In 2006, Yamanaka et al. reported the generation of induced pluripotent stem cells (iPSc) from somatic tissue with the introduction of just 4 transcription factors: Oct4, Sox2, Klf4, and c-Myc.1 The finding, with its promise of autologous therapy, was the beginning of a vast and unprecedented effort to better understand the epigenetic basis of pluripotency and of “stemness.”2 SALL4 is a zinc finger transcription factor critical for the establishment of pluripotency in embryonic stem cells.3 Within a few years, several groups found that SALL4 enhanced the reprogramming of somatic cells to iPSc by upregulating OCT4 expression via binding to the OCT4 promoter.4 A stem cell gene, SALL4, is downregulated in differentiated adult cells, yet essential in maintaining self-renewal in hematopoietic stem cells.4
SALL4 has also been studied as a diagnostic marker of malignancy in cancer.5 This past decade has produced mounting evidence of SALL4 oncogenic potential. SALL4 is constitutively expressed in human AML and is capable of inducing AML in mice.6 More recently, high SALL4 was shown to be a marker of an aggressive subtype of hepatocellular carcinoma in humans.5 Because SALL4 promotes cell survival via transcriptional repression of PTEN, blocking the oncogenic role of SALL4 has emerged as a potential therapeutic strategy.5
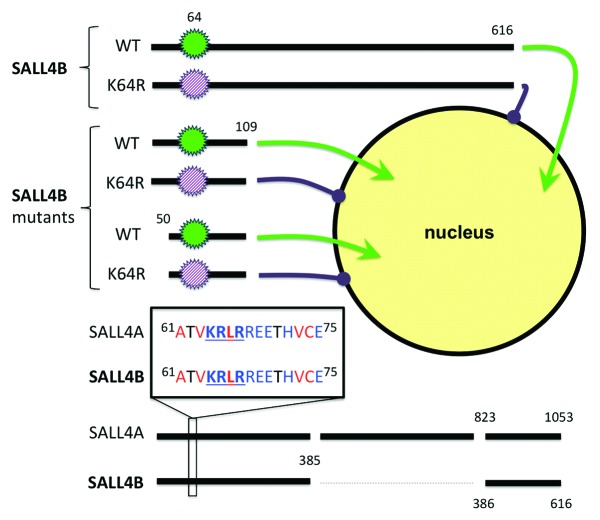
SALL4B is a splicing variant product of SALL4 with full transcriptional activity that undergoes post-translational modifications, including sumoylation. In this study, Wu et al. investigated the mechanism of SALL4B nuclear localization.7 In order to identify the necessary SALL4B regions, Wu et al. generated mutants of the SALL4 wild-type protein labeled with HA tags for a loss-of-function study. Mutants contained either unique, large lysine-rich domain deletions or unique individual point mutations, with lysine residues replaced with arginine. SALL4B mutant proteins were visualized in HeLa cells with a radioactive anti-HA tag with Hoechst nuclear labeling. Surprisingly, only a single point mutation that replaced lysine 64 with arginine (K64R) disrupted nuclear localization. K64R was visualized in the cytosol, whereas all the other mutants were localized to the nucleus, including, intriguingly, the 4R mutant designed to prevent SUMO-modification. The authors thus identified a SALL4 nuclear localization mechanism, independent of sumoylation, that required lysine 64.
Fractionation of HeLa cytoplasmic and nuclear fractions confirmed that, indeed, the K64R mutant was primarily present in the cytosol compared with only trace amounts of wild-type SALL4B. However, K64R was not limited to the cytosol, and a band against both the HA tag and SALL4B were clearly seen in the nuclear fraction. Wu et al. repeated the experiment in HEK293T cells, with similar results, only the K64R mutant was visualized outside the nucleus, and K64R was the only mutant detected via western blot in the cytoplasmic fraction. However, K64R was still detected in the in nuclear fraction and chromatin. Compared with the HeLa cells, the HEK293T cells stained for K64R appear to have more clearly delineated nuclei, opening the possibility that factors in nuclear localization vary between cell lines, potentially due to differences in nuclear porosity.
Now that it was clear that lysine 64 played a key role in localization, Wu et al. generated both K64R and wild-type mutants with truncated amino acid sequences and tested for nuclear localization. The truncated mutants were smaller than the original SALL4B 616 amino acid sequence, with one mutant truncated after position 109, and the other truncated before 50 and after 109. Both wild-type mutants were able to specifically localize to the nucleus, whereas all the K64R mutants were highly visualized in the cytoplasm. The results suggested that a localization signal existed between positions 50 and 109. A conserved motif across 7 species, including human, was identified at amino acid positions of 64, 65, and 67, reading 64KRxR67. The human SALL4B 64KRLR67 sequence matched the canonical nuclear localization signal. Via alignment, we confirmed that 64KRLR67 was also found in the SALL4A isoform (Fig. 1).
Figure 1. The KRLR sequence plays a role in SALL4 nuclear localization. A “K to R” substitution at position 64 (K64R) results in impaired SALL4 translocation into the nucleus. This remains true for SALL4B mutants with large truncation of lysine-rich domains.7 Our sequence alignment confirms that the KRLR sequence is also found in SALL4A. Charged residue, blue; hydrophobic, red.
Finally, the authors asked: does nuclear localization play a role in SALL4B-mediated OCT4 upregulation? In an OCT4-promoter luciferase assay, luciferase signal was stronger with wild-type SALL4B compared with K64R, suggesting a functional role for 64KRLR67 nuclear localization.
In this study, Wu et al. characterize a nuclear localization signal that is independent of sumoylation. The finding suggests a new regulatory mechanism of SALL4, and, by extension, of “stemness” and the establishment of pluripotency epigenetic networks.
Wu M, et al. Cell Cycle. 2014;13:1456–62. doi: 10.4161/cc.28418.
References
- 1.Takahashi K, et al. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Lengner CJ, et al. Cell Cycle. 2008;7:725–8. doi: 10.4161/cc.7.6.5573. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, et al. Nat Cell Biol. 2006;8:1114–23. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 4.Tsubooka N, et al. Genes Cells. 2009;14:683–94. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- 5.Yong KJ, et al. N Engl J Med. 2013;368:2266–76. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Y, et al. Blood. 2006;108:2726–35. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu M, et al. Cell Cycle. 2014;13 doi: 10.4161/cc.28418. [DOI] [PMC free article] [PubMed] [Google Scholar]