microRNAs are small noncoding RNAs that reduce protein output from complementary target mRNA transcripts. They are a conserved family of gene regulators in multicellular organisms, targeting much of the transcriptome.1 This pervasiveness is at odds with their generally modest repressive capacity, and suggests that microRNAs might not be cellular decision makers; rather they might ensure the stability or precision of cellular decisions.2 This attribute becomes critical when such decisions are iterated across millions of cells in a tissue or across individuals in a population. Further, microRNAs are frequently embedded in negative feedback loops, and other types of gene network stabilizing motifs. Presumably then, variability resulting from any perturbation that affects mRNA concentrations—stochastic transcription, environmental fluctuation, or genomic polymorphism—might be mitigated from influencing protein expression by homeostatic microRNA activity. This idea is particularly relevant in today’s personal genome era. The path from DNA polymorphisms with disease risk to actual disease manifestation is convoluted by “buffering” factors that potentially correct for genetic discrepancies. Therefore understanding the link between genotype and phenotype is of central importance.
Our recent study addressed the question of whether microRNAs alter the relationship between the genome and the phenome.3 To test this, we chose a robust quantitative phenotype of scutellar sensory organ (SSO) cell fate acquisition in the genetically tractable fruit fly. SSO fate is determined by high expression of a transcription factor Senseless (Sens).4 Each SSO is selected from its parent cluster of 20–30 competent cells. Subtle initial differences in Sens expression are magnified into binary outcomes of either SSO or epidermal fate.5 This is achieved by 2 mechanisms: (1) creation of a biochemical limit on Sens protein concentration that must be exceeded for a cell to acquire SSO fate; and (2) Sens inhibition in non-selected cells. Ectopic SSOs arise if either process is impaired. Thus, restricting fluctuations of Sens from erroneously triggering ectopic fate decisions is critical to restrict variation in outcome at the organismal level. Since SSO fate is determined in a strictly stereotypical fashion, variability resulting from experimental perturbations can be directly quantified as number of SSOs generated in the adults.
Sens transcripts are regulated by a conserved anti-neural microRNA miR-9a via 2 binding sites in the 3′ untranslated region. Consequently, to perturb miR-9a-mediated regulation we employed 2 parallel strategies: (1) reduction of microRNA concentration by creating miR-9a genomic heterozygotes; and (2) replacement of endogenous Sens with Sens transgenes containing mutated miR-9a binding sites. Loss of miR-9a regulation by either method led to increased variability in the number of SSOs specified in the adult population. Phenotype variability increased with progressive mutation of miR-9a binding sites on Sens. This suggested that most of the variability arose specifically due to miR-9a’s regulatory loss over Sens.
Clearly, impairing microRNA regulation uncovered pre-existing sources of biological heterogeneity. How much of this heterogeneity was a function of genomic diversity? Since such variants are heritable by nature, we performed multigenerational selection experiments to investigate this. In such selection experiments, populations progressively evolve, and the rate of evolution is a measure of the contribution that genomic diversity has on phenotypic variation.6 Previously, Drosophila subjected to selection for the most SSOs were found to transmit a heritable tendency for their offspring to have more SSOs.7 Therefore, we performed selection using animals with uniform genomic landscapes and either populations with reduced miR-9a concentration or mir-9a-resistant Sens. Both populations displayed a higher rate of evolution when compared with wild-type populations.3 Furthermore, as revealed by whole-genome sequencing, landscapes selected under reduced miR-9a conditions had 2.5-fold more variants that changed allelic frequencies due to selection. Thus, pre-existing genomic variants were more potent phenotype modifiers when miR-9a buffering of Sens was impaired. Consequently, greater enrichment of these variants was observed under a positive selection regime. Our study has therefore provided the first molecular mechanism for buffering of the genome by direct regulation of a transcription factor by a microRNA (Fig. 1).
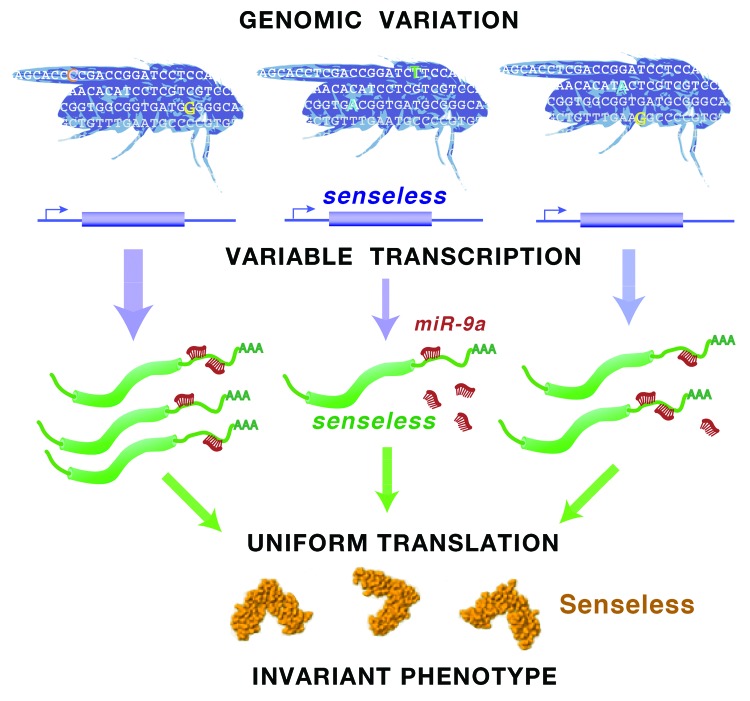
Figure 1. Cells that potentially form sensory organs transcribe Sens mRNA. This occurs through a complex gene regulatory network. Individuals bear abundant genomic diversity, some of which affects the level of Sens mRNA. Thus, Sens mRNA abundance varies between individuals. The microRNA miR-9a represses production of Sens protein by associating with Sens mRNA, normalizing Sens protein levels between individuals. This ensures a reliable outcome in choice of sensory cells, which does not vary between individuals.
The demonstrated capacity of a microRNA to effectively conceal genomic variation, and the unmasking of these variants under miR-9a heterozygosity, has implications for disease outcome. For a disease that is influenced by a person’s genome, might buffering mechanisms as we have described decouple the predictability that certain genome variants lead to disease onset? We think that this possibility is worth exploring. Consider also the progression of diseases such as cancer. Many studies confirm that loss of microRNA activity is associated with increased tumorigenic potential as well as increased resistance to therapy.8 Might this be a result of enhanced phenotypic heterogeneity in the absence of microRNAs that accelerates tumor evolution? Certain cancers, such as prostate cancer, show remarkable phenotypic tumor heterogeneity. This is coupled with genotypic heterogeneity and possibly even impaired buffering mechanisms, such as the one that we have described. It will be interesting to determine if microRNA-mediated buffering of the genome is broken when cancer ensues.
Cassidy JJ, et al. Cell. 2013;155:1556–67. doi: 10.1016/j.cell.2013.10.057.
References
- 1.Friedman RC, et al. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebert MS, et al. Cell. 2012;149:515–24. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassidy JJ, et al. Cell. 2013;155:1556–67. doi: 10.1016/j.cell.2013.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nolo R, et al. Cell. 2000;102:349–62. doi: 10.1016/S0092-8674(00)00040-4. [DOI] [PubMed] [Google Scholar]
- 5.Jafar-Nejad H, et al. Genes Dev. 2003;17:2966–78. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falconer DS, et al. (Harlow, 1996). [Google Scholar]
- 7.Rendel JM. Evolution. 1959;13:425–39. doi: 10.2307/2406126. [DOI] [Google Scholar]
- 8.Kumar MS, et al. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]