33:2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- 85.Daftuar L, Zhu Y, Jacq X, Prives C. Ribosomal proteins RPL37, RPS15 and RPS20 regulate the Mdm2-p53-MdmX network. PLoS One. 2013;8:e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin Y, Zeng SX, Dai MS, Yang XJ, Lu H. MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. J Biol Chem. 2002;277:30838–30843. doi: 10.1074/jbc.M204078200. [DOI] [PubMed] [Google Scholar]
- 88.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 89.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 92.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 93.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 94.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dai MS, Jin Y, Gallegos JR, Lu H. Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia. 2006;8:630–644. doi: 10.1593/neo.06334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 97.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 98.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 99.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 100.Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 101.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1 p following 486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 102.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 2011;30:4921–4930. doi: 10.1038/emboj.2011.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, Dundr M, Levens D. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30:846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- 107.Zhang X, Berger FG, Yang J, Lu X. USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 2011;30:2177–2189. doi: 10.1038/emboj.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benassi B, Flavin R, Marchionni L, Zanata S, Pan Y, Chowdhury D, Marani M, Strano S, Muti P, Blandino G, et al. MYC is activated by USP2a-mediated modulation of microRNAs in prostate cancer. Cancer Discov. 2012;2:236–247. doi: 10.1158/2159-8290.CD-11-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Priolo C, Tang D, Brahamandan M, Benassi B, Sicinska E, Ogino S, Farsetti A, Porrello A, Finn S, Zimmermann J, et al. The isopeptidase USP2a protects human prostate cancer from apoptosis. Cancer Res. 2006;66:8625–8632. doi: 10.1158/0008-5472.CAN-06-1374. [DOI] [PubMed] [Google Scholar]
- 110.Sun XX, Challagundla KB, Dai MS. Positive regulation of p53 stability and activity by the deubiquitinating enzyme Otubain 1. EMBO J. 2012;31:576–592. doi: 10.1038/emboj.2011.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4:517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Edelmann MJ, Iphöfer A, Akutsu M, Altun M, di Gleria K, Kramer HB, Fiebiger E, Dhe-Paganon S, Kessler BM. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 113.Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Soares L, Seroogy C, Skrenta H, Anandasabapathy N, Lovelace P, Chung CD, Engleman E, Fathman CG. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat Immunol. 2004;5:45–54. doi: 10.1038/ni1017. [DOI] [PubMed] [Google Scholar]
- 115.Li S, Zheng H, Mao AP, Zhong B, Li Y, Liu Y, Gao Y, Ran Y, Tien P, Shu HB. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J Biol Chem. 2010;285:4291–4297. doi: 10.1074/jbc.M109.074971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP. OTUB1 enhances TGFβ signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun. 2013;4:2519. doi: 10.1038/ncomms3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Juris SJ, Shah K, Shokat K, Dixon JE, Vacratsis PO. Identification of otubain 1 as a novel substrate for the Yersinia protein kinase using chemical genetics and mass spectrometry. FEBS Lett. 2006;580:179–183. doi: 10.1016/j.febslet.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 118.Edelmann MJ, Kramer HB, Altun M, Kessler BM. Post-translational modification of the deubiquitinating enzyme otubain 1 modulates active RhoA levels and susceptibility to Yersinia invasion. FEBS J. 2010;277:2515–2530. doi: 10.1111/j.1742-4658.2010.07665.x. [DOI] [PubMed] [Google Scholar]
- 119.Stanisić V, Malovannaya A, Qin J, Lonard DM, O'Malley BW. OTU Domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1) deubiquitinates estrogen receptor (ER) alpha and affects ERalpha transcriptional activity. J Biol Chem. 2009;284:16135–16145. doi: 10.1074/jbc.M109.007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 121.Goncharov T, Niessen K, de Almagro MC, Izrael-Tomasevic A, Fedorova AV, Varfolomeev E, Arnott D, Deshayes K, Kirkpatrick DS, Vucic D. OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 2013;32:1103–1114. doi: 10.1038/emboj.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Juang YC, Landry MC, Sanches M, Vittal V, Leung CC, Ceccarelli DF, Mateo AR, Pruneda JN, Mao DY, Szilard RK, et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sato Y, Yamagata A, Goto-Ito S, Kubota K, Miyamoto R, Nakada S, Fukai S. Molecular basis of Lys-63-linked polyubiquitination inhibition by the interaction between human deubiquitinating enzyme OTUB1 and ubiquitin-conjugating enzyme UBC13. J Biol Chem. 2012;287:25860–25868. doi: 10.1074/jbc.M112.364752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zulkifle N. Systemmatic yeast two-hybrid analysis of human E2 Ubiquitin-conjugating enzyme and deubiquitin [DUB] protein interaction. Int J Biol Chem. 2013;7:14. [Google Scholar]
- 126.Li Y, Sun XX, Elferich J, Shinde U, David LL, Dai MS. Monoubiquitination is critical for ovarian tumor domain-containing ubiquitin aldehyde binding protein 1 (Otub1) to suppress UbcH5 enzyme and stabilize p53 protein. J Biol Chem. 2014;289:5097–5108. doi: 10.1074/jbc.M113.533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 128.Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 129.Wiener R, DiBello AT, Lombardi PM, Guzzo CM, Zhang X, Matunis MJ, Wolberger C. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol. 2013;20:1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 131.Popowicz GM, Czarna A, Wolf S, Wang K, Wang W, Dömling A, Holak TA. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery. Cell Cycle. 2010;9:1104–1111. doi: 10.4161/cc.9.6.10956. [DOI] [PubMed] [Google Scholar]
- 132.Graves B, Thompson T, Xia M, Janson C, Lukacs C, Deo D, Di Lello P, Fry D, Garvie C, Huang KS, et al. Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci USA. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reverdy C, Conrath S, Lopez R, Planquette C, Atmanene C, Collura V, Harpon J, Battaglia V, Vivat V, Sippl W, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chem Biol. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 134.Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KG, Konietzny R, Fischer R, Kogan E, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 135.Devine T, Dai MS. Targeting the ubiquitin-mediated proteasome degradation of p53 for cancer therapy. Curr Pharm Des. 2013;19:3248–3262. doi: 10.2174/1381612811319180009. [DOI] [PMC free article] [PubMed] [Google Scholar]


