The ATPases associated with various cellular activities (AAA+ ATPases) form a broad family of multimeric ATP-driven motors that are ubiquitous and perform a vast variety of functions in living cells. These ATPases form ring-shaped structures with catalytic sites created in the interface between 2 neighboring motor subunits. Despite decades of structural and biochemical research on AAA+ and other multimeric ATPases, it remains largely unknown how their typical ring geometries enable them to perform different mechanical work on their target macromolecules. Undoubtedly the required force is produced via structural changes that are fueled by nucleotide binding, hydrolysis, and release, but knowing the distinct conformational states that are possible in these ring ATPases, and when they appear during target interaction, is required to understand the mechanism of these biological motors.
Enhancer binding proteins (EBPs) are unique bacterial transcription activators that are required to initiate transcription by σ54-RNA polymerase holoenzyme (σ54-RNAP). They have an AAA+ motor domain that directly interacts with the σ54-factor via conserved loops (GAFTGA loops). The AAA+ domains of EBPs remodel σ54-RNAP to enable its promoter-melting function. Previous structural work on EBPs characterized closed-ring and helical ensembles of the ATPase domain and showed that the ATPase subunits are highly responsive to changes of the nucleotide status of the active sites.1-4 Moreover, the functional GAFTGA loops were shown to move dramatically upon interaction with ATP.4 The structures seen in these studies displayed high levels of symmetry and thus left unanswered the question: how does an EBP ring ensemble remodel an asymmetric target complex of σ54-RNAP bound to promoter DNA?
Recently, a combination of small-angle X-ray scattering (SAXS), X-ray crystallography, and electron microscopy (EM) was used to establish yet another stable configuration of an EBP—NtrC1 from Aquifex aeolicus.5 In this state, the apo NtrC1 oligomer, a heptamer of identical monomers, gains a dramatic asymmetry upon sub-saturating binding by ATP—it forms split hexamers (Fig. 1). In the split hexamers the crucial GAFTGA loops responsible for interacting with the σ54-RNAP form a spiral staircase. The asymmetrically poised loops guide contact between the ATPase ring and a complex of σ54 and promoter DNA. It is also likely that this asymmetry directs the chemical reaction of ATP hydrolysis in particular sites of the ring oligomer.
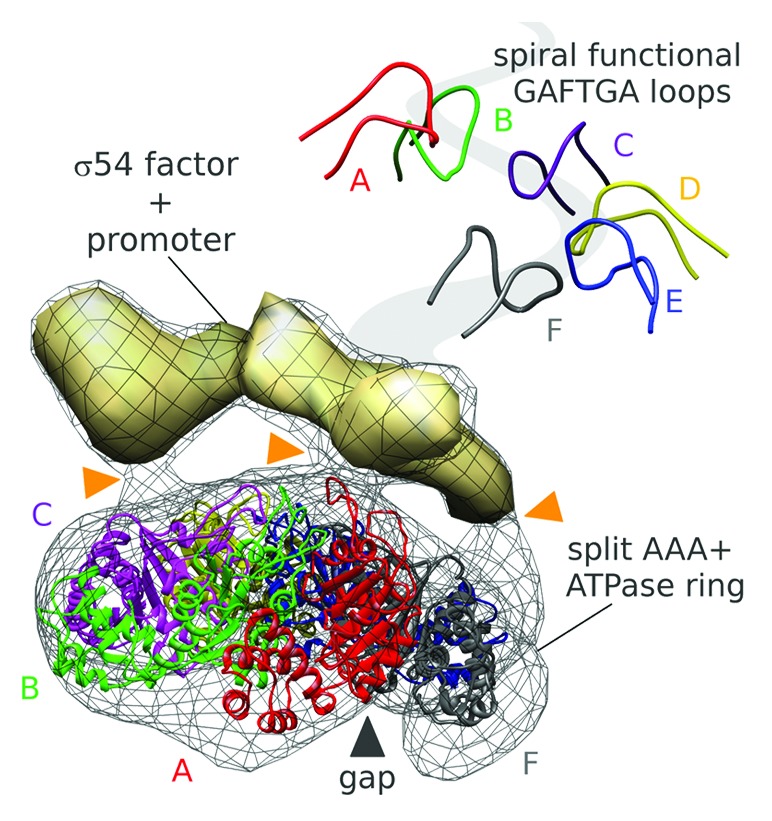
Figure 1. The functional role of asymmetry in a AAA+ ATPase homo-hexameric motor. ATP binding to the transcription activator NtrC1 (wire mesh and colored ribbons) breaks a rotational symmetry and directs interactions (orange marks) with the σ54-promoter complex (solid density). In the split hexamer the functional pore loops (top–right panel) are arranged as a spiraling staircase.
Intriguingly, the asymmetry and general features of the bacterial activator strongly resemble those in some helicases that are evolutionarily very distant—the AAA+ E1 helicase of human papillomavirus and the RecA-type Rho termination factor of Escherichia coli.6,7 The similar asymmetries in these different ATPases together with their common use of pore loops (that arise from different places in the ATPase domain fold) to bind to their respective target macromolecules may reflect convergent evolution to a general mode of action of ring-shaped ATPases.
While in case of the helicases, the functional pore loops are known to contact target a nucleic acid strand to thread it through the pore, the GAFTGA loops of the transcription activator interact with σ54-holoenzyme. The role of the pore in this case is still unclear. In the EM reconstruction of the complex of NtrC1, σ54, and promoter DNA, the σ54-factor appears to interact with at least 3 NtrC1 subunits within the split hexamer with no apparent density in the ring pore. That is suggestive of lateral binding of σ54 to the activator ring. Nevertheless, the dimensions of the pore in the NtrC1 ring and the spacing of the pore GAFTGA loops correlate closely with parameters of a protein α-helix. Currently it is not possible to rule out threading of a protein or DNA strand into the pore of the transcription activator.5 It is clear though that the asymmetry induced by non-uniform nucleotide occupancy results in loop and catalytic site specializations that provide unique interactive surface and underlying hydrolytic capacity for remodeling σ54-RNAP.
The new insights about multimeric ATPases create awareness of the allosteric complexity in macromolecular machines. Though long suspected, this complexity is only now being accessed by modern research tools. Establishing new conformational states and linking them with biochemical steps will teach us how these motors integrate their catalytic cycles with contact to macromolecular targets. The new capacity for time-resolved SAXS will aid in this endeavor, just as it gave us a glimpse at the structural transitions of the NtrC1 ATPase ring. Complementary information will become available via single-molecule approaches like high-speed AFM, dynamic TEM, targeted FRET studies, MD simulations, and eventually single-molecule diffraction. This synergetic approach will allow us to establish the underlying mechanisms of these fascinating ATP-driven macromolecular machines.
Sysoeva TA, et al. Genes Dev. 2013;27:2500–11. doi: 10.1101/gad.229385.113.
References
- 1.Lee SY, et al. Genes Dev. 2003;17:2552–63. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rappas M, et al. Science. 2005;307:1972–5. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B, et al. Structure. 2007;15:429–40. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen B, et al. Structure. 2010;18:1420–30. doi: 10.1016/j.str.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sysoeva TA, et al. Genes Dev. 2013;27:2500–11. doi: 10.1101/gad.229385.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enemark EJ, et al. Nature. 2006;442:270–5. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 7.Thomsen ND, et al. Cell. 2009;139:523–34. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]


