Abstract
Depletion of spindle assembly checkpoint (SAC) genes in Drosophila epithelial tissues leads to JNK-dependent programmed cell death and additional blockade of the apoptotic program drives tumorigenesis. A recent report proposes that chromosomal instability (CIN) is not the driving force in the tumorigenic response of the SAC-deficient tissue, and that checkpoint proteins exert a SAC-independent tumor suppressor role. This notion is based on observations that the depletion of CENP-E levels or prevention of Bub3 from binding to the kinetochore in Drosophila tissues unable to activate the apoptotic program induces CIN but does not cause hyperproliferation. Here we re-examined this proposal. In contrast to the previous report, we observed that depletion of CENP-E or Nsl1—the latter mediating kinetochore targeting of Bub3—in epithelial tissues unable to activate the apoptotic program induces significant levels of aneuploidy and drives tumor-like growth. The induction of the JNK transcriptional targets Wingless, a mitogenic molecule, and MMP1, a matrix metaloproteinase 1 involved in basement membrane degradation was also observed in these tumors. An identical response of the tissue was previously detected upon depletion of several SAC genes or genes involved in spindle assembly, chromatin condensation, and cytokinesis, all of which have been described to cause CIN. All together, these results reinforce the role of CIN in driving tumorigenesis in Drosophila epithelial tissues and question the proposed SAC-independent roles of checkpoint proteins in suppressing tumorigenesis. Differences in aneuploidy rates might explain the discrepancy between the previous report and our results.
Keywords: CIN, mitotic checkpoint, tumorigenesis, JNK
Introduction
A recent systematic analysis of 43 205 human tumors found that 68% of solid tumors show aneuploidy,1 an abnormal number of chromosomes. While the impact of chromosomal structural changes on cancer progression is widely accepted, the effect of chromosome numerical changes is highly debated (reviewed in ref. 2). Several mouse models mutant for or overexpressing mitotic checkpoint genes have been generated to address the role of chromosomal instability (CIN) in tumor progression. Many of these CIN models display increased susceptibility to spontaneous tumorigenesis in lung epithelial cells (reviewed in ref. 3), enhanced tumor formation,4 tumor relapse,5 and induced loss of heterozygosity of tumor suppressor genes.6 These observations thus reinforce the role of CIN in cancer development. However, aneuploidy has been shown to inhibit tumorigenesis in tissues prone to tumor formation,7 and high rates of CIN exert a tumor suppressor function.8
In the last few years, the contribution of CIN to tumorigenesis has also been analyzed in Drosophila cancer models. CIN per se does not induce tumorigenesis in brain9 or epithelial tissues.10 Since aneuploidy can be detrimental for cells, additional mutations might contribute to the survival of these cells and to CIN-induced tumorigenesis. Indeed, CIN leads to p53-dependent apoptosis of mammalian cells,11,12 and mutations in the p53 tumor suppressor gene increase the frequency of spontaneous tumorigenesis observed in CIN mouse models.11 In Drosophila epithelial tissues, CIN leads to Drosophila p53 (dp53)-independent apoptosis and maintenance of aneuploid cells in the tissue through additional blockade of PCD causes tissue overgrowth, basement membrane (BM) degradation, and host invasiveness.10 The tumorigenic response of the tissue relies on aneuploid cells delaminating from the epithelium and activating a JNK-dependent transcriptional program that triggers the expression of the Matrix Metaloproteinase 1 (MMP1) and the mitogenic molecule Wingless (Wg). Expression of Wg in the delaminating cell population plays a fundamental role in the growth of the tissue.
A recent report in Drosophila proposes, however, that CIN is not the driving force in the tumorigenic response of the SAC-deficient epithelial tissue upon additional blockade of the PCD pathway.13 This notion is based on observations that depletion of CENP-E (which mediates precise interactions between kinetochores and microtubules of the mitotic spindle14) or Nsl1 (which targets Bub3 to the kinetochore15) induces CIN but does not cause hyperproliferation.13 The authors suggest a SAC-independent tumor suppressor role of Bub3, on the basis that several mitotic checkpoint proteins can play additional roles such as transcriptional regulation16 or nuclear import.17 Since these results contrast with the findings that the depletion of several SAC genes or genes involved in spindle assembly, chromatin condensation, and cytokinesis induces CIN and tumorigenesis upon PCD blockade,10,13 we have revisited this proposal. We have used 2 different Gal4 drivers and several dsRNA forms to deplete CENP-E or Nsl1 in epithelial tissues unable to activate the apoptotic program. Depletion of any of these 2 genes leads to significant levels of aneuploidy and drives tumor-like overgrowth. This overgrowth is accompanied by the expression of MMP1 and Wg in delaminating cells both in larval tissues as well as in allograft transplants. These results reinforce the tumorigenic role of CIN in Drosophila epithelial tissues and question the proposed SAC-independent roles of checkpoint proteins in suppressing tumorigenesis. We propose that the discrepancy between previous results13 and those reported in this work is attributable to the levels of aneuploidy induced in the tissue.
Results
The Drosophila primordia of adult wings (wing imaginal discs) are epithelial monolayers that actively proliferate during larval development. These structures have proved useful as model systems to elucidate the molecular mechanisms underlying tumorigenic growth,18,19 including the contribution of CIN to tumorigenesis.10 The depletion of genes involved in the spindle assembly checkpoint (bub3, rough deal), spindle assembly (abnormal spindle [asp]), chromatin condensation (orc2), and cytokinesis (diaphanous [dia])—all known to recapitulate the genomic defects most frequently associated with human cancer, including chromosome rearrangements and aneuploidy9—induce CIN and PCD in wing imaginal disc cells.10,13 Additional blockade of the PCD pathway, by expressing the baculovirus protein p35, known to bind and repress the effector caspases DrICE and Dcp-1,20 or by the generation of cells mutant for the pro-apoptotic genes hid, reaper, and grim, leads to tissue overgrowth.10 Similarly, depletion of CENP-E or Nsl1 has also been shown to induce CIN and PCD in wing disc cells.13 Surprisingly, however, blocking apoptosis in these tissues was reported to not causing overgrowth,13 questioning the role of CIN in driving tumorigenesis.13
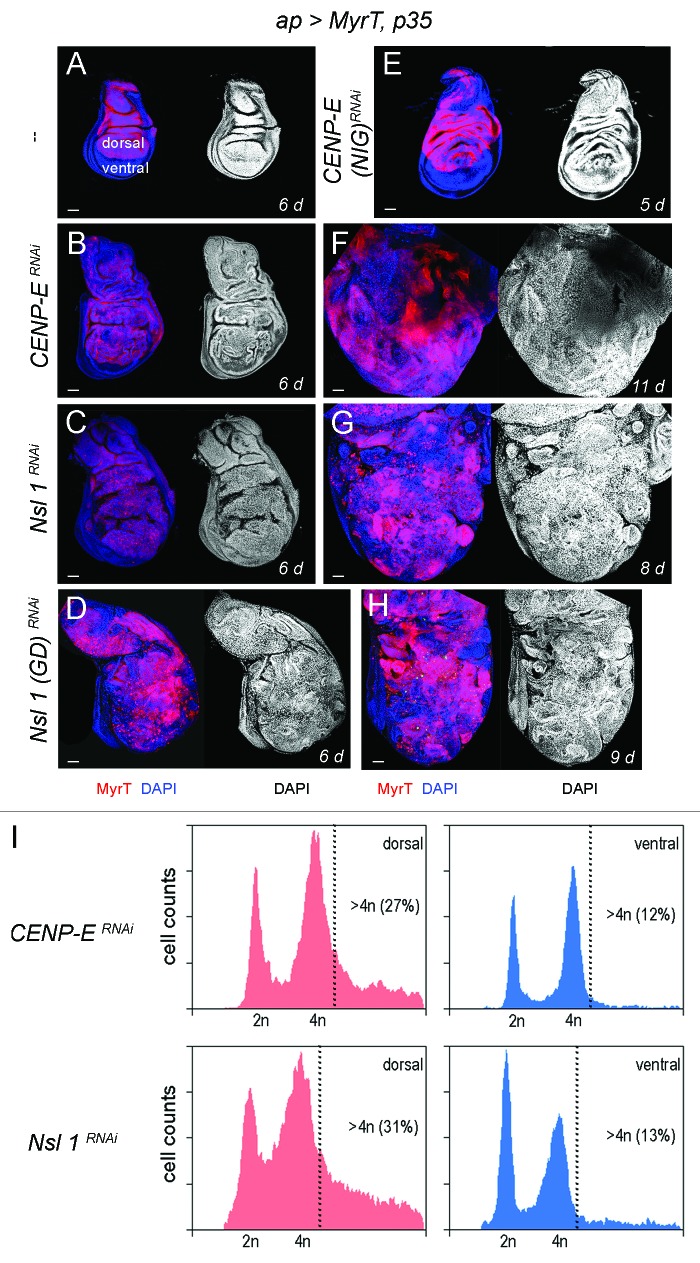
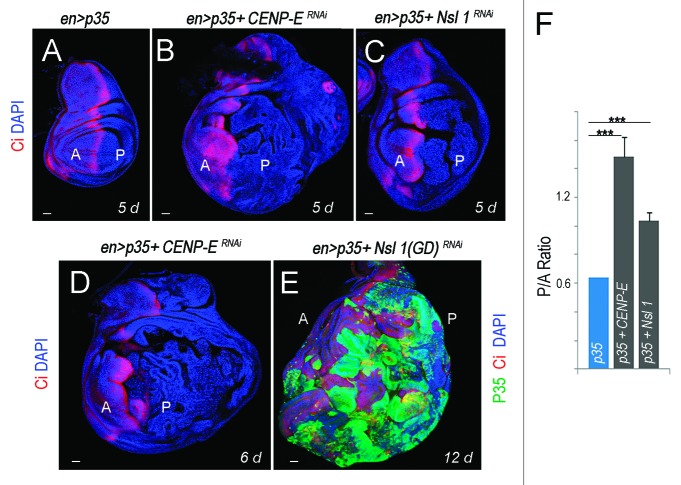
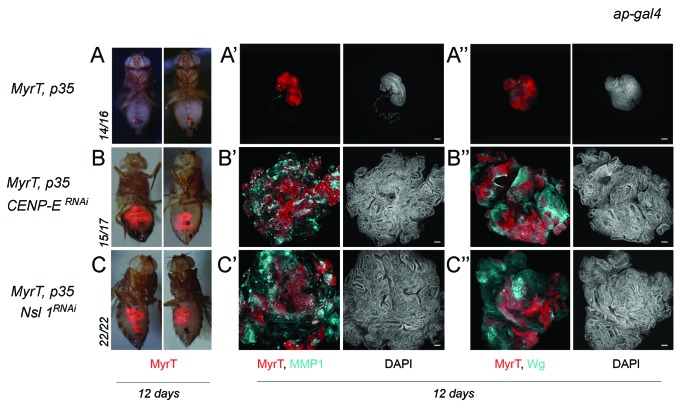
Here we have revisited the impact of CENP-E or Nsl1 depletion in tumorigenesis. For this purpose, we expressed p35 and dsRNA forms of CENP-E or Nsl1 in specific territories of the wing primordia. We first used the ap-gal4 driver to induce transgene expression in the dorsal compartment (Fig. 1A). The DNA content profile of dissociated cells subject to CENP-E or Nsl1 depletion and expressing p35 revealed a high percentage with DNA content higher than 4n (up to 30%) when compared with control cells (Fig. 1I). This percentage is very similar to that observed upon depletion of bub3 in wing disc cells (around 30%10). In contrast to the previous report,13 we observed tissue overgrowth upon CENP-E or Nsl1 depletion (compare Fig. 1A and B–E). Larvae expressing dsRNA forms of CENP-E or Nsl1 together with p35 kept growing for longer than p35-expressing control larvae and ultimately died (data not shown). Interestingly, the resulting wing primordia were massively overgrown, and the cell population subjected to genetically induced CIN (labeled in red) invaded the neighboring wild-type (unlabeled) territory (Fig. 1F–H). To better quantify the tissue overgrowth caused by CENP-E or Nsl1 depletion, we used the en-gal4 driver to induce transgene expression in the posterior compartment, whereby the anterior compartment serves as a control. Also, in this case, strong overgrowth occurred in the posterior compartment when compared with the neighboring wild-type tissue and with age-matched p35-expressing wing discs (Fig. 2A–C, quantification in Fig. 2F). Similarly to what happened with the ap-gal4 driver, larvae expressing dsRNA forms of CENP-E or Nsl1 together with p35 under control of en-gal4 kept growing for longer than p35-expressing control larvae and ultimately died (data not shown). The resulting wing primordia were massively overgrown, and the cell population subjected to genetically induced CIN (unlabeled in Fig. 2D or labeled in green in Fig. 2E) invaded the neighboring, wild-type territory (labeled in red, Fig. 2D and E). To further characterize the growth potential of the tissue subjected to CENP-E or Nsl1 depletion and PCD blockade, wing tissue expressing p35, MyrTomato (MyrT), and the corresponding dsRNA transgenes was transplanted into the abdomen of adult females and maintained for 12 days. While p35-expressing tissue scarcely grew after implantation (Fig. 3A–A”), tissue depleted of CENP-E or Nsl1 and expressing p35 grew several times larger than the controls expressing p35 alone and showed disorganized tissue architecture with extensive folding (Fig. 3B–B” and C–C”).
Figure 1.CENP-E or Nsl1 depletion induces aneuploidy and massive tissue overgrowth upon additional blockade of the apoptotic pathway. (A–H) Wing discs expressing MyrT, p35, and dsRNA forms of the indicated genes under the control of the ap-gal4 driver (labeled in red), and stained for DAPI (blue or white). Larvae were 6 d (A–D), 5 d (E), 11 d (F), 8 d (G) and 9-d-old (H). All pictures were taken at the same magnification. Bar in (A–H) represents 50 µm. (I) DNA content analysis by fluorescence-activated cell sorting (FACS) of dorsal (left panels) and ventral cells (right panels) of wing primordia shown in (B and C), respectively. Percentage of cells with DNA content higher than 4n is indicated. Genotypes: ap-gal4,UAS-myrT/+; UAS-p35/+ (A), ap-gal4,UAS-myrT/+; UAS-CENP-ERNAi/UAS-p35 (B and F),ap-gal4,UAS-myrT/UAS-Nsl1RNAi; UAS-p35/+ (C and G), ap-gal4,UAS-myrT/UAS–Nsl1RNAi(GD); UAS-p35/+ (D and H), ap-gal4,UAS-myrT/+; UAS-p35/ UAS-CENP-ERNAi(NIG) (E). d, days.
Figure 2.CENP-E- or Nsl1-depleted tissues overgrow and invade neighboring tissues upon additional blockade of the apoptotic pathway. (A–E) Wing primordia expressing the indicated transgenes in posterior cells under the control of the en-gal4 driver, and stained for DAPI (blue), Ci (red) and p35 (green). Larvae were 5 d (A–C), 6 d (D), and 12-d-old (E). All pictures were taken at the same magnification. Bar in (A–E) represents 50 µm. Ci labels the anterior (A) compartment. (F) Histogram plotting the P/A size ratio of wing primordia shown in (A–C). Genotypes: en-gal4,UAS-p35/+ (A); en-gal4,UAS-p35/+; UAS-CENP-ERNAi/+ (B and D); en-gal4,UAS-p35/UAS-Nsl1RNAi (C); en-gal4,UAS-p35/UAS–Nsl1RNAi(GD) (E). d, days.
Figure 3. Depletion of CENP-E or Nsl1 drives tumor-like growth in allograft transplants. (A–C) Micrographs of adult flies carrying MyrT-labeled (red) implants of the indicated genotypes. Pictures were taken 12 d after implantation, and ratios indicating the reproducibility of the phenotype are shown. (A’–C”) Allograft transplants of the indicated genotypes stained to visualize MMP1 (a'–c', blue), Wg (A”–C”, blue), MyrT (red) expression, and DAPI (blue or white). Transplants were extracted 12 d after implantation. Bar in (a’–c”) represents 50 µm. Genotypes: ap-gal4,UAS-myrT/+; UAS-p35/+ (A–A”); ap-gal4,UAS-myrT/+; UAS-CENP-ERNAi/UAS-p35 (B–B”); ap-gal4,UAS-myrT/UAS-Nsl1RNAi; UAS-p35/+ (C–C”). d, days.
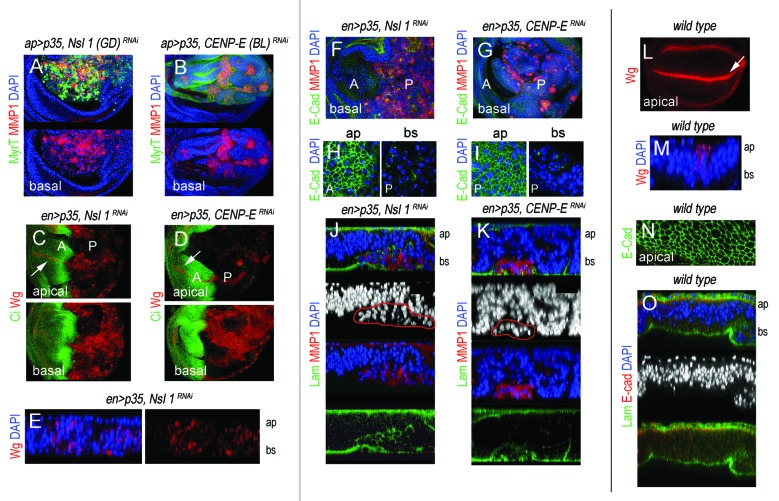
The JNK pathway plays a critical role in the tumorigenic behavior of Drosophila epithelial cells,21-24 and CIN-induced tumorigenesis is not an exception.10 CIN-induced tumorigenesis relies on highly aneuploid cells delaminating from the main epithelium, activating JNK, and inducing the expression of JNK targets Wg and MMP1. While Wg is an absolute requirement for CIN-induced tissue overgrowth,10 MMPs are well-known to contribute to BM degradation both in flies and mammals.24-26 We thus monitored the expression of these 2 JNK targets in tissues depleted of CENP-E or Nsl1. In wild-type wing primordia, Wg is expressed in a stripe that corresponds to the future wing margin (Fig. 4L, white arrow) and MMP1 is not expressed (data not shown). Both Wg and MMP1 were induced in the cell population subjected to genetically induced CIN (Fig. 4A–D, F, and G, the CIN-induced cell population is labeled in green; in Fig. 4A and B, it is labeled by the absence of Ci expression; in green in Fig. 4C and D, and it is not labeled in Fig. 4F and G). Most interestingly, expression of these 2 JNK targets was also observed in wing allografts depleted of CENP-E or Nsl1 and expressing p35 (Fig. 3B” and C”). Ectopic expression of Wg and MMP1 was mainly observed in delaminated cells located on the basal side of the epithelium (Fig. 4A–E, compare Fig. 4E with Fig. 4M), and the BM was clearly disrupted in the cell population subjected to CIN (labeled with Laminin-γ, Fig. 4J and K, compare with Fig. 4O). Dissociation of Drosophila epithelial cells upon CIN is thought to be a consequence of E-Cadherin mis-localization.10 Consistent with this, E-Cadherin lost its tight junctional localization in the delaminating cells of CENP-E- or Nsl1-depleted tissues (Fig. 4H and I, compare with Fig. 4N). These results present unequivocal evidence that CENP-E or Nsl1 depletion promotes tumorigenic behavior of epithelial cells—in terms of invasiveness of the neighboring tissue, cell delamination, and BM degradation—upon additional blockade of the PCD pathway.
Figure 4. Cell delamination, Wingless, and MMP1 expression, and basement membrane degradation in CENP-E- or Nsl1-depleted tissues. (A and B) Wing primordia expressing the indicated transgenes under the control of the ap-gal4 driver, and stained for MyrT (green), MMP1 (red) and DAPI (blue). Basal sides of the epithelia are shown. (C and D) Wing primordia expressing the indicated transgenes under the control of the en-gal4 driver, and stained for Wg (red), and Ci (green). Ci labels the anterior (A) compartment. Ectopic expression of Wg was mainly observed in the basal side of the epithelium. White arrows indicate the endogenous expression of Wg along the future wing margin. (E) Cross-section of a wing pirmordium expressing the indicated transgenes under the control of the en-gal4 driver and stained for DAPI (blue) and Wg (red). ap, apical, and bs, basal. (F and G) Wing primordia expressing the indicated transgenes under the control of the en-gal4 driver, and stained for DAPI (blue), E-cad (green), and MMP1 (red). (H and I) Magnification of the apical (ap) and basal (bs) sections of the wing primordial are shown in (F and G). In (C, D, and F–I), anterior (A) and posterior (P) compartments are indicated. (J and K) Cross-sections of the posterior compartment of wing primordia expressing the indicated transgenes under the control of the en-gal4 driver and stained for DAPI (blue), laminin-γ (labels the BM, in green), and MMP1 (red). Expression of MMP1 is restricted to delaminated cells (marked in red). ap, apical; bs, basal. (L) Wild-type wing primordium stained for Wg (red). White arrow indicates the endogenous expression of Wg along the future wing margin. (M) Cross-section of a wild-type wing pirmordium stained for DAPI (blue), and Wg (red). ap, apical; bs, basal. (N) Magnification of the apical section of a wild-type wing pirmordium stained for E-cad (green) expression. (O) Cross-section of a wild-type wing pirmordium stained for DAPI (blue), laminin-γ (green), and E-cad (red). ap, apical; bs, basal. Genotypes: ap-gal4,UAS-myrT/UAS–Nsl1RNAi(GD); UAS-p35/+ (A), ap-gal4,UAS-myrT/+, UAS-p35/ UAS-CENP-ERNAi(BL) (B), en-gal4,UAS-p35/UAS-Nsl1RNAi(C, E, F, H, and J); en-gal4,UAS-p35/+; UAS-CENP-ERNAi/+ (D, G, I, and K), wild–type (L–O).
Discussion
Several mitotic checkpoint proteins exert non-SAC functions, such as transcriptional regulation16 or nuclear import.17 A recent report proposes a SAC-independent tumor suppressor role of mitotic checkpoint genes in Drosophila epithelial tissues upon the inhibition of apoptosis.13 According to this report, depletion of the SAC gene bub3 caused overgrowth, with a remarkable 100% penetrance, of wing disc cells unable to enter apoptosis (see also ref. 10). In contrast, the penetrance of tissue overgrowth upon depletion of other SAC genes, such as Mad2 or BubR1, was much lower (10–30%), and CENP-E or Nsl1 depletion does not cause any detectable growth phenotype. Similar observations were made in allograft transplants of the corresponding genotypes.13 Although the authors state that these results present evidence that aneuploidy resulting from an impaired SAC is not sufficient to drive tumorigenesis, the levels of aneuploidy—measured by cellular DNA content analysis or by karyotypic analysis of mitotic cells—was relatively heterogenous in the different genetic backgrounds tested.13 Depletion of Bub3 induced by far the most significant levels of aneuploidy (around 30% in mitotic cells) when compared with Mad2-, BubR1-, CENP-E-, or Nsl1-depleted cells (12–19%).13 These observations suggest that tissue overgrowth occurs only above a particular percentage of aneuploid cells present in the tissue. Consistent with this proposal, we found that the levels of aneuploidy (around 30%)—measured by cellular DNA content analysis—in CENP-E- or Nsl1-depleted tissues were very similar to those obtained in bub3-depleted ones,10 and the response of the tissue to depletion of any of these 3 genes was exactly the same. These results, together with those previously reported upon depletion of other SAC genes (rod), genes involved in the spindle assembly (abnormal spindle [asp]), chromatin condensation (orc2), and cytokinesis (diaphanous [dia]),10 all causing significant levels of aneuploidy (around 30%), reinforce the proposal that CIN plays a major tumorigenic role in Drosophila epithelial cells upon additional blockade of the PCD pathway and question the SAC-independent tumor suppressor role of mitotic checkpoint genes.13 While high rates of CIN exert a tumor suppressor function in mouse models,8 our results indicate that a minimum rate of CIN is required in the tissue to cause tumor-like growth. This model system provides an ideal genetic experimental set up to identify new molecular elements involved in CIN-induced tumorigenesis in epithelial tissues. Remarkably, one common feature in the spectrum of spontaneous tumors in the various CIN mouse models analyzed so far is the predominant formation of tumors in lung epithelial cells.3 Whether JNK activity or Wnt expression also play a crucial role in this context remains to be elucidated.
Materials and Methods
Fly stocks
The following dsRNA forms of CENP-E and Nsl1 were used in this work: UAS–CENP-ERNAi (ID 35081, VDRC); UAS–CENP-ERNAi(BL) (P[TRiP.HMJ21427]attP40, ID 54004 Bloomington Stock Center); UAS–CENP-ERNAi(NIG) (ID 6392 R-2, National Institute of Genetics NIG-Fly); UAS–Nsl1RNAi (ID 106889, VDRC); UAS–Nsl1RNAi(GD) (ID 19530, VDRC).
Immunohistochemistry
Rabbit anti-p35 (1:200) (IMG5740, IMGENEX); rat anti-Ci (1:10) (2A1, DSHB); mouse anti-Wg (1:20) (4D4, DSHB); mouse anti-MMP1 (1:20) (14A3D2, DSHB); rabbit anti-Laminin-γ (1:50) (ab47651, AbCam); rat anti-E-cadherin (1:50) (DCAD2, DSHB) were used at the indicated dilutions.
Quantification of tissue growth
Sizes of the A and P compartments in experimental wing primordia were measured using the ImageJ Software (NIH). At least 10 wing discs per genotype were scored. The average P/A ratio and the corresponding standard deviations were calculated and t test analysis was performed.
Flow cytometry analysis
Wing imaginal discs of each genotype were dissected in cold PBS, dissociated with trypsin-EDTA at 32 °C for 45 min, and fixed with 4% formaldehyde for 20 min. Cells were centrifuged at 2000 rpm for 2 min, resuspended in 1 ml of 70% ethanol, and incubated for 1 h at room temperature (RT). After centrifugation, the pellet was resuspended in PBS with DAPI 1 mg/ml and RNase 100 mg/ml and incubated for 1 h at RT. DAPI fluorescence was determined by flow cytometry using a FacsAria I SORP sorter (Beckton Dickinson). Excitation with the blue line of the laser (488 nm) permits the acquisition of scatter parameters. Green laser (532 nm) was used for MyrT excitation, and a UV laser (350nm) for DAPI excitation. Doublets were discriminated using an integral/peak dotplot of DAPI fluorescence. Cell cycle histograms were obtained on each sample according to the MyrT fluorescence, and cell cycle analysis was done on DAPI fluorescence collected at 440 nm. DNA analysis (ploidy analysis) on single fluorescence histograms was done using Summit software (Dako).
Allograft transplants
Wing discs expressing MyrT and p35, or MyrT, p35, and the corresponding UAS-transgenes that cause CIN were dissected from third instar larvae and transferred to cold PBS. Discs were fragmented into small pieces using electrolytically sharpened tungsten needles and implanted into the abdomens of virgin w1118 females, as described in reference 27. Tumors were recovered 12 d after implantation for analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Bloomington (USA), VDRC (Austria), and National Institute of Genetics (Japan) stock centers for flies, the Developmental Studies Hybridoma Bank (USA) for antibodies, T Yates for help in text editing, and two anonymous reviewers for helpful comments. M.M. is an ICREA Research Professor and M.M.’s laboratory was funded by grants from the Spanish Ministerio de Ciencia e Innovación (BFU2010-21123 and Consolider CSD2007-00008), the Generalitat de Catalunya (2005 SGR 00118), and the EMBO Young Investigator Programme.
References
- 1.Duijf PH, Schultz N, Benezra R. Cancer cells preferentially lose small chromosomes. Int J Cancer. 2013;132:2316–26. doi: 10.1002/ijc.27924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–15. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janssen A, Medema RH. Genetic instability: tipping the balance. Oncogene. 2013;32:4459–70. doi: 10.1038/onc.2012.576. [DOI] [PubMed] [Google Scholar]
- 4.Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, Jhanwar-Uniyal M, Dai W. Colonic tumorigenesis in BubR1+/-ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci U S A. 2005;102:4365–70. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–40. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci U S A. 2013;110:E4134–41. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellanos E, Dominguez P, Gonzalez C. Centrosome dysfunction in Drosophila neural stem cells causes tumors that are not due to genome instability. Curr Biol. 2008;18:1209–14. doi: 10.1016/j.cub.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 10.Dekanty A, Barrio L, Muzzopappa M, Auer H, Milán M. Aneuploidy-induced delaminating cells drive tumorigenesis in Drosophila epithelia. Proc Natl Acad Sci U S A. 2012;109:20549–54. doi: 10.1073/pnas.1206675109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:14188–93. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci U S A. 2005;102:11296–301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morais da Silva S, Moutinho-Santos T, Sunkel CE. A tumor suppressor role of the Bub3 spindle checkpoint protein after apoptosis inhibition. J Cell Biol. 2013;201:385–93. doi: 10.1083/jcb.201210018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown KD, Coulson RM, Yen TJ, Cleveland DW. Cyclin-like accumulation and loss of the putative kinetochore motor CENP-E results from coupling continuous synthesis with specific degradation at the end of mitosis. J Cell Biol. 1994;125:1303–12. doi: 10.1083/jcb.125.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkei Z, Przewloka MR, Glover DM. Drosophila Mis12 complex acts as a single functional unit essential for anaphase chromosome movement and a robust spindle assembly checkpoint. Genetics. 2011;187:131–40. doi: 10.1534/genetics.110.119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon YM, Baek KH, Jeong SJ, Shin HJ, Ha GH, Jeon AH, Hwang SG, Chun JS, Lee CW. WD repeat-containing mitotic checkpoint proteins act as transcriptional repressors during interphase. FEBS Lett. 2004;575:23–9. doi: 10.1016/j.febslet.2004.07.089. [DOI] [PubMed] [Google Scholar]
- 17.Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–63. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- 18.Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302:1227–31. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- 19.Brumby AM, Richardson HE. Using Drosophila melanogaster to map human cancer pathways. Nat Rev Cancer. 2005;5:626–39. doi: 10.1038/nrc1671. [DOI] [PubMed] [Google Scholar]
- 20.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–9. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 21.Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006;16:1139–46. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- 22.Igaki T, Pastor-Pareja JC, Aonuma H, Miura M, Xu T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in Drosophila. Dev Cell. 2009;16:458–65. doi: 10.1016/j.devcel.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003;22:5769–79. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlirova M, Bohmann D. JNK- and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006;25:5294–304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Srivastava A, Pastor-Pareja JC, Igaki T, Pagliarini R, Xu T. Basement membrane remodeling is essential for Drosophila disc eversion and tumor invasion. Proc Natl Acad Sci U S A. 2007;104:2721–6. doi: 10.1073/pnas.0611666104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaucher M, Hersperger E, Page-McCaw A, Shearn A. Metastatic ability of Drosophila tumors depends on MMP activity. Dev Biol. 2007;303:625–34. doi: 10.1016/j.ydbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 27.Ursprung H. In vivo culture of Drosophila imaginal discs. T.Y. Crowell, New York, 1967. [Google Scholar]