Protein degradation in eukaryotic cells is mediated mainly by the ubiquitin (Ub)-proteasome system. One fundamental question in this field is about specific features in a protein that trigger its ubiquitylation and subsequent proteasomal degradation? In other words, the problem of degradation signals (degrons).1
The first degradation signals to be identified were N-degrons, which comprised destabilizing N-terminal residues of protein substrates.2 Detailed analysis of the N-degrons gave rise to the N-end rule, which enables the nature of N-terminal residue and its modifications to dictate the in vivo half-life of a protein. In eukaryotes, the N-end rule pathway comprises 2 major branches, the Arg/N-end rule pathway and the Ac/N-end rule pathway.3,4
The Arg/N-end rule pathway was the first specific pathway of the Ub system to be discovered.2 This pathway involves, in particular, the N-terminal arginylation of a protein and N-recognins (E3 Ub ligases in eukaryotes). The E3 N-recognins specifically target unacetylated basic (Arg, Lys, His) or bulky hydrophobic (Leu, Phe, Tyr, Trp, Ile) N-terminal residues. The Arg/N-end rule pathway has been implicated in a wide range of functions, including the sensing of small peptides or nitric oxide, protein quality control, DNA repair, meiosis, cardiovascular development, etc.3,4
The other branch, called the Ac/N-end rule pathway, targets for degradation proteins through their Nα-terminally acetylated (Nt-acetylated) residues. In eukaryotes, Nt-acetylation is one of the most common protein modifications, observed in up to 90% of human proteins.5 Despite its abundance and pervasiveness, the biological role of Nt-acetylation in a majority of proteins had been largely unknown. Nt-acetylation was mostly thought to protect a protein from degradation. In contrast, it was discovered in 2010 that Nt-acetylation creates specific degrons for the Ac/N-end rule pathway.6
This finding has shown that the Nt-acetyl group at the N-termini of many cellular proteins is an entirely new class of degradation signals, termed Ac/N-degrons. Nt-acetylation occurs largely cotranslationally and apparently irreversibly.5 In other words, nearly every protein obtains a specific Ac/N-degron from the moment of its birth. Thus, formed Ac/N-degrons were therefore suggested to be conditionally active, only if they were accessible to Ac/N-recognins, when an Ac/N-degron is not masked through rapid intramolecular folding of a protein or intermolecular assembly with other proteins.6 In accordance with this assumption, the Ac/N-degron of Cog1, a subunit of the Golgi-localized COG complex, was shown to be sterically shielded by its interactions with Cog2, Cog3, and Cog4 in the COG complex, and thereby to monitor and control the levels of Cog1 in a cell, relative to those of its binding partners.7 This study provided not only the first example of physiological functions of the Ac/N-end rule pathway, but also a simple scheme for the control of protein quality and stoichiometries.
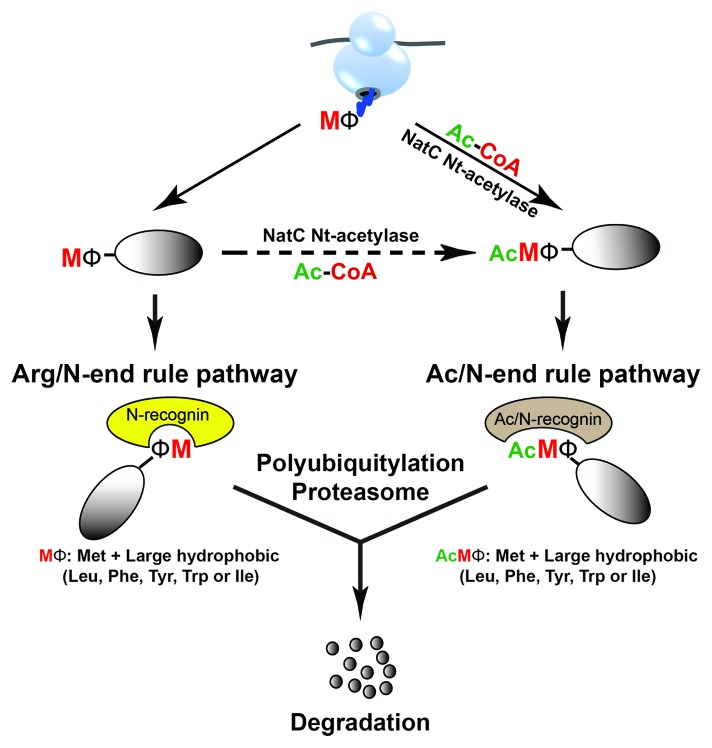
The conditionality of the built-in Ac/N-degrons in the broad range of substrates opened up a new question: might there be a proteolytic system to process non-Nt-acetylated proteins by targeting unmodified “stabilizing” N-terminal residues, which were previously classified in the Arg/N-end rule pathway?3,4 To address this unanswered question, we paid attention to N-terminal methionine (Met), because it is present in virtually all nascent polypeptides. We found that Saccharomyces cerevisiae Ubr1 E3 N-recognin and its mouse counterparts UBR1 and UBR2 of the Arg/N-end rule pathway recognized proteins with unacetylated N-terminal Met, if it is followed by a large second-position Ф residue (Leu, Ile, Phe, Tyr, Trp)8 (Fig. 1). We further demonstrated that Ubr1 mediated degradation of not only quasi-randomly chosen natural proteins (Msn4, Sry1, Arl3, Pre5), but also misfolded proteins by targeting their Met-Ф residues. Very importantly, the discovery of Met-Ф degrons greatly expands the substrate range of the Arg/N-end rule pathway, because approximately 15% of all DNA-encoded proteins, in both yeast and mammals, belong to this class of proteins. Furthermore, our finding reveals mechanical and functional cross-talks between the Ac/N-end rule pathway and the Arg/N-end rule pathway. Specifically, proteins with Met-Ф N termini, if Nt-acetylated, can be removed by the Ac/N-end rule pathway or, if they escape Nt-acetylation, by the Arg/N-end rule pathway8 (Fig. 1).
Figure 1. Crosstalk between the Arg/N-end rule pathway and the Ac/N-end rule pathway. Nearly all nascent proteins bear N-terminal Met due to the AUG start codon. The Arg/N-end rule pathway targets for degradation proteins with unacetylated N-terminal Met if it is followed by a large hydrophobic Ф residue (Leu, Ile, Phe, Tyr, Trp) at position 2. Nt-acetylase catalyzes Nt-acetylation of the Met-Ф proteins either cotranslationally or posttranslationally, and thereby creates the Ac/Met-Ф degrons for the Ac/N-end rule pathway.
The discovery that N-terminal Met acts as a degron suggests that nearly all 20 amino acids are likely to be destabilizing N-terminal residues according to specific sequence context and their states of Nt-acetylation.8 The dual targeting mechanism of the 2 branches of the N-end rule pathway may be true for conditional degradation of many full-length proteins or proteolytic fragments in a cell. The current and further understandings of the N-end rule pathway may, therefore, account for extensive degradation of most cellular proteins by the ubiquitin–proteasome system, including relatively long-lived proteins.
Kim HK, et al. Cell. 2014;156:158–69. doi: 10.1016/j.cell.2013.11.031.
References
- 1.Ravid T, et al. Nat Rev Mol Cell Biol. 2008;9:679–90. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmair A, et al. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. Protein Sci. 2011;20:1298–345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sriram SM, et al. Nat Rev Mol Cell Biol. 2011;12:735–47. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 5.Starheim KK, et al. Trends Biochem Sci. 2012;37:152–61. doi: 10.1016/j.tibs.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Hwang CS, et al. Science. 2010;327:973–7. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shemorry A, et al. Mol Cell. 2013;50:540–51. doi: 10.1016/j.molcel.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HK, et al. Cell. 2014;156:158–69. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]