Abstract
Telomeres are repetitive nucleoprotein structures at the ends of chromosomes. Like most genomic regions consisting of repetitive DNA, telomeres are fragile sites prone to replication fork stalling and generation of chromosomal instability. In particular, abrogation of the TRF1 telomere binding protein leads to stalled replication forks and aberrant telomere structures known as “multitelomeric signals”. Here, we report that TRF1 deficiency also leads to the formation of “ultra-fine bridges” (UFB) during mitosis, and to an increased time to complete mitosis mediated by the spindle assembly checkpoint proteins (SAC). We find that topoisomerase IIα (TopoIIα), an enzyme essential for resolution of DNA replication intermediates, binds telomeres in a TRF1-mediated manner. Indeed, similar to TRF1 abrogation, TopoIIα downregulation leads to telomere fragility and UFB, suggesting that these phenotypes are due to decreased TopoIIα at telomeres. We find that SAC proteins bind telomeres in vivo, and that this is disrupted upon TRF1 deletion. These findings suggest that TRF1 links TopoIIα and SAC proteins in a pathway that ensures correct telomere replication and mitotic segregation, unveiling how TRF1 protects from telomere fragility and mitotic defects.
Keywords: SAC, Shelterin, TRF1, TopoII, fragility, mitosis
Introduction
Telomeres are specialized nucleoprotein structures at the termini of linear chromosomes composed of tandem repeats of the DNA TTAGGG sequence bound by associated proteins.1-3 Telomere binding proteins include a 6-subunit complex known as shelterin3 that protects the end of chromosomes from degradation, exonucleolytic attack, homologous recombination, and end-to-end fusions.3-5 The 6 subunits of shelterin encompass TRF1, TRF2, TIN2, RAP1, TPP1, and POT1. TRF1, the first mammalian telomere-binding protein to be identified,6,7 binds to the double-stranded DNA portion of telomeres as a homodimer. TRF1 has been shown to lead to telomere shortening when overexpressed, whereas its inhibition by a dominant-negative form caused telomere elongation.8 Ubiquitous deletion of the Trf1 gene in mice results in early embryonic lethality,9 and ES cells conditionally deleted for TRF1 showed increased chromosomal aberrations.10 More recently, conditional depletion of TRF1 in mouse was found to lead to a massive DNA damage response at telomeres along with multitelomeric signals (MTS), an aberration suggested to be due to telomere fragility.10-12
In order to maintain genetic information with each cell division, accurate genome duplication is necessary. However, there are regions within the mammalian genome that present a challenge to the replication process, such as fragile sites. These are defined as loci that display breaks on mitotic chromosomes upon partial inhibition of DNA synthesis.13 It is believed that the observed breaks occur due to under-replicated sequences upon chromosome condensation at the onset of mitosis. The delay in replication was believed to be a result of replication forks encountering secondary structures that form more frequently in areas rich in AT repeats. However, other regions rich in guanine tracts that are capable of forming secondary structures known as G-quadruplexes (G4) can also cause replication problems. It has been suggested that replication forks arrested at G4 quadruplexes are remodeled into recombination intermediates to be more easily processed and repaired.14 Telomeres are very G-rich and prone to G4 formation. Shelterins are able to bind these structures and regulate them throughout the cell cycle.15
An important characteristic of telomeric chromatin is that, like centromeric chromatin, it is a highly repetitive DNA sequence that has the potential to become an obstacle for the replication machinery. Many studies have implied that replication forks can pause or stall naturally at telomeres in yeast16-18 and in human primary fibroblasts.19 In agreement with these thoughts, there is mounting evidence suggesting that telomeres behave like fragile sites.11,12,20-23 In particular, increased replication stress either by ATR deficiency,20 or by drugs such as aphidicolin and hydroxyurea, which render replication suboptimal,11,12 leads to telomere fragility as indicated by formation of multitelomeric signals (MTS).
Strikingly, shelterins have been shown to protect from telomere fragility. In particular, abrogation of shelterin components such as TPP121 and Rap124 also lead to formation of multitelomeric signals. In the case of TRF1, it has been shown that TRF1 deficiency causes slower replication rates at telomeres and increased MTS, thus reinforcing the notion that shelterins aid in telomere replication and prevent fragility at telomeres.11,12
Common fragile sites are sequences that are intrinsically difficult to replicate. Advancement of replication forks at these sites generates vast topological stress that must be solved by DNA topoisomerases.25 An important player in ensuring replication fork progression as well as mitotic segregation of cells with dysfunctional telomeres is TopoIIα. In particular, yeast mutants for the telomere binding protein Taz1 show severe mitotic defects, which can be corrected by a catalytically dead topo2–191 mutant, a temperature-sensitive TopoIIα mutant that causes chromosome missegragation in S. pombe.26 Topoisomerase 2 is a homodimeric enzyme that is able to modulate DNA topology and, thus, maintain chromosome integrity. It does so by creating transient DSBs in the DNA in order to promote and allow the disentanglement of the duplex. TopoIIα has also been found to be able to relax super coiled DNA and resolve or promote catenation between DNA circles.27-30 Topoisomerases do not act randomly. In fact, they are able to preferentially distinguish between different types of DNA and collaborate with other factors capable of specifically targeting their activity, such as SMCs during chromosome compaction (reviewd in ref. 31). Although there has been much progress in understanding the function and role of topoisomerases in various cellular functions, including replication and chromosome segregation, transcription, and recombination and repair (reviewed in ref. 31), a specific role at telomeres has not been described. In a recent study, TopoIIα was suggested to act in the same telomere protection pathway as TRF2 and Apollo,32 directly linking TopoIIα to telomere maintenance for the first time. Studies have shown that treatment with ICRF193, a TopoIIα inhibitor, leads to mitotic arrest.33 TopoIIα has also been found to be important for the resolution of thin DNA threads that connect kinetochores of sister chromatids during anaphase.34 It is interesting to note that BLM, topoismoerase IIIα, and hRMI have been shown to associate with these threads.35 These proteins have been found to form a complex that is required for the resolution of recombination and replication intermediates.36-38 Another important player in the resolution of DNA threads is PICH (Plk1-interacting checkpoint helicase), a protein recently identified as a key component of the spindle assembly checkpoint (SAC). PICH was found to localize to kinetochores and inner centromeres as well as decorating ultrafine threads of DNA.34 Further studies discovered that TopoIIα is fundamental for the resolution of these threads during anaphase, allowing the complete separation of sister chromatids.39 It is believed that the PICH-BLM-coated DNA threads originated from incompletely replicated DNA or catenated duplexes.35
Precise segregation of sister chromatids during mitosis is essential for the maintenance of euploidy and genomic stability. The spindle assembly checkpoint (SAC) is fundamental for ensuring the correct attachment of spindle microtubules to kinetochores during prometaphase, and for delaying progression to anaphase in the event of incomplete or incorrect attachment. The core components of this checkpoint are products of the mitotic arrest-deficient (MAD) and the budding uninhibited by benzimidazole (BUB) genes and a handful of other proteins (reviewed in ref. 40). All known SAC transducers, including BubR1, Mad1, and Mad2, associate with unattached kinetochores in prometaphase. It is believed that kinetochore-localized Mad1/Mad2 heterodimers catalyze the conversion of soluble inactive Mad2 to an active form that is able to stably bind Cdc20 and inhibit activation of the anaphase-promoting complex/cyclosome (APC/C) and, hence, the progression to anaphase.41 However, there is evidence to suggest that SAC signaling is not solely dependent on kinetochores. In mammalian cells and yeast strains that lack functional kinetochores the mitotic checkpoint complex (Mad2/BubR1 bound to Cdc20) was detected in interphase cells.42,43 BubR1 and Mad2 are required not only to signal the presence of unattached kinetochores, but also to specify the minimum duration of mitosis.44 Moreover, cells lacking BubR1 do not proliferate and, instead, die within a week. This phenotype can be rescued by ectopic expression of a fragment of BubR1 that binds to Cdc20 but does not localize to kinetochores.45 These data imply a kinetochore-independent function for BubR1, whereby it may act as an inhibitor of APC/C (Cdc20) during interphase to prevent premature degradation of specific APC/C substrates and unscheduled anaphase.
A precise role for TRF1 in mitotic regulation has not been thoroughly explored to date. However, a direct interaction between TRF1 and both the SAC protein Mad1 and the mitotic kinase Nek2 was detected by yeast 2-hybrid screen assay.46 In addition, Mad2 and BubR1 were imaged to colocalize in vivo with TRF1 at telomeres in keratinocytes overexpressing TRF1.47 Remarkably, TRF1 was found to cosediment with SA1-cohesion complex and was found to play a role in telomere cohesion.48 More recently, BubR1 SUMOylation has been associated with both centromeric and telomeric cohesion.49 The above data implicate both TRF1 and BubR1 in telomere cohesion. It is possible that they may be acting together to ensure telomere cohesion resolution for anaphase onset. Together, these results suggest a putative link between TRF1 and the SAC.
To this end, we closely examined the role of TRF1 both in TopoIIα regulation of stalled replication and SAC coordination, thus ensuring correct mitotic segregation using TRF1-deficient cells. We demonstrate that TopoIIα, BubR1, and Mad1, are able to bind to telomeres in vivo in a TRF1-dependent manner, and that cells lacking TRF1 have a severe mitotic phenotype, coincidental with occurrence of DNA threads.
Results
TopoIIα localizes to telomeres in a TRF1-dependent manner
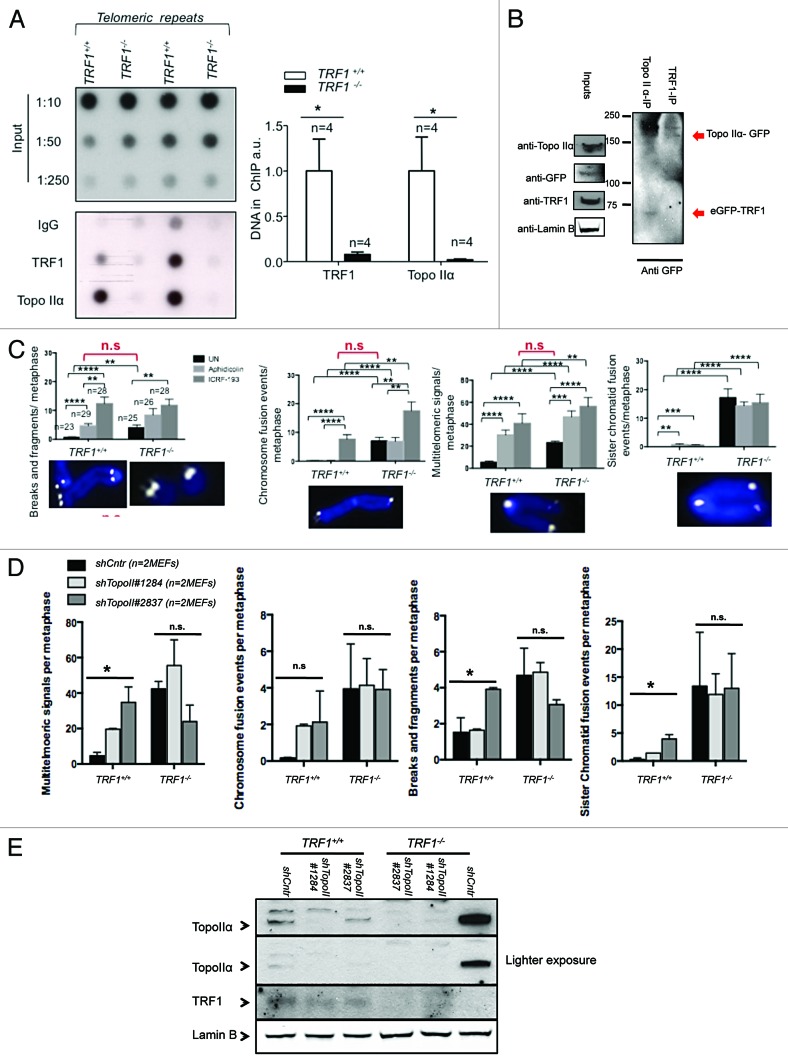
Previous work has shown that dysfunction of a single shelterin, such as TRF1, is sufficient to cause replication-dependent defects akin to fragile sites.11,12 A recent study implicated TopoIIα in the resolution of topological stress during telomere replication.32 Moreover, TopoIIα was shown to colocalize with telomeres in a cell cycle-specific manner and play an important role in protecting telomeres from replicative damage.32 The recruitment of TopoIIα to telomeres was found to be dependent on the levels of another shelterin component, TRF2.32 To establish if TopoIIα telomere binding was altered in the absence of TRF1, and therefore could be the possible cause of replication stalling in these cells, we first performed ChIP using specific antibodies against TopoIIα. We confirmed that TopoIIα was present at mouse telomeres (Fig. 1A). Interestingly the amount of TopoIIα bound to telomeres was significantly reduced in TRF1-null cells (Fig. 1A). These findings suggest the absence of TopoIIα at telomeres owing to TRF1 deficiency could be associated to the fragile telomere phenotype observed. We also detected an in vitro interaction between TopoIIα and TRF1. In particular, upon overexpression of tagged versions of both TRF1 and TopoIIα in 293T cells, we could co-immunoprecipitate TopoIIα when we pulled down TRF1, and TRF1 when we pulled down TopoIIα (Fig. 1B). This suggests that these proteins are able to interact either directly or in a complex.
Figure 1. TopoIIα binds to telomeres via TRF1 (A) ChIP of Trf1+/+ p53−/− and Trf1 −/− p53−/− MEFs for TopoIIα and TRF1. The amount of immunoprecipitaded telomere repeats was normalized to the amount of telomere repeats present in the chromatin fraction unbound to preimmune serum. n, independent MEFs used. Bars represent the average between replicates. (Right) Quantification. (Left) Representative dot blot. (B) 293T cells transfected with eGFP-TopoIIα and eGFP-TRF1 were collected and lysed. Equal amounts of lysate were immunoprecipitated with TRF1 IgG or TopoIIα IgG, immunoprecipitaes were blotted for GFP and cell lysates for TRF1 or TopoIIα. (C) Aberration frequency in metaphase spreads of the indicated genotypes untreated (UN) or treated with indicated drugs. At least 10 metaphases from 2 independent MEFs per genotype were analyzed. Statistical comparisons using Student t test are shown. (D) Aberration frequency in metaphase spreads of the indicated genotypes untreated (UN) or infected with indicated shRNAs against TopoIIα. At least 10 metaphases from 2 independent MEFs per genotype were analyzed. Statistical comparisons using Student t test are shown. (E) Western blot demonstrating knockdown of TopoIIα in cells of indicated genotypes.
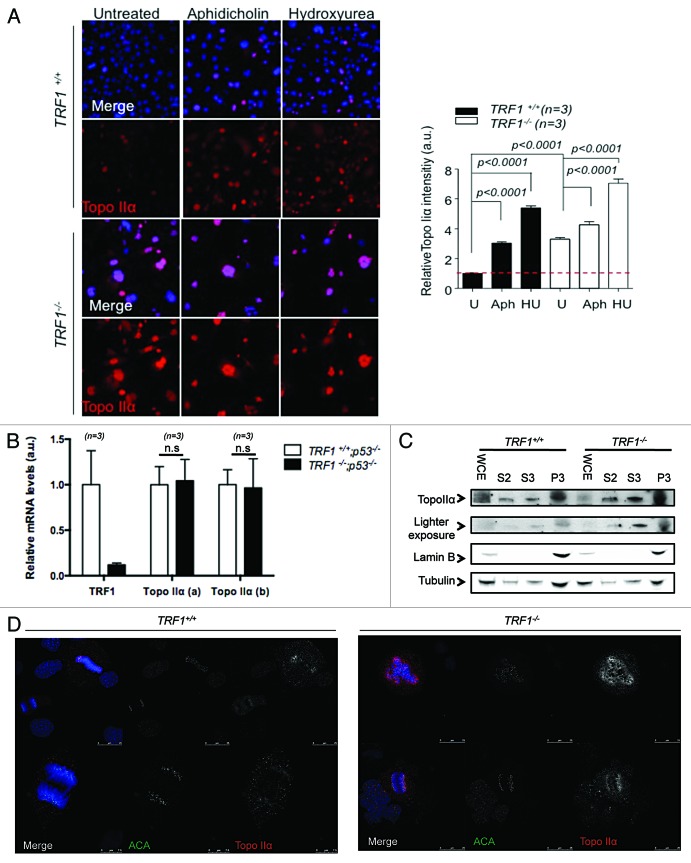
To confirm the hypothesis that TopoIIα may play a role in the resolution of telomere fragile sites, we speculated that the inhibition of TopoIIα could result in increased multitelomeric signals (MTS), a type of telomere aberration previously shown to be an indicator of telomere replication problems.11,12 Using replication-stalling agent aphidicolin and the TopoIIα-specific inhibitor ICRF-193, we treated both TRF1 wild-type and knockout cells and quantified the presence of MTS and of other chromosomal aberrations (Fig. 1C). We found that both multitelomeric signals (MTS) and chromosome fusions were increased in ICRF-193-treated wild-type cells to similar levels seen in untreated TRF1-knockout cells (Fig. 1C). Interestingly, treatment with ICRF-193 further increased these aberrations in TRF1-deficient cells. Previous reports have suggested that treatment with ICRF-193 may also affect TopoIIβ,50 which may explain the additive effect in TRF1-deficient cells. To circumvent this, we used 2 specific shRNAs against TopoIIα in both Trf1+/+p53−/− and Trf1−/−p53−/− and checked for chromosome aberrations. Upon specific depletion of TopoIIα, as confirmed by western blot (Fig. 1E), we detected a significant increase in MTS, breaks, and fragments, as well as sister chromatid fusions in TRF1-proficient cells but not in TRF1-deleted cells (Fig. 1D). This strongly supports the notion that TRF1-dependent recruitment of TopoIIα to telomeres is important for telomere fragile site resolution. Surprisingly, TRF1 knockout MEFS appeared to have higher TopoIIα levels as detected by western blot (Fig. 1E). Higher TopoIIα levels have recently been reported to occur in coincidence with high DNA damage burden.51We confirmed this by immunofluorescence using replication stalling agents aphidicolin and hydroxyurea as controls for TopoIIα induction, and quantified the nuclear fluorescence intensity of TopoIIα. We found that untreated Trf1−/−p53−/− MEFs harbored approximately 3-fold more TopoIIα than wild-type cells, and that levels of TopoIIα increased in both cell types upon treatment (Fig. 2A). This increase in TopoIIα levels was not due to increased mRNA, as seen by Q-RT PCR using 2 distinct sets of primers (Fig. 2B), in agreement with previous findings by Eguren et al.51 The subcellular localization of TopoIIα was affected in the absence of TRF1, with more TopoIIα accumulating in the nucleoplasmic fraction (Fig. 2C). By using immunofluorescence on cells undergoing mitosis, we did not see any aberrant localization (Fig. 2D). Together, these findings are in agreement with recent results linking high levels of TopoIIα with increased DNA damage.51 Therefore, the increased nuclear fluorescence of TopoIIα in the absence of TRF1 may result from a higher DNA damage burden in these cells.
Figure 2. TRF1 deletion increasesTopoIIα expression. (A) Representative images of Trf1+/+ p53−/− and Trf1 −/− p53−/− MEFs untreated (U) treated with aphidicolin (Aph) and hydroxyurea (HU) stained with an antibody against TopoIIα (red). Nuclei are counterstained with Dapi (blue). Quantification of mean TopoIIα intensity per nucleus normalized to Trf1+/+ untreated control of 3 independent MEFs. Student t test was used for statistical analysis, and P values are indicated (*P < 0.05, **P < 0.002, ***P < 0.0005, ****P < 0.0001). All error bars represent standard error. Bars = 50 μM. (B) TRF1 and TOPOIIα transcription levels in MEFs transduced with or without Cre, as estimated by qRT-PCR. The mRNA fold change relative to levels in wild-type MEFs 3 d after transduction using the indicated primers is depicted. n, independent MEF clones used per condition. Error bars: s.e.m. Statistical analysis: 2-sided Student t-test. (C) Subcellular fractionation of Trf1+/+ p53−/− and Trf1−/− p53−/− MEFs. Tubulin was used as a loading control and Lamin B for the chromatin-bound fraction. Whole-cell extract (WCE), cytoplasmic fraction (S2), nucleoplasmic fraction (S3), chromatin fraction (P3). (D) Representative images of Trf1+/+ p53−/− and Trf1 −/− p53−/− MEFs stained with an antibody against TopoIIα (red) showing that TopoIIα localization during mitosis is not altered. Nuclei are counterstained with Dapi (blue).
TRF1 abrogation leads to the appearance of TopoIIα-dependent DNA threads
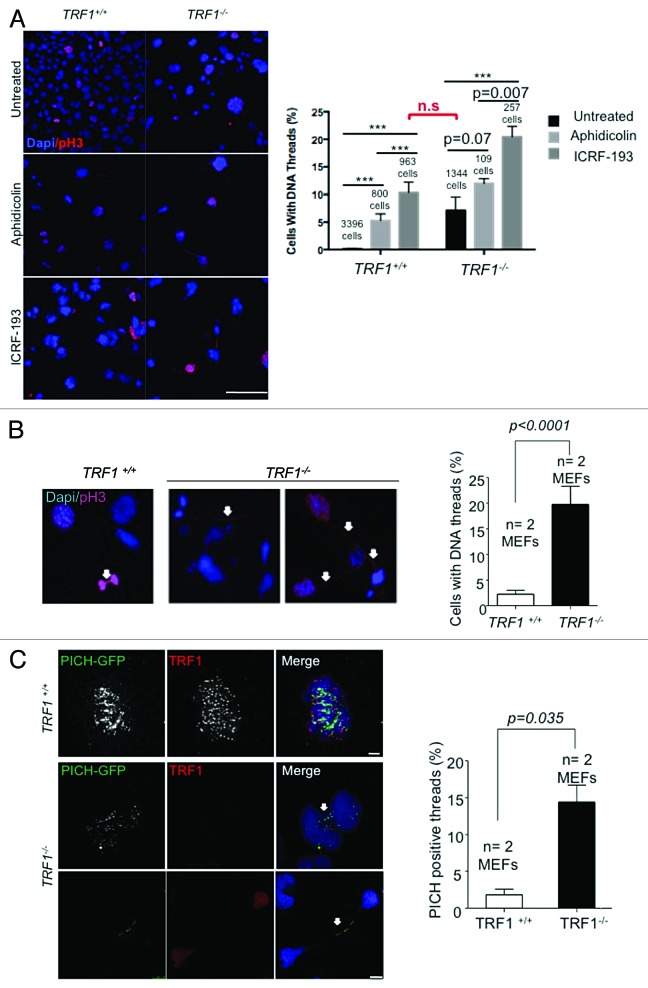
In addition to its role in topologically modifying DNA for correct chromosome segregation during mitosis,52 TopoIIα is also an important factor in the resolution of thin DNA threads that appear at anaphase onset, which have been proposed to represent catenated DNA.34 These threads stretch under tension until decatenation by TopoIIα resolves them before the end of anaphase.34 DNA threads, which can persist throughout anaphase, have been suggested to comprise incompletely replicated DNA or unresolved recombination intermediates.35 The findings described above prompted us to investigate whether decreased TopoIIα at telomeres owing to TRF1 deficiency could also lead to persistent DNA threads due to the incomplete resolution of telomeric replication intermediates. To this end, we performed immunofluorescence with a pH3 antibody to visualize chromatin threads in wild-type and TRF1-deficient cells upon catalytic inhibition of TopoIIα. In particular, we treated both TRF1 wild-type and knockout cells with aphidicolin and ICRF-193 and quantified the presence of DNA threads (Fig. 3A). We found a significant increase in DNA threads in both genotypes upon aphidicolin treatment. The percentage of DNA threads was further increased upon specific inhibition of TopoIIα. Remarkably, the catalytic inhibition of TopoIIα in wild-type cells resulted in a similar percentage of DNA threads to that found in untreated TRF1-deficient cells (Fig. 3A). Upon quantification, we observed that 20% of untreated TRF1-knockout cells were connected by pH3-positive DNA threads compared with only <5% of wild-type controls (Fig. 3B). PICH (Plk1-interacting checkpoint helicase), a helicase-like protein found to bind to Plk1, localizes to centromeres, but it is also present at DNA threads.34 PICH-positive threads have been found to require TopoIIα activity after anaphase onset for their resolution.39 Thus, we transfected wild-type and TRF1-deficient cells with a GFP-PICH construct. In this case, we found that 15% of TRF1-abrogated cells were connected by GFP-PICH-positive threads compared with only <3% of the wild-type counterparts (Fig. 3C). These results suggest that TRF1 deficiency results in incorrectly decatenated DNA leading to threads, and that the severity of this phenotype is similar to that of TopoIIα inhibition. In turn, DNA catenates are likely to be the products of incompletely replicated DNA or unresolved recombination intermediates in the absence of TRF1.12 Taken together these data support a model whereby absence of TRF1 results in decreased TopoIIα at telomeres and a defective resolution of replication intermediates, thus leading to the appearance of thin DNA threads.
Figure 3. Catalytic inhibition of TopoIIα increases thin DNA threads. (A) (Left) Representative images of Trf1+/+ and Trf1−/− MEFs treated with aphidicolin and ICRF-193 stained pH3 (red) and DNA threads connecting various nuclei. Nuclei are counterstained with Dapi (blue). Arrows indicate DNA threads. Bars = 100 μM. (Right) Quantification of the number of cells with threads. Student t test was used for statistical analysis, and P values are indicated. (B) Representative images of Trf1+/+ and Trf1−/− untreated MEFs stained with pH3 (red) and DNA threads (arrow) connecting various nuclei in Trf1−/− MEFs. Nuclei are counterstained with Dapi (blue). Quantification of the number of cells that display threads. (C) (Left panel) Representative immunofluorescence images of Trf1+/+ and Trf1−/− MEFs stained with GFP antibody to detect PICH-GFP (green) and TRF1 (red). Arrows indicate DNA threads. Bars = 10 μM. (Right panel) Quantification of the percentage of cells displaying PICH-positive DNA threads connecting more than one nucleus. Fisher exact test was used for statistical analysis, and P values are indicated. All error bars represent standard error.
TRF1 interacts with SAC proteins and is important for their telomeric localization
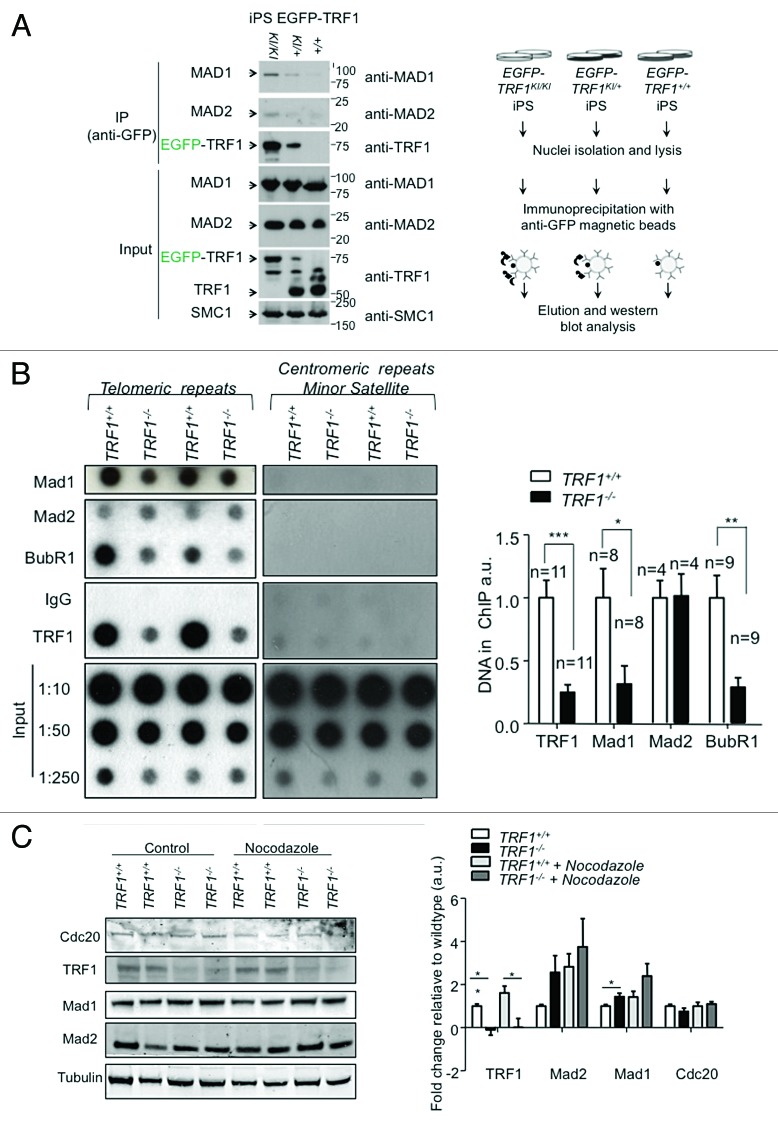
PICH has been identified as a crucial component of the spindle assembly checkpoint (SAC), a checkpoint that ensures bipolar attachment of chromosomes to the mitotic spindle and correct segregation during mitosis.34 Interestingly, there are multiple reports that suggest a direct interaction of spindle assembly checkpoint (SAC) proteins and TRF1.46,47 To explore if this interaction occurred in vivo, we performed immunoprecipitation experiments using cells with a knock-in TRF1 allele, in which TRF1 is fused to the eGFP protein previously generated by us.53 Using GFP antibodies to immunoprecipitate extracts from heterozygous eGFP-TRF1+/KI, homozygous eGFP-TRF1KI/KI, and wild-type induced pluripotent stem (iPS) cells. We chose iPS as a platform to discover TRF1-interacting proteins, as pluripotent cells have been previously described by us and others to have endogenously higher levels of TRF1 compared with differentiated cells.53,54 As positive control, we previously showed that eGFP-TRF1 selectively co-immunoprecipitated the known components of the shelterin complex.53 Interestingly, we found here that eGFP-TRF1 also co-immunoprecipitates the SAC components Mad1 and Mad2 (Fig. 4A), providing an in vivo demonstration for the previously suggested associations between TRF1 and both Mad146 and Mad2.47
Figure 4. TRF1 interacts with SAC components. (A) Western blot of the co-immnuoprecipitation of MAD1 and MAD2 proteins with TRF1 protein with anti-GFP antibody in EGFP-TRF1 induced pluripotent stem cells (left panel). Note that only the iPS carrying the GFP-tagged version of TRF1 (EGFP-TRF1KI/KI, EGFP-TRF1KI/+) immunoprecipitate Mad1 and Mad2. Cohesin SMC1 was used as loading control. Schematic representation of the immuneprecipitation experiment (right panel). (B) ChIP of Trf1+/+ and Trf1−/−p53−/− MEFs using indicated antibodies The amount of immunoprecipitaded telomere repeats was normalized to the amount present in the chromatin fraction unbound to preimmune serum. (Left) Duplicate blot against centromeric DNA. Relative association of the indicated proteins with telomeric DNA was calculated by normalizing the telomeric DNA recovered in each ChIP to that recovered in corresponding wild-type cells. n, independent MEFs used. Bars represent the average between replicates; standard error is used. (Right) Quantification of ChIP. A Student t test was used to calculate statistical significance. (C) Western blot of whole cell extracts of asynchronous and Nocodazole treated Trf1+/+ and Trf1−/− p53−/− MEFs of Mad1, Mad2, and TRF1.
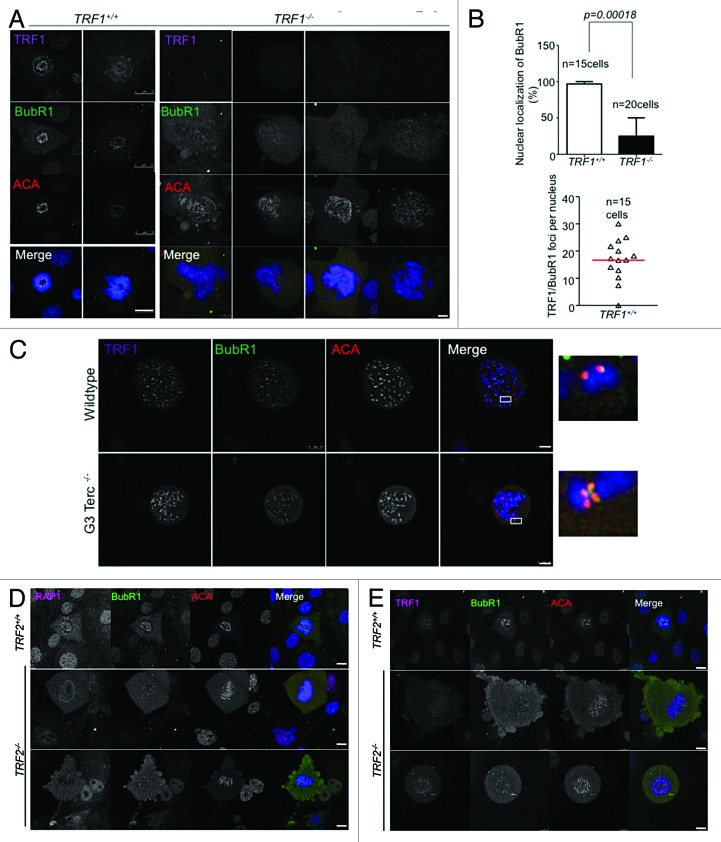
TRF1 has also been described to co-localize with BubR1 at telomeres in murine keratinocytes.47 Thus, we set out to study whether Mad1, Mad2, and BubR1 could directly bind to telomeres in vivo by using chromatin immunoprecipitation (ChIP) analyses in wild-type and TRF1-deficient mouse embryonic fibroblasts (MEFs) previously generated by us.11 As a positive control, we confirmed strong TRF1 binding to telomeres by ChIP, which was abrogated in TRF1-deficient cells (Fig. 4B). Interestingly, we found that both Mad1 and BubR1 showed significant binding to telomeres in vivo, and that the interactions were considerably decreased in the absence of TRF1 (Fig. 4B). Mad2 showed lower affinity for telomeric DNA that was not affected by the abrogation of TRF1. Using centromeric Minor and Major probes confirmed binding specificity of the antibodies (Fig. 1A; Fig. S1A). Decreased binding of SAC proteins to telomeres in the absence of TRF1 was independent of the total amount of cellular protein. In particular, when whole-cell extracts of both wild-type and TRF1-deficient MEFs were checked for alterations in the total amount of SAC proteins, we found a small but significant increase in the total amount of Mad1 in TRF1-null cells (Fig. 4C). Moreover, we see that the levels of both Mad1 and Mad2 in wild-type cells arrested with nocodazole is comparable to that seen in untreated TRF1 knockout cells, suggesting that these cells have a delay in mitosis (Fig. 4C). To ensure that the alteration in telomeric binding of these proteins was due specifically to the depletion of TRF1, we examined the effect of TRF2 depletion by using TRF2-deficient MEFs.55 In the absence of TRF2, neither Mad1 nor BubR1 binding to telomeric DNA was affected, as seen by ChIP (Fig. S1B). Remarkably, the only protein affected by the absence of TRF2 was TRF1 (Fig. S1B). In particular, in 3 independent MEFs, we found a consistent and significant decrease of telomeric DNA pulled down by TRF1 as seen by ChIP (Fig. S1B). Previous reports state that TRF1 is still present at telomeres in the absence of TRF2,55 consistent with our observation that the decrease in TRF1 was not sufficient to alter the telomeric binding of the SAC proteins. These data strongly suggest that Mad1 and BubR1 SAC proteins are bound to telomeres, and that TRF1, but not TRF2, is important for their telomeric localization. Remarkably, immunofluorescence with BubR1 antibodies in prometaphase cells showed that the previously described colocalization of BubR1 with TRF1 at telomeres,47 as well as the well characterized localization of BubR1 at kinetochores, was significantly decreased and in some cases completely lost in the absence of TRF1 (Fig. 5A). In particular, Trf1+/+ cells in prometaphase showed a BubR1 pattern consisting of discrete nuclear foci that was largely abolished in Trf1−/− cells (Fig. 5B). To confirm that this delocalization was specific to TRF1 depletion, and not to the severe telomere dysfunction and DNA damage caused by TRF1 deficiency, we studied BubR1 localization in MEFs derived from third generation (G3) telomerase-deficient mice (Terc−/−), which also show telomeric dysfunction owing to the presence of critically short telomeres.56 In this instance, however, the normal discrete foci pattern of BubR1 was not affected and colocalized with the centromeric marker ACA (Fig. 5C). The unaltered localization of BubR1 in G3 Terc−/− cells suggests that the presence of dysfunctional telomeres, per se, is not the cause of the above-noted delocalization of BubR1; instead, this should be attributed to the depletion of TRF1. To further ensure the observed phenotype was specific to the absence of TRF1 and not of other shelterin components, we examined the localization of BubR1 in the cells lacking another TRF2 (Fig. 5D and E). Previous reports have shown that depletion of TRF2 affects the telomeric binding of Rap1.57 We used the absence of Rap1 nuclear foci as readout for cells lacking TRF2, and found that BubR1 centromeric localization, as seen by its co-localization with ACA, was not affected (Fig. 5D). We also assessed TRF1 localization in these cells and found that it was not affected, and that we could still find BubR1 colocalizing with TRF1 foci (Fig. 5E).
Figure 5. BubR1 delocalizes in the absence of TRF1. (A) Representative images of BubR1 (green), TRF1 (purple), ACA (anti centromere antibody) (red) and Dapi (blue) in Trf1+/+ p53−/− and Trf1−/− p53−/− MEFs. (B) (Left) Quantification of the percentage of cells that display the punctuate centromeric and telomeric localization of BubR1. More than 15 cells from 3 independent MEFs per genotype were scored. Statistical significance was calculated using the Fisher exact test; P values are indicated. (Right) Quantification of the number of TRF1/BubR1 foci that colocalize in Trf1+/+ p53−/− MEFs. (C) Representative images of BubR1 (green) and ACA (red) in G3 Terc −/− MEFs. (D) Representative images of Rap1 (pink), BubR1 (green), and ACA (red) in Trf2−/− p53−/− MEFs. (E) Representative images of TRF1 (pink), BubR1 (green), and ACA (red) in Trf2−/− p53−/− MEFs. Bars = 10 μM.
Together, these results indicate that members of the SAC can localize to telomeres in a TRF1-dependent manner. In addition, we find that localization of BubR1 to the kinetochores is also dependent on TRF1 but not on telomere length or the TRF2 shelterin components, thus suggesting a specific role for TRF1 in the proper localization of SAC proteins in the cell.
TRF1 is important for the correct completion of mitosis
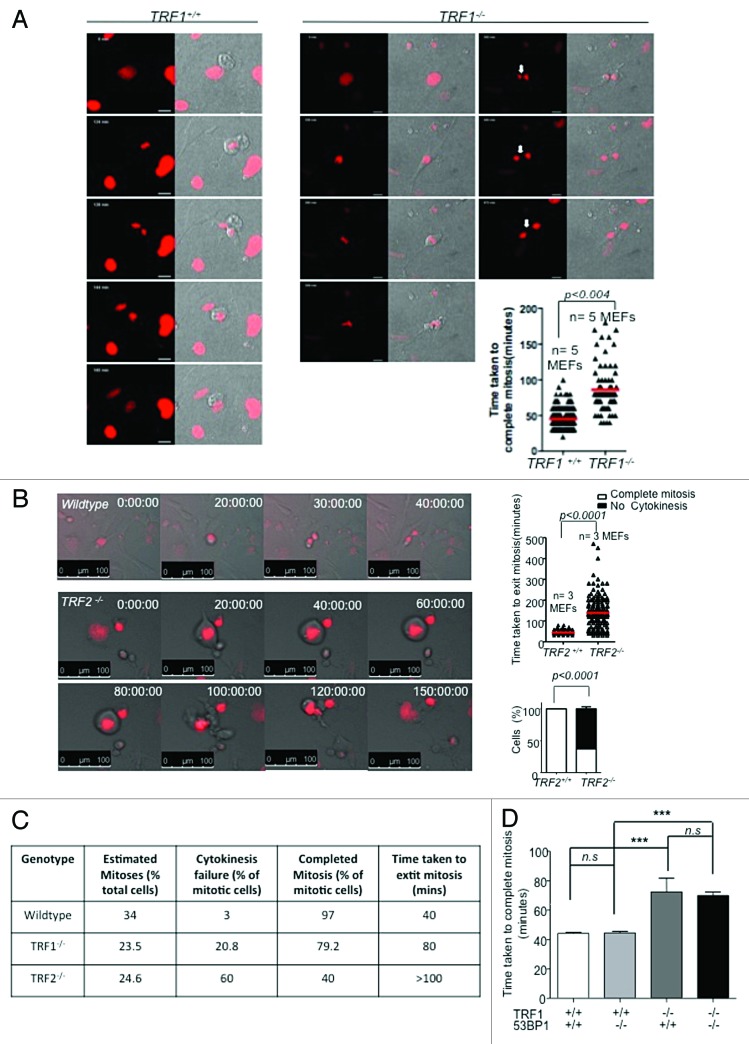
A study by Malureanu et al. suggests that the main function of BubR1 is during interphase and not through kinetochore binding.45 In particular, during interphase, BubR1 is proposed to act as a pseudosubstrate inhibitor of APC/CCdc20, thus preventing unscheduled degradation of specific APC/C substrates.45 Intriguingly, BubR1 deficiency was found to interfere with a cell’s ability to cope with genotoxic stress.58 Cells heterozygous for BubR1 were unable to undergo significant mitotic arrest in the presence of DNA damage.58 However, there are studies that have shown that persistent telomere damage signaling due to telomere uncapping occurs when TRF2 is deleted from Lig4−/− (NHEJ-deficient) cells55 as well as in POT1a/b doubly deficient cells,59 causing these cells to bypass mitosis. To further explore possible mitotic defects in the absence of TRF1, we used time-lapse live-cell imaging of asynchronously growing Trf1−/−p53−/− MEFs as well as the corresponding wild-type controls. During the 2-d imaging session, we found that, whereas control cells divided normally, taking on average 40 min to complete mitosis, TRF1-deleted cells also divided but took approximately 80 min to complete mitosis, and a large fraction divided into multiple nuclei remaining connected by chromatin bridges (Fig. 6A). We observed nuclear envelope breakdown and chromosome condensation in H2B-Cherry-expressing cells in the absence of TRF1 (Fig. 6A).
Figure 6. (A) Time-lapse images of Trf1+/+ and Trf1−/− or (B) Trf2+/+p53−/− and Trf2−/− p53−/− MEFs treated with Cre. Selected time points are shown. Cells are transfected with H2B-CHERRY (red). Arrow indicates chromatin bridge. (Right panel) Quantification of the time taken to exit mitosis of 5 independent Trf1+/+ and Trf1−/− MEFs. Error bars represent standard error. (B) Quantification of the time taken to exit mitosis (top) and the percentage of cells that complete cytokinesis (bottom) of 3 independent Trf2+/+p53−/− and Trf2−/−p53−/− MEFs. (C) Table of comparison of percentages of mitotic cells and time taken to exit mitosis in wild-type, Trf1−/−, and Trf2−/− MEFs. (D) Quantification of the time taken to exit mitosis of 2 independent Trf1+/+ and Trf1−/− MEFs either wild-type or knockout for 53BP1. Error bars represent standard error. Student t test was used for all statistical analysis, and P values are indicated. ***Represents a P value of < 0.0001; n.s., non-significant.
Based on the above-described role for TRF1 in the localization of SAC proteins to telomeres and the observed mitotic delay, together with previous reports suggesting that disruption of the telomeric structure causes not only mitotic arrest but also mitotic abnormalities,11,21,47,59 We more closely examined the effect of TRF1 abrogation. To this end, we deleted TRF1 in Trf1Lox/Lox MEFs simultaneously deficient in p53 (Trf1Lox/Loxp53−/− cells) to avoid the potential S-phase arrest caused by the DNA damage response (DDR) induced by TRF1 deletion.11,12,60,61 Interestingly, Trf1−/−p53−/− MEFs showed polyploidization as characterized by FACS profiles, showing a discrete 8n peak (Fig. S2A, left panel). The polyploid cell fraction (defined here as the fraction of cells with a DNA content > 4n) increased from a basal level of 9–10% to 25–30% at day 8 after TRF1 deletion (Fig. S2A, right panel). In addition, 40% of TRF1-knockout cells were multinucleated and accumulated supernumerary centrosomes (Fig. S2B–E). These results suggest that TRF1 depletion may result in abnormal mitosis.
As overexpression of TRF1 is known to cause mitotic spindle defects,47 and TRF1 is reported to associate with the mitotic spindle in HeLa cells,62 we next examined spindle integrity by using α-tubulin to label the spindle in control and Trf1−/−p53 −/− MEFs. We found that TRF1-knockout cells had multiple centrosomes, and these were able to initiate spindle formation resulting in multi-polar spindles (Fig. S2E), thus suggesting that TRF1 was not necessary for spindle formation.
To address whether the delay in mitosis observed in the absence of TRF1 is due to increased telomere damage, as previously described for Pot1a/1b double deficiency,59 we examined the time taken to complete mitosis in cells with increased telomere damage owing to telomerase deficiency and presence of critically short telomeres, such as late generation (G3) Terc−/− MEFs (G3) (Fig. S4),4,56 as well as in cells deficient for TRF2, TRF2−/− p53−/− MEFs55 (Fig. 6B). We found that cells lacking telomerase took on average 50 min to complete mitosis compared with 40 min in the case of the wild-type controls, and most of the cells (>90%) were able to complete mitosis (Fig. 6B; Fig. S4). Surprisingly, TRF2−/− p53−/− MEFs also entered mitosis, rounding up and displaying nuclear envelope breakdown, but, in the majority of the cases, the cells flattened out again after an average of >100 min, with approximately 60% of the cells being unable to undergo cytokinesis, resulting in their nuclei becoming enlarged (Fig. 6B and C). Although the percentage of cells that enter mitosis is very similar between TRF1−/− p53−/− and TRF2−/− p53−/− MEFs, (23.5% and 24.6%, respectively), the percentage of these that do not undergo cytokinesis is markedly higher in the absence of TRF2 (60% vs. 20.8%) (Fig. 6C). Cells lacking TRF2 did not show the appearance of chromatin bridges as seen in the absence of TRF1 (data not shown). The severity of the mitotic phenotype in TRF2 and TRF1 knockout cells vs. Terc-deficient MEFs suggests that the mitotic delay phenotype is likely to be related to the increased telomere damage in TRF1- and TRF2-deficient cells compared with the late-generation Terc−/− mice (see below).
In order to address whether the mitotic phenotype was related to the previously described increase in telomere fusions upon TRF1 deletion,11,12 we analyzed by time-lapse microscopy the mitotic duration in cells doubly deficient for the NHEJ pathway component 53BP1 and for TRF1.63 The absence of 53BP1 was not able to rescue the delay in mitosis observed in TRF1-knockout cells (Fig. 6D), implying that the delay is not due to fusions per se, but could be related to the induction of the DNA damage response at dysfunctional telomeres.
The SAC is responsible for mitotic delay associated to TRF1 ablation
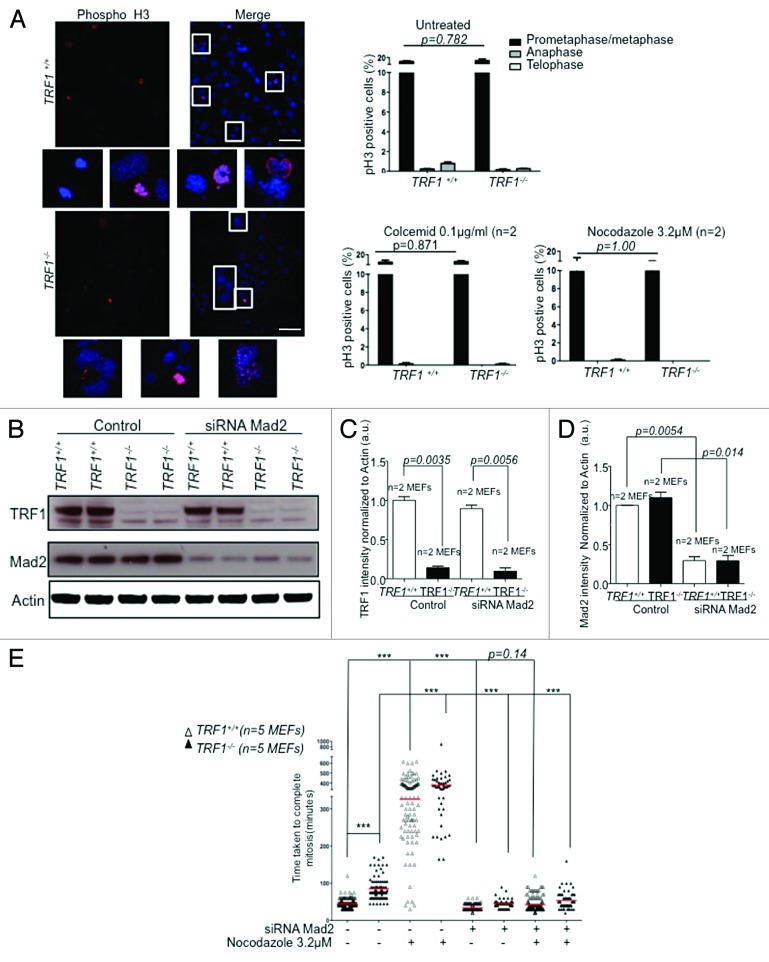
TRF1 depletion causes extensive DNA damage, resulting in a rapid DDR.11,12 Previous studies have reported that massive DNA damage causes a p53-independent delay in the division process.64 The authors found that the observed transient metaphase block was due to activation of the SAC, and that the delay could be rescued by Mad2 depletion.64 To further analyze the mechanisms by which TRF1 deficiency results in extended time to complete mitosis, we analyzed SAC activation in TRF1-deficient cells. To this end, we treated wild-type and TRF1−/−p53−/− MEFs with the microtubule depolymerizing agents nocodazole and colcemid, which activate the SAC, leading to mitotic arrest. We first stained cells with pH3 and determined the mitotic index and found no significant differences between wild-type and TRF1-knockout cells in their ability to activate the SAC in response to the mitotic poisons, as indicated by a similar percentage of cells arresting in prometaphase in wild-type and TRF1-deficient MEFs (Fig. 7A). Next, we set out to address whether abnormal SAC activation in TRF1-deficient cells was responsible for the increase duration of mitosis. We disrupted the SAC in both wild-type and TRF1-null cells by using an siRNA against the essential SAC component Mad2 (Fig. 7B). We achieved a 70% decrease in Mad2 protein levels as indicated by western blot (Fig. 7B–D). To address whether the increased time to exit mitosis in the absence of TRF1 was mediated by the SAC, we analyzed the time taken to complete mitosis in wild-type and TRF1−/−p53−/− MEFs as compared with cells in the presence of nocodazole alone, with the siRNA against Mad2 alone, and with a combination of both siRNA and nocodazole. We found that TRF1-knockout cells responded to treatment with nocodazole, as did control cells, activating the SAC due to spindle depolymerization and delayed completion of mitosis. This confirmed that the SAC is fully functional in the absence of TRF1. When the SAC was disrupted by the siRNA against Mad2, the observed delay in mitosis in TRF1-deficient cells was rescued, and cells completed mitosis at the same rate as their wild-type counterparts even in the presence of the same amounts of chromosomal aberrations and genome instability. As control, the delay in mitosis in the presence of nocodazole was also reversed upon knockdown of Mad2 (Fig. 7E). Taken together, these data suggest that TRF1 deficiency causes activation of SAC-dependent mitotic arrest, and this is responsible for the delay in the completion of mitosis of TRF1-deficient cells. This is in agreement with the studies performed by Mikhailov et al.64 It is relevant to point out that the abnormal localization of SAC proteins in the absence of TRF1 (lower amounts at telomeres) does not affect SAC function (see “Discussion”).
Figure 7. TRF1 knockout cells have a functional SAC. (A) Trf1−/− MEFs display an efficient mitotic assembly checkpoint in the presence of colcemid or nocodazole. (Right) Percentages of pH3-positive cells after indicated treatments. n refers to independent MEF cultures. Bars = 50 μM (Left) Representative images of Trf1+/+ and Trf1−/− MEFs pH3-positive cells. Nuclei are counterstained with Dapi (blue). Two-way ANOVA used to determine statistical significance. P values are shown. (B) Western blot of Trf1+/+ and Trf1−/− MEFs treated with scrambled oligo (control) or with an siRNA against Mad2. (C) Quantification of TRF1 and Mad2 (D) normalized to Actin. Student t test was used to determine significance. P values are shown. (E) Trf1+/+ and Trf1−/− MEFs transfected with either scrambled siRNA or an siRNA against Mad2 and subjected to mitotic arrest by nocodazole and analyzed by time-lapse video microscopy. Dot plot of elapsed time (min) between NEBD and anaphase onset for individual cells. Analysis performed on >50 cells per condition from 5 independent MEFs. Statistical analysis using the Student t test. ***Represents a P value of < 0.0001 all other values are indicated.
Unlike for Pot1/1b double deficiency, TRF1-deficient cells show normal APC/Cdc20 activity
If cells are to proceed from metaphase to anaphase, then cohesin must be cleaved due to the activation of the ubiquitin ligase APC/Cdc20. The mechanism behind the delay observed in vertebrate cells with DNA damage is as yet unclear. However, a proposed hypothesis is that DNA damage is able to prevent the activation of APC as is the case in Drosophila.65 To examine the status of the mitotic checkpoint, we studied APC/Cdc20 mitotic targets in synchronized Trf1−/−p53−/− MEFs. Upon release from G1/S blockage, both control and TRF1 knockout cells entered S phase, as shown by FACs profile (Fig. S3A), but, as expected, TRF1 knockout cells exhibited an increase in the number of 4n cells and in polyploid cells compared with control cells. We first examined by western blot the protein levels of Geminin, a negative regulator of replication and an APC/Cdc20 target, and Aurora B, a member of the chromosomal passenger complex, and saw no significant alteration in the stabilization of these proteins in the absence of TRF1, contrary to what has been reported for the POT1a/b doubly deficient cells (Fig. S3C–E).59 We also confirmed a normal localization of Aurora B in the absence of TRF1. In particular, we found Aurora B to localize to the centromere during prometaphase, the midbody in anaphase/telophase, and at the cleavage furrow in both wild-type and TRF1-deleted cells (Fig. S3D and E). Upon closer examination, we also found no alterations in the kinetics of other APC/Cdc20 targets such as Cyclin B1 or Securin. Interestingly we were also unable to detect any differences among Aurora A levels, which were previously reported to cause mitotic abnormalities that are regulated by TRF166 (Fig. S3A–C). These studies reveal that the observed mitotic delay induced upon TRF1 abrogation is not correlated with an inability to activate APC.
Discussion
The repetitive nature of telomeres confers them a high risk of DNA replication stalling, a phenomenon that is known as fragility. Indeed, both in yeast and mammals replication forks were found to stall at telomeres.16-19 In particular, replication stress induced by drugs that make replication suboptimal, or by decreasing the amounts of ATR, causes increased fragility at telomeres.11,12,20 Furthermore, shelterin proteins, which are important for telomere protection, have been also recently found to prevent telomere fragility.11,12,20-23 Here, we are the first to show that TopoIIα is able to directly bind to telomeres during mitosis in a TRF1-dependent manner. Furthermore, we find that TopoIIα catalytic inhibition results in increased telomere fragility as measured by abundance of MTS and UFB, comparable to that seen upon abrogation of TRF1. Interestingly, previous reports suggested a role for TopoIIα during telomere replication,26,32 which is in line with the previously described role for TRF1 in allowing proper telomere replication.11,12 Thus, our findings suggest that unreplicated DNA or unresolved homologous recombination in the absence of TRF1 maybe the consequence, at least in part, of decreased TopoIIα binding to TRF1-depleted telomeres.
We also confirm the previously reported association of TRF1 with SAC proteins46,47 at telomeres, and that this is strictly dependant on the presence of TRF1. The SAC plays a crucial role in ensuring correct microtubule attachment before cell division, preventing cells from progressing through to anaphase if attachment is incomplete or incorrect. Here, we are the first to show that the localization of SAC proteins to telomeric chromatin in vivo is disrupted upon TRF1 deletion. In the absence of TRF1, cells show a significantly increased time to complete mitosis, which is dependent on SAC function. We postulate that TRF1 links TopoIIα and SAC proteins at telomeres, ensuring correct telomere replication and mitotic segregation. These findings clarify the molecular mechanism by which TRF1 deficiency results in telomere fragility and mitotic defects.
A TRF1-dependent Topoisomerase IIα telomeric localization
The absence of TRF1 has been shown to cause telomeres to behave as fragile sites as a result of inaccuracies during telomere replication.11,12 TRF2, another shelterin component, has been suggested to work in a complimentary manner with Apollo and TopoIIα in relieving topological stress during telomere replication.32 In this study, the authors postulate that perhaps TRF1, and possibly other shelterin components, are enriched at telomeric regions encountering replication problems and suggest that telomeres are more sensitive than the rest of the genome to the lack of topoisomerase 2 activity. Our data supports this hypothesis, as we are able to detect TopoIIα at telomeres in wild-type cells. However, this is significantly impaired in the absence of TRF1. This could explain the presence of increased multitelomeric signals detected in the absence of TRF1 and the fragile site-like behavior observed.11,12 Upon inhibition of TopoIIα with a catalytic inhibitor and, more specifically, with shRNAs against TopoIIα, we see an increase in the presence of DNA threads, chromosomal fusions, and multitelomeric signals in wild-type cells comparable to that seen in TRF1-deleted cells (Figs. 1 and 3). It is interesting to note that studies using catalytically inactive mutants of TopoIIα found that it was still able to bind chromosomal DNA throughout the cell cycle like the wild-type form.67 The authors intimate that the catalytic ability of TopoIIα can be independent of its structural function.67 Strikingly the catalytic inhibition of TopoIIα in wild-type cells led to an increase of these aberrations to levels equivalent to those seen in untreated TRF1 knockout cells. Of note, the increase in DNA threads is proportional to the increase in multitelomeric signals seen per metaphase. This is in agreement with earlier reports, where the frequency of UFBs was found to correlate with telomere replication defects.68 Our data strongly implies a role for TopoIIα in the resolution of MTS, and that multitelomeric signals are caused by incomplete resolution of catenated DNA that, if unresolved, leads to DNA threads. Interestingly, there is evidence to suggest that the inhibition of TopoIIα by ICRF-193 is able to induce DNA damage and prolong the duration of mitosis in certain cell types.64 This may be in part due to differences in telomere capping and protection. Those cell lines that have short, deprotected telomeres and therefore less TRF1 are more sensitive to replication stalling agents, resulting in a delayed exit from mitosis. Of note, although the telomeric localization of TopoIIα is impaired, there is an overall increase in the amount of TopoIIα in the nucleus of TRF1 knockout cells (Figs. 1E and 2A). This could be a result of cells not being able to repair the stalled forks and overcompensating by increasing the total amount of TopoIIα.
TopoIIα has been implicated in the resolution of centromeric catenates via a pathway involving BLM helicase and PICH.69,70 The absence of either BLM or PICH helicases results in a lack of recruitment of TopoIIα to centromeres and the formation of ultrafine bridges.70 It is believed that PICH and BLM collaborate to keep anaphase DNA threads free of nucleosomes, providing a window of opportunity for TopoIIα, and possibly other enzymes, to resolve these aberrant DNA threads.69 In general, UFBs do not contain histones and cannot be visualized by Dapi. In cells depleted for either BLM or PICH, these DNA threads were found to contain histones69 much like the threads we see in the absence of TRF1. However, we were still able to see localization of PICH to these threads, albeit through the use of a PICH–GFP plasmid. This suggests that appearance of these histone-rich threads could be a result of impaired BLM localization. PICH- and BLM-positive threads have been shown to evolve from inner centromeres39 as well as those that extend from telomeric foci.68 BLM has been found to be important for telomere maintenance in cells using alternative lengthening of telomeres,71,72 and more recently in the suppression of telomere fragility.12,22 It is not unreasonable to believe that TRF1 recruits TopoIIα to telomeres, allowing it to function in the same or parallel pathway to PICH and BLM in the presence of replication intermediates, thus resolving telomeric DNA catenates.
TRF1 interacts with SAC proteins and is important for their telomeric localization
PICH has also been found to be an essential component of the spindle assembly checkpoint (SAC). This led us to more closely examine previously suggested interactions of TRF1 with the SAC protein Mad1,46 as well as with Mad2, another member of the SAC. Using a cellular system in which TRF1 is endogenously abundant (i.e., iPS cells) and by using co-immunoprecipitation, we find that TRF1 does appear to interact with both Mad1 and Mad2. Furthermore, by using chromatin immunoprecipitation we found binding of Mad1 and BubR1, another important component of SAC, and to a lesser extent, Mad2 to telomeres in vivo. The fact that we were unable to detect any alteration in Mad2 binding to telomeres may suggest that its localization to telomeres is not mediated by TRF1. Alternatively, the interaction between TRF1 and Mad2 may be weak and not reliably detectable by chromatin immunoprecipitation. Indeed, we previously reported co-localization of TRF1 and both Mad2 and BubR1 at telomeres only upon overexpression of TRF1.47
Importantly, here we make the unprecedented finding that binding of Mad1 and BubR1 to telomeres in vivo is largely dependent on TRF1. We confirmed this finding by immunofluorescence in the case of BubR1. In particular, the characteristic BubR1 punctuate staining is abolished in the absence of TRF1. Interestingly, the defective BubR1 localization was not observed in cells with telomere damage owing to TRF2 deficiency or telomerase deficiency and presence of critically short telomeres, suggesting that the phenotype is specific for cells with TRF1 abrogation. Fascinatingly, in Drosophila HOAP mutants with uncapped telomeres, BubR1 is found both at kinetochores and telomeres.73 In the case of Drosophila, the accumulation of BubR1 at uncapped telomeres was mediated by proteins associated to the DDR, and proposed to activate the SAC either through the negative regulation of Cdc20 or other APC/C subunits causing a metaphase arrest.73 However, the situation in mammals seems to be different, as we see decreased levels of both Mad1 and BubR1 at TRF1-abrogated telomeres, and this is concomitant with SAC activation and SAC-mediated delay in the time to complete mitosis. BubR1 has been proven to be an important protein not only for ensuring correct distribution of genetic material, but also in lifespan. Mice hypomorphic for BubR1 were found to develop many age-related phenotypes, such as short lifespan, growth retardation, sarcopenia, and subdermal fat loss and infertility.74,75 It is interesting to note that these phenotypes are very similar to those observed in late generation mice deficient for telomerase.76,77 The similarity in phenotypes between the above-mentioned mouse models and the increased severity in the BubR1 mouse may be due to increased telomere fragility resulting in increased rates of telomere attrition resulting from a lack of surveillance. However, more detailed analysis of the telomeric phonotype of the BubR1 mutant mouse is necessary to confirm this hypothesis.
BubR1 is an essential protein, as the knockout is embryonic lethal.78 This may be due to its multiple functions as a mitotic checkpoint control protein, an active kinase at unattached kinetochores, and as a cytosolic inhibitor of APC/Ccdc20.44,79-81 These functions have been uncoupled, and it has been proven that the N terminus of BubR1 is essential for APC/Ccdc20 inhibition during interphase45 and that CDK1 phosphorylation of BubR1 is important for kinetochore–microtubule attachements.82 However, the GLEBS domain is essential for both functions. Mutations in this domain resulting in faulty binding to Bub3 result in reduced accumulation at kinetochores and undetectable phosphorylation culminating in congression and checkpoint defects.82 Cells lacking TRF1 show reduced BubR1 accumulation at kinetochores as well as at telomeres and mitotic timing defects. This could be due to impairment of BubR1–Bub3 binding through modification of the GLEBS domain or structural changes in the protein resulting in our observed phonotype. Of note, Mad1 was also found to have a SAC-independent function in Drosophila.83 We find both Mad1 and BubR1 at telomeres, where they may be either exerting a non-SAC function and ensuring correct chromosome segregation, or an undefined novel function. However, in order to ascertain this, more detailed analysis of BubR1 and Mad1 structure and binding abilities in the presence and absence of telomere binding needs to be done.
TRF1 is important for the correct completion of mitosis
A recent report by Davoli et al. shows that the absence of p53 in a persistent telomere damage scenario owing to Pot1a/1b double deficiency can result in a bypass of mitosis leading to tetraploidization.59 They also find that endoreduplication and mitotic failure occurs in human fibroblasts during telomere crisis.84 Our results indicate that deletion of TRF1 also induces a sustained and persistent telomere damage signal. In particular, cells simulatenously deficient in TRF1 and p53 become multinucleated over time and display multiple spindle forming centromeres, in agreement with persistent telomere damage being an important factor in tetraplodization. However, in contrast to Davoli et al., cells lacking TRF1 did not bypass mitosis. In fact they were able to complete mitosis, although with increased mitotic duration and the occurrence of multinucleated cells. In agreement with this, and in contrast with previous reports, we were unable to see the stabilization of APC targets such as Geminin, Securin, or Cyclin B.59 This, along with our observation that in other telomere damage scenarios we did not see the same effect on mitosis, proffers a specific role for TRF1 in the completion of mitosis. Our data support the hypothesis that telomeres may act as a mitotic-duration checkpoint responsible for eliminating cells that fail to complete mitosis properly.85 The differences we observe are most likely due to the different systems used to induce telomere destabilization. The authors in this study induced telomere deprotection through prolonged mitotic arrest using microtubule poisons or by RNAi of cohesin, sororin, and shugoshin, whereas we directly induced telomere damage through removal of TRF1. The authors saw a significant reduction of TRF2 bound to telomeres; however, although there was a slight reduction in the levels of TRF1, they did not suspect a role for TRF1 in this phenotype, as its overexpression did not rescue the TIF phenotype.85 Our data suggests that TRF1 plays an important role in the mitotic checkpoint and for the correct completion of mitosis.
The SAC is responsible for mitotic delay associated to TRF1 ablation
Further supporting the idea that incorrect resolution of stalled telomere replication is the cause for the activation of the SAC and delay in mitosis, inhibition of TopoIIα by ICRF-193 (and therefore failure to decatenate DNA) was shown to cause a metaphase arrest that is independent of DNA damage and, above all, is independent of the recruitment of kinetochore-associated SAC proteins Mad2 and Bub1.33 However, though the arrest is not due to the recruitment of Mad2 to kinetochores, it does depend on Mad2 function, which we also see. Upon depletion of Mad2 by siRNA, we were able to see a rescue in the delay in mitosis observed in the absence of TRF1 and induced by treatment with nocodazole (Fig. 6B–E). Taken together our results strengthen the hypothesis that there is a metaphase checkpoint that allows entry into anaphase only after TopoIIα-dependent DNA decatenation, and that in the case of telomeres, this is dependent on the recruitment of TopoIIα by TRF1.
Experimental Procedures
Isolation of primary MEFs
TRF1lox/lox; p53−/− and TRF1+/+; p53−/−, TRF2+/+ p53−/−, TRF2lox/lox p53−/− and G3 Terc−/− primary MEFs were isolated from E13.5 embryos. TRF1lox/lox; p53−/− and TRF1+/+; p53−/− and TRF2+/+ p53−/−, TRF2lox/lox p53−/− MEFs were infected once with retroviral Cre-recombinase as previously described,11 or with Puromycin (2 Μg/mL) selection was added 48 h after the infection. Other cell types used are listed in Table 1.
Table 1. Cells used.
| Cell type | Relevant genotype | Experiments |
|---|---|---|
| Mouse Embryonic Fibroblast (MEF)11 | TRF1+/+;p53−/−, TRF1Lox/Lox;p53−/− |
Figure 1A, C, and D Figure 2 Figure 3B and C Figure 4A Figure 5A and C Figure 6 Figure S1A Figure S2 Figure S3 |
| Mouse Embryonic Fibroblast (MEF)4 | G3 TERC−/−, G3 TERC+/+ |
Figure 4B Figure S4 |
| Mouse Embryonic Fibroblast (MEF) | TRF2+/+;p53−/−, TRF2Lox/Lox;p53−/− |
Figure 4C Figure 5B and C Figure S1B |
| Mouse Embryonic Fibroblast (MEF)63 | TRF1+/+;p53−/−; 53BP1+/+, TRF1Lox/Lox;p53−/−;53BP1−/− |
Figure 5D |
| 293T | Overexpressing TopoIIα-EGFP;EGFP-TRF1 |
Figure 1B |
| Induce Pluripotent Stem cell (iPS)53 | EGFP-TRF1 +/+, EGFP-TRF1 KI/+, EGFP-TRF1 KI/KI, |
Figure 3A |
TRF1 and TRF2 deletion in Cre-infected MEFs was checked by PCR with: Forward primer: E1-popout 5′-ATAGTGATCA AAATGTGGTC CTGGG-3′; Reverse primer: SA1 5′-GCTTGCCAAA TTGGGTTGG-3′ and excision of TRF2 was confirmed as in.55
Retroviral infections
Retroviral supernatants were produced in 293T cells (5 × 105 cells per 100 mm diameter dish) transfected with the ecotropic packaging plasmid pCL-Eco and either pBabe-Cre, or pBabe,86 as described.11 Deletion of Trf1 was confirmed by PCR with primers described in.11
Preparation of nuclear extracts and immunoprecipitation
EGFP-TRF1 KI/KI, KI/+, and +/+ iPS cells were generated as described in Schneider et al.53 30 × 106 per sample were resuspended in 5 mL of osmotic buffer (10 mM Hepes pH 7.5, 210 mM manitol, 70 mM sucrose, 1 mM EDTA, 1× protease inhibitor cocktail [Sigma], 1 mM NaVO4, 5 mM β-glycerophosphate, 0.1 M PMSF, 5 mM NaF), and cytoplasmic membrane was broken using a Dounce homogenizer. Nuclei were pelleted by centrifugation at 800 g for 10 min. at 4 °C, resuspended in 1 mL of RIPA buffer (150 mM NaCl, 10 mM Tris pH 7.5, 0.1% SDS, 1% Triton, 1% deoxycholate, 5 mM EDTA), and lysed for 30 min in a rotating wheel for 30 min at 4 °C. Nuclear lysate were sheared by sonication for 15 min (30 s ON and 30 s OFF) and then cleared by centrifugation at 13 000 rpm for 10 min. Total nuclear lysate were immnunoprecipitad using ΜMACS™ GFP Tagged Protein Isolation Kit (Miltenyibiotec; 130-091-125), following the manufactures instructions.
293T cells were transfected using X-tremeGENE transfection reagent (Roche) as per the manufacturer’s protocol. Four micrograms of pBabe-EGFP-TRF1 and 4 μg pEGFP-C3 TopoII α were used for transfection. Cells were collected 48 h after transfection, and nuclear extracts were prepared as described in reference 87. Immunoprecipitation was performed on nuclear extracts using antibodies specific for topoisomerase II α, TopoII α (ABCAM-ab45175) 4 μl, and against TRF1, rabbit anti-mouse TRF1 serum (generated in our lab) 4 μl. Equal amounts of protein (500 μg) were analyzed by gel electrophoresis followed by western blotting. Anti-Lamin B (Santa Cruz, sc-6216) was used as loading control.
Western blotting
Whole-cell and nuclear protein extracts, as described by reference 11, were used for western blot analysis. Protein concentration was determined using the Bradford assay (Sigma). Up to 50 µg of protein per extract were separated in SDS–polyacrylamide gels by electrophoresis. After protein transfer onto nitrocellulose membrane (Whattman), the membranes were incubated with the indicated antibodies. Antibody binding was detected after incubation with a secondary antibody coupled to horseradish peroxidase using chemiluminescence with ECL detection KIT (GE Healthcare) or by antibodies coupled to Alexa680 (Invitrogen) emitting light at 700 nm upon excitation.
Primary antibodies
MAD1 (Santa Cruz sc-137026), MAD2 (MBL K0167-3), TRF1 (Abcam; ab-10579), GFP (Clontech; JL-8 632380), SMC1 (Bethyl; A300-055A) p55 Cdc20 (H-175) (Santa Cruz sc-8355), Geminin (FL-209) (Santa Cruz sc-13015), Cyclin B1 (Chemicon International MAB3684), Securin (abcam ab3305), Aurora B (abcam ab2254), Aurora A (abcam ab13824), TOP II α (TopoGEN, Inc, TG2011-1), TOP II α (ABCAM-ab45175), Actin (Sigma; a2228), Tubulin Clone GTU-88 (Sigma; T6557), BubR1 (Abcam ab28193).
ChIP assay
ChIP assays were performed as previously described88 in colcemid-arrested cells. In brief, after cross-linking and sonication, chromatin from 4 × 106 cells were used per each immunoprecipitation the following antibodies: 8 μl of anti Mad1 (Santa Cruz sc-137026), 8 μl of anti Mad2 (MBL K0167-3), 8 μl of anti BubR1 (Abcam ab28193), 8 μl of anti-TopoIIα (EPY1102Y) (Abcam ab52934), and 8 μl of polyclonal rabbit anti-TRF1 serum at (homemade and described in ref. 89). The immunoprecipitated DNA was transferred to a Hybond N+ membrane using a dot blot apparatus. The membrane was then hybridized with either a telomeric probe containing TTAGGG repeats or a probe recognizing major or minor satellite sequences, which is characteristic of pericentric heterochromatin. Quantification of the signal was performed with ImageJ software (NIH). In all cases, ChIP values are represented as percentages of the total input DNA, therefore correcting for differences in the numbers of telomere repeats.
Quantitative real-time PCR
Total RNA from cells was extracted with Trizol (Life Technologies). Samples were treated with DNase I before reverse transcription, using random priming and iScript™ (BioRad) according to the manufacturer’s protocols or using Ready-To-Go You-Prime First-Strand Beads Kit (GE Healthcare). Quantitative real-time PCR was performed using an ABI PRISM 7700 (Applied Biosystems), using DNA Master SYBR-Green I mix (Applied Biosystems) according to the manufacturers protocol. All values were obtained in triplicates. Primers used are as follows: mTRF1-F: 5′-GTCTCTGTGC CGAGCCTTC-3′; mTRF1-R: 5′-TCAATTGGTA AGCTGTAAGT CTGTG-3′; Gapdh-F, 5′-TTCACCACCA TGGAGAAGGC-3′; Gapdh-R, 5′-CCCTTTTGGC TCCACCCT-3′, TopoIIα (a)_Forward 5′-TGGTCAGTTT GGAACCAGGC-3′ TopoIIα (a)_Reverse 5′-TCAGGCTCAA CACGTTGGTT-3′ TopoIIα (b)_Forward 5′-AACGAGAGAC ACATCATTGT CAG-3′ TopoIIα (a)_Reverse 5′-TCACCTTCCC TATCACAGTC C-3′
Antimitotic drugs and quantification of mitotic index
TRF1lox/lox; p53−/− and TRF1+/+; p53−/− primary MEFs were treated overnight with nocodazole (0.2 μg/ml; Sigma) or colcemid (33 nmol; Sigma) and fixed in PBS-buffered 4% paraformaldehyde at room temperature for 10 min, followed by permeabilization with PBS-0.1% Triton X-100 for 10 min. Cells were then blocked with 2% bovine serum albumin (Sigma) in PBS for 1 h at room temperature and incubated for 1 h at room temperature with a rabbit anti-phospho-histone H3 (Ser10) antibody (Upstate; 1:500). After being labeled, cells were rinsed with PBS-0.1% Triton X-100 and incubated with anti-rabbit antibody-Alexa 488 (Molecular Probes), and DNA was counterstained with DAPI. The mitotic index was calculated by scoring the ratio of positive cells for phospho-histone H3.
Time-lapse analysis
For time-lapse videomicroscopy, MEFs of the indicated genotypes expressing Histone H2B-Cherry were plated on glassbottom dishes (24 well plates; MatTek) and transfected with siGenome SMART pool against Mad2 (Thermo Scientific Dharmacon), mouse MAD2L1 M-059314-01-0005) where indicated. After 24 h, cells were followed by time-lapse microscopy in fresh medium or in the presence of nocodazole for 48 h. Image acquisition was performed using a CCD fluorescence microscope (Leica) equipped with a selective filter for Cherry emission. Images were acquired at 10 and 4 min intervals using LAS AF (Leica) software for analysis.
Flow cytometric analysis
MEFs (1 × 106) were washed twice with PBS and fixed/permeabilized with ice-cold 70% ethanol. Fixed cells were washed with PBS and resuspended in 1 ml PBS containing 0.2 μg propidium iodide and 100 μg RNase. The samples were incubated overnight at 4 °C, and the samples were acquired on a FACS Canto-II or a LSRII Fortessa (for SSEA-1) (BD) using pulse processing to exclude cell aggregates and debris. At least 20 000 events were collected per sample. Data was analyzed using FlowJo Software v.9.1 (Treestar).
Immunfluorescence
Cells were grown on coverslips or glassbottom dishes (24-well plates; MatTek), washed twice with PBS, and fixed for 10 min in PHEM buffer (20 mM PIPES pH 6.8, 0.2% Triton X-100, 10 mM EGTA, and 1 mM MgCl2) containing 4% paraformaldehyde. For immunofluorescence staining, coverslips were washed twice with PBS, and then blocked 100% goat serum albumin for 1 h before the indicated primary antibody was applied. Cells were then washed with PBS and incubated with Alexa Fluor 488-, 555-, or 647-conjugated secondary antibody (Molecular Probes). Cell nuclei were stained with DAPI (Sigma). After washing, coverslips were mounted onto glass microscope slides with Prolong antifade (Invitrogen).
Microscopy
All the fluorescence stainings were acquired in a confocal high-resolution microscope Leica TCS-SP5 (AOBS). Type, magnification, and numerical aperture of the objective lenses: 40 × 1.25 oil UV plan apochromat; 63 × 1.4 oil UV plan apochromat. Temperature: room temperature. Imaging medium: Prolong Antifade Kit (Invitrogen). Fluorochromes: AlexaFluor 488, AlexaFluor 555/568, AlexaFluor 647 (Invitrogen). Photomultiplier tubes (PMT). Stacks were taken with a step size of 0.8 Μm and maximum projected by the LAS AF (Leica) software for analysis.
Analysis of fluorescence intensities of all the staining that were analyzed was performed with Definiens Developer XD1.2 or XD 1.5 software (Definiens). Nuclei were segmented with cellenger and developer algorithms. Intensity results were exported with a developer ruleset into Excel.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Aknowledgements
M.S.d’A. is the recipient of a “Juan de la Cierva” Contract from the Spanish Ministry of Economy and Competitiveness. Research in the Blasco lab was funded by the Spanish Ministry of Economy and Competitiveness Projects SAF2008-05384 and CSD2007-00017, the European Union FP7 Projects 2007-A-201630 (GENICA), and 2007-A-200950 (TELOMARKER), the European Research Council (ERC) Project TEL STEM CELL (GA#232854), the Körber Foundation, the AXA Research Fund, “Fundación Botín” and “Fundación Lilly” (Spain). We thank Mónica Álvarez-Fernández, Manuel Eguren, Guillermo de Cárcer Díez and Scott Lowe for reagents.
References
- 1.Blackburn EH. Telomeres: structure and synthesis. J Biol Chem. 1990;265:5919–21. [PubMed] [Google Scholar]
- 2.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–34. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/S0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 5.O’Connor MS, Safari A, Xin H, Liu D, Songyang Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc Natl Acad Sci U S A. 2006;103:11874–9. doi: 10.1073/pnas.0605303103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–7. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 7.Zhong Z, Shiue L, Kaplan S, de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol. 1992;12:4834–43. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–3. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 9.Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23:6533–41. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto K, Iwano T, Tachibana M, Shinkai Y. Distinct roles of TRF1 in the regulation of telomere structure and lengthening. J Biol Chem. 2008;283:23981–8. doi: 10.1074/jbc.M802395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martínez P, Thanasoula M, Muñoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–75. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 14.Ribeyre C, Lopes J, Boulé JB, Piazza A, Guédin A, Zakian VA, Mergny JL, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juranek SA, Paeschke K. Cell cycle regulation of G-quadruplex DNA structures at telomeres. Curr Pharm Des. 2012;18:1867–72. doi: 10.2174/138161212799958404. [DOI] [PubMed] [Google Scholar]
- 16.Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–96. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makovets S, Herskowitz I, Blackburn EH. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol. 2004;24:4019–31. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–8. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 19.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–20. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 20.McNees CJ, Tejera AM, Martínez P, Murga M, Mulero F, Fernandez-Capetillo O, Blasco MA. ATR suppresses telomere fragility and recombination but is dispensable for elongation of short telomeres by telomerase. J Cell Biol. 2010;188:639–52. doi: 10.1083/jcb.200908136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tejera AM, Stagno d’Alcontres M, Thanasoula M, Marion RM, Martinez P, Liao C, Flores JM, Tarsounas M, Blasco MA. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–89. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149:795–806. doi: 10.1016/j.cell.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 23.Vannier JB, Sandhu S, Petalcorin MI, Wu X, Nabi Z, Ding H, Boulton SJ. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science. 2013;342:239–42. doi: 10.1126/science.1241779. [DOI] [PubMed] [Google Scholar]
- 24.Martinez P, Thanasoula M, Carlos AR, Gómez-López G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12:768–80. doi: 10.1038/ncb2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–36. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Germe T, Miller K, Cooper JP. A non-canonical function of topoisomerase II in disentangling dysfunctional telomeres. EMBO J. 2009;28:2803–11. doi: 10.1038/emboj.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baldi MI, Benedetti P, Mattoccia E, Tocchini-Valentini GP. In vitro catenation and decatenation of DNA and a novel eucaryotic ATP-dependent topoisomerase. Cell. 1980;20:461–7. doi: 10.1016/0092-8674(80)90632-7. [DOI] [PubMed] [Google Scholar]
- 28.Goto T, Wang JC. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J Biol Chem. 1982;257:5866–72. [PubMed] [Google Scholar]
- 29.Hsieh T, Brutlag D. ATP-dependent DNA topoisonmerase from D. melanogaster reversibly catenates duplex DNA rings. Cell. 1980;21:115–25. doi: 10.1016/0092-8674(80)90119-1. [DOI] [PubMed] [Google Scholar]
- 30.Liu LF, Liu CC, Alberts BM. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980;19:697–707. doi: 10.1016/S0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- 31.Vos SM, Tretter EM, Schmidt BH, Berger JM. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol. 2011;12:827–41. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye J, Lenain C, Bauwens S, Rizzo A, Saint-Léger A, Poulet A, Benarroch D, Magdinier F, Morere J, Amiard S, et al. TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell. 2010;142:230–42. doi: 10.1016/j.cell.2010.05.032. [DOI] [PubMed] [Google Scholar]
- 33.Skoufias DA, Lacroix FB, Andreassen PR, Wilson L, Margolis RL. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol Cell. 2004;15:977–90. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Baumann C, Körner R, Hofmann K, Nigg EA. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell. 2007;128:101–14. doi: 10.1016/j.cell.2006.11.041. [DOI] [PubMed] [Google Scholar]
- 35.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–4. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 37.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–73. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seki M, Nakagawa T, Seki T, Kato G, Tada S, Takahashi Y, Yoshimura A, Kobayashi T, Aoki A, Otsuki M, et al. Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol. 2006;26:6299–307. doi: 10.1128/MCB.00702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang LH, Schwarzbraun T, Speicher MR, Nigg EA. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma. 2008;117:123–35. doi: 10.1007/s00412-007-0131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 41.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 42.Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–59. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Malureanu LA, Jeganathan KB, Hamada M, Wasilewski L, Davenport J, van Deursen JM. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–31. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prime G, Markie D. The telomere repeat binding protein Trf1 interacts with the spindle checkpoint protein Mad1 and Nek2 mitotic kinase. Cell Cycle. 2005;4:121–4. doi: 10.4161/cc.4.1.1351. [DOI] [PubMed] [Google Scholar]
- 47.Muñoz P, Blanco R, de Carcer G, Schoeftner S, Benetti R, Flores JM, Malumbres M, Blasco MA. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol Cell Biol. 2009;29:1608–25. doi: 10.1128/MCB.01339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187:165–73. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang F, Huang Y, Dai W. Sumoylated BubR1 plays an important role in chromosome segregation and mitotic timing. Cell Cycle. 2012;11:797–806. doi: 10.4161/cc.11.4.19307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang KC, Gao H, Yamasaki EF, Grabowski DR, Liu S, Shen LL, Chan KK, Ganapathi R, Snapka RM. Topoisomerase II poisoning by ICRF-193. J Biol Chem. 2001;276:44488–94. doi: 10.1074/jbc.M104383200. [DOI] [PubMed] [Google Scholar]
- 51.Eguren M, Alvarez-Fernandez M, Garcia F, Lopez-Contreras AJ, Fujimitsu K, Yaguchi H, et al. A Synthetic Lethal Interaction between APC/C and Topoisomerase Poisons Uncovered by Proteomic Screens. Cell Rep 2014. [DOI] [PubMed] [Google Scholar]
- 52.Ishida R, Sato M, Narita T, Utsumi KR, Nishimoto T, Morita T, Nagata H, Andoh T. Inhibition of DNA topoisomerase II by ICRF-193 induces polyploidization by uncoupling chromosome dynamics from other cell cycle events. J Cell Biol. 1994;126:1341–51. doi: 10.1083/jcb.126.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider RP, Garrobo I, Foronda M, Palacios JA, Marión RM, Flores I, Ortega S, Blasco MA. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat Commun. 2013;4:1946. doi: 10.1038/ncomms2946. [DOI] [PubMed] [Google Scholar]
- 54.Boué S, Paramonov I, Barrero MJ, Izpisúa Belmonte JC. Analysis of human and mouse reprogramming of somatic cells to induced pluripotent stem cells. What is in the plate? PLoS One. 2010;5:5. doi: 10.1371/journal.pone.0012664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–8. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 56.Herrera E, Samper E, Martín-Caballero J, Flores JM, Lee HW, Blasco MA. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J. 1999;18:2950–60. doi: 10.1093/emboj/18.11.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–67. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang Y, Liu T, Wang X, Yang YM, Deng H, Kunicki J, Traganos F, Darzynkiewicz Z, Lu L, Dai W. BubR1 is involved in regulation of DNA damage responses. Oncogene. 2006;25:3598–605. doi: 10.1038/sj.onc.1209392. [DOI] [PubMed] [Google Scholar]
- 59.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–28. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carder P, Wyllie AH, Purdie CA, Morris RG, White S, Piris J, Bird CC. Stabilised p53 facilitates aneuploid clonal divergence in colorectal cancer. Oncogene. 1993;8:1397–401. [PubMed] [Google Scholar]
- 62.Nakamura M, Zhou XZ, Kishi S, Lu KP. Involvement of the telomeric protein Pin2/TRF1 in the regulation of the mitotic spindle. FEBS Lett. 2002;514:193–8. doi: 10.1016/S0014-5793(02)02363-3. [DOI] [PubMed] [Google Scholar]
- 63.Martínez P, Flores JM, Blasco MA. 53BP1 deficiency combined with telomere dysfunction activates ATR-dependent DNA damage response. J Cell Biol. 2012;197:283–300. doi: 10.1083/jcb.201110124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mikhailov A, Cole RW, Rieder CL. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr Biol. 2002;12:1797–806. doi: 10.1016/S0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- 65.Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat Cell Biol. 2000;2:90–5. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 66.Ohishi T, Hirota T, Tsuruo T, Seimiya H. TRF1 mediates mitotic abnormalities induced by Aurora A overexpression. Cancer Res. 2010;70:2041–52. doi: 10.1158/0008-5472.CAN-09-2008. [DOI] [PubMed] [Google Scholar]
- 67.Mo YY, Beck WT. Association of human DNA topoisomerase IIalpha with mitotic chromosomes in mammalian cells is independent of its catalytic activity. Exp Cell Res. 1999;252:50–62. doi: 10.1006/excr.1999.4616. [DOI] [PubMed] [Google Scholar]
- 68.Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res. 2012;40:7358–67. doi: 10.1093/nar/gks407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ke Y, Huh JW, Warrington R, Li B, Wu N, Leng M, Zhang J, Ball HL, Li B, Yu H. PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. EMBO J. 2011;30:3309–21. doi: 10.1038/emboj.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rouzeau S, Cordelières FP, Buhagiar-Labarchède G, Hurbain I, Onclercq-Delic R, Gemble S, Magnaghi-Jaulin L, Jaulin C, Amor-Guéret M. Bloom’s syndrome and PICH helicases cooperate with topoisomerase IIα in centromere disjunction before anaphase. PLoS One. 2012;7:e33905. doi: 10.1371/journal.pone.0033905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–50. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 72.Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002;11:3135–44. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 73.Musarò M, Ciapponi L, Fasulo B, Gatti M, Cenci G. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat Genet. 2008;40:362–6. doi: 10.1038/ng.2007.64. [DOI] [PubMed] [Google Scholar]
- 74.Baker DJ, Dawlaty MM, Wijshake T, Jeganathan KB, Malureanu L, van Ree JH, Crespo-Diaz R, Reyes S, Seaburg L, Shapiro V, et al. Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat Cell Biol. 2013;15:96–102. doi: 10.1038/ncb2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 76.Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–74. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 77.Samper E, Flores JM, Blasco MA. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc-/- mice with short telomeres. EMBO Rep. 2001;2:800–7. doi: 10.1093/embo-reports/kve174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–66. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 80.Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/S0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- 81.Mao Y, Desai A, Cleveland DW. Microtubule capture by CENP-E silences BubR1-dependent mitotic checkpoint signaling. J Cell Biol. 2005;170:873–80. doi: 10.1083/jcb.200505040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elowe S, Dulla K, Uldschmid A, Li X, Dou Z, Nigg EA. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J Cell Sci. 2010;123:84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- 83.Emre D, Terracol R, Poncet A, Rahmani Z, Karess RE. A mitotic role for Mad1 beyond the spindle checkpoint. J Cell Sci. 2011;124:1664–71. doi: 10.1242/jcs.081216. [DOI] [PubMed] [Google Scholar]
- 84.Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–76. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, Karlseder J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol. 2012;19:387–94. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 87.Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–12. doi: 10.1128/MCB.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.García-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet. 2004;36:94–9. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 89.Muñoz P, Blanco R, Flores JM, Blasco MA. XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet. 2005;37:1063–71. doi: 10.1038/ng1633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.