Abstract
Isthmin 1 (ISM1) constitutes the founder of a new family of secreted proteins characterized by the presence of 2 functional domains: thrombospondin type 1 repeat (TSR1) and adhesion-associated domain in MUC4 and other proteins (AMOP). ISM1 was identified in the frog embryo as a member of the FGF8 synexpression group due to its expression in the brain midbrain–hindbrain boundary (MHB) or isthmus. In zebrafish, ISM1 was described as a WNT- and NODAL-regulated gene. The function of ISM1 remains largely elusive. So far, ISM1 has been described as an angiogenesis inhibitor that has a dual function in endothelial cell survival and cell death. For a better understanding of ISM1 function, we examined its spatiotemporal distribution in mouse and chick using RT-PCR, ISH, and IHC analyses. In the mouse, ISM1 transcripts are found in tissues such as the anterior mesendoderm, paraxial and lateral plate mesoderm, MHB and trunk neural tube, as well as in the somites and dermomyotome. In the newborn and adult, ISM1 is prominently expressed in the lung and brain. In addition to its putative role during embryonic and postnatal development, ISM1 may also be important for organ homeostasis in the adult. In the chick embryo, ISM1 transcripts are strongly detected in the ear, eye, and spinal cord primordia. Remarkable differences in ISM1 spatiotemporal expression were found during mouse and chick development, despite the high homology of ISM1 orthologs in these species.
Keywords: ISM1, embryo, newborn, adult, mouse, chick
Introduction
Isthmin 1 (ISM1) is the founder member of a novel family of proteins first discovered in the frog embryo.1 By secretion cloning, ISM1 was identified as a secreted 60-KDa protein that was named after its prominent expression in the midbrain–hindbrain boundary or isthmus of the brain. ISM1 has strong maternal expression in the Xenopus egg, and other expression domains in the embryo include the blastopore lip, paraxial mesoderm, neural folds, cranial neural crest, and ear placode. As this pattern closely resembles that of FGF8, the authors described ISM1 as part of the FGF8 synexpression group, which also includes SPRY and SEF genes. It was thus suggested that ISM1 might be involved in the same biological processes as the members of the synexpression group. Subsequent work in the zebrafish embryo has identified ISM1 as a new target of the WNT/β-CATENIN signaling.2 Gain- and loss-of-function of WNT/β-CATENIN signaling leads to expansion or loss of ISM1 expression in the nascent mesoderm, respectively. In addition, ISM1 has also been described as a NODAL-regulated gene in the zebrafish embryo.3 In this study, it was found that ISM1 expression throughout the margin in embryos at shield stage is lost in the absence of NODAL signaling. Whether WNT/β-CATENIN and NODAL signaling pathways act independently or have a cooperative effect on ISM1 expression during zebrafish gastrulation remains undetermined. Besides its mesodermal expression, ISM1 is found in other regions of the zebrafish embryo at later stages, such as MHB, notochord, and tail bud.4
The ISM family of proteins is unique to chordates and it is characterized by the presence of 2 conserved domains: TSR1 located centrally and AMOP at the C-terminal. The TSR1 domain contains about 60 amino acids, and it was initially identified in thrombospondin 1 (THBS1).5 This domain is a common motif within the genome, and it is present in secreted or extracellular proteins, such as THSB, SPO, CCN, ADAMT, and C complement pathway proteins. The main functions reported for the TSR1 domain include regulation of cell–cell and cell–extracellular matrix interactions, cell guidance, and inhibition of angiogenesis.6 The AMOP domain is a novel extracellular domain of about 160 amino acids that was originally found in some splice variants of MUC4.7 It is uncommon in the genome, and it has been described in only 4 proteins so far: MUC4, SUSD2, ISM1, and ISM2. The function of AMOP domain remains unknown, although it has been suggested that it may have a role in cell adhesion.
The biological function of ISM1 remains largely elusive. So far, ISM1 has been described as an angiogenesis inhibitor.4 ISM1 inhibits angiogenesis in vitro and in vivo, during tumor progression as well as during generation of the trunk intersegment vessels in the zebrafish embryo. Surprisingly, the authors found that such function of ISM1 in angiogenesis is not mediated by the TSR1 but AMOP domain instead. Further work from the same group has revealed a dual function of ISM1 in endothelial cells through interaction with its cell surface receptor αVβ5 integrin.8 ISM1 exerts pro-survival effect on endothelial cells when immobilized, whereas soluble ISM1 induces endothelial cell apoptosis. How such roles of ISM1 contribute to the development of the vascular system in other species or whether ISM1 has such dual function in other cell types has not being address yet.
In our lab, we are aiming at understanding the function of ISM1 during embryonic development and adult homeostasis. In this work, a first step toward such knowledge, we have isolated and characterized ISM1 in mouse and chick. We have examined ISM1 distribution in the mouse, from early embryonic stages to adulthood. ISM1 transcripts are found in tissues such as the anterior mesendoderm, paraxial and lateral plate mesoderm. ISM1 is also found in the MHB and trunk neural tube. Additionally, ISM1 is present in the somites and dermomyotome. In the newborn and adult mouse, ISM1 is prominently expressed in the lung and brain. ISM1 is found in the epithelium of bronchi and alveoli of the lung as well as in the cerebellum, cerebral cortex and hippocampus of the brain. ISM1 may be particularly relevant for the development and homeostasis of these organs. In the chick embryo, ISM1 transcripts are strongly detected in tissues such as the anterior head mesoderm, otic and optic vesicles, lateral plate mesoderm, trunk neural tube, and dermomyotome. Despite their high protein sequence homology, we found remarkable differences in ISM1 spatiotemporal expression during mouse and chick development.
Results
Isolation and characterization of ISM1 in mouse and chick
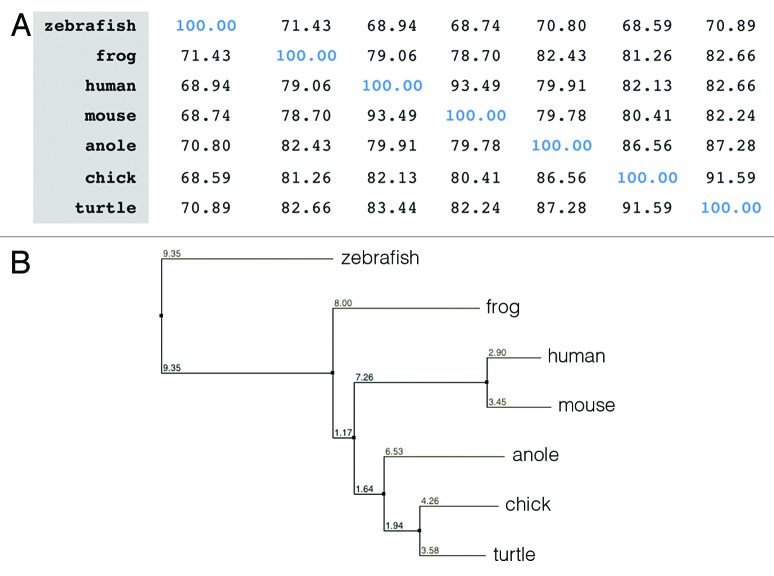
In this study, we have first identified ISM1 in both mouse and chick. In the mouse, ISM1 is located at chromosome 2, whereas in the chick, it is located on chromosome 3. The genomic sequence for ISM1 spans over 80 Kb, and it contains 6 exons and 5 introns. We have cloned the mouse full-length cDNA and the chick partial cDNA corresponding to exons 2 to 6. Their deduced amino acid sequences were used for further analysis. Mouse ISM1 encodes a 461-amino acid protein with a predicted size of 52 KDa. By Clustal Omega analysis,9 we have compared the ISM1 protein sequence between human, mouse, chick, zebrafish, frog, anole, and turtle (Figs. 1 and 2A) and found that there is a high homology between ISM1 orthologs. Mouse ISM1 shares 93, 80, and 78 percent of identity with human, chick, and frog, respectively. Chick ISM1 shares 82 and 81 percent of identity with human and frog, respectively. The most divergent region of ISM1 protein corresponds to its N terminus (from amino acid 1 to 214) and the percentage of identity between mouse and human, chick, and frog is 87, 65, and 64, respectively. Among the orthologs analyzed, ISM1 from zebrafish is the one that shows less homology. A neighbor-joining tree using percentage of identity was constructed using Jalview,10 and it shows the evolutionary relationship of ISM1 orthologs (Fig. 2B). Additional bioinformatics analyses allowed us to point out some other features of the ISM1 proteins. SignalIP3.011 identified a highly conserved signal peptide located at positions 1–29 (highlighted in blue in Fig. 1). NetNGly1.0 identified 2 putative N-glycosylation sites at positions 39 and 282. The TSR1 contains one consensus sequence for O-fucosylation.12 ISM1 proteins have a total of 14 conserved cysteine residues, 6 of which are located in the TSR1 domain (amino acids 215–159, highlighted in green in Fig. 1), whereas the other 8 are in the AMOP domain (amino acids 286–449, highlighted in orange in Fig. 1). These residues may be involved in the formation of disulphide bridges and thus contribute to the structural organization of ISM1 protein.
Figure 1. Analysis of the ISM1 protein sequence. (A) Amino acid sequence alignment of ISM1 orthologs by Clustal Omega analysis. Asterisk (*) indicates identical residues, colon (:) indicates conserved substitution of residues, and period (.) indicates semi-conserved substitution. The absence of amino acid at a particular position is indicated by dash. The signal peptide sequence is highlighted in blue, TSR1 domain in green, and AMOP domain in orange. The ISM1 protein sequences that were used are: human (H. sapiens, NP_543016.1), mouse (M. musculus, NP_001263418.1), chick (G. gallus, XP_415036.4), zebrafish (D. ranio, NM_001012376.1), anole (A. carolinensis, XM_003215294.1), turtle (C. pitca bellii, XP_005287456.1), frog (X. laevis, NP_001082228.1).
Figure 2. Analysis of the ISM1 protein sequence. (A) Percentage identity matrix of ISM1 orthologs. (B) Molecular phylogenetic analysis of the ISM1 orthologs. A neighbor-joining tree using percentage of identity was constructed using Jalview. The ISM1 protein sequences that were used are human (H. sapiens, NP_543016.1), mouse (M. musculus, NP_001263418.1), chick (G. gallus, XP_415036.4), zebrafish (D. ranio, NM_001012376.1), anole (A. carolinensis, XM_003215294.1), turtle (C. pitca bellii, XP_005287456.1), frog (X. laevis, NP_001082228.1).
Dynamic ISM1 expression during early mouse development
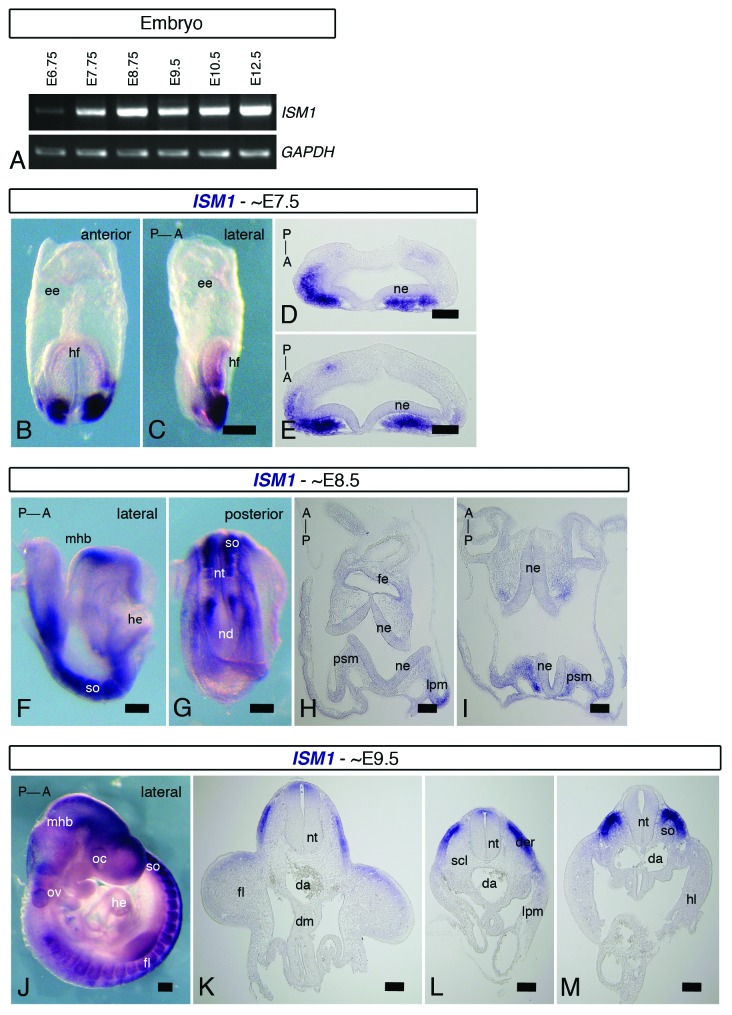
In both frog and zebrafish, ISM1 is expressed since early stages of embryonic development.1,4 In order to examine if this also occurs in the mouse, we have first conducted a temporal analysis of ISM1 expression by RT-PCR using total RNAs from embryos at several developmental stages from E (embryonic day) 6.75 to E12.5 (Fig. 3A). We found that ISM1 transcripts could be readily detected from E7.75 onwards, although low levels could already be observed in E6.75 embryos. We have next investigated the spatial distribution of ISM1 transcripts by whole mount ISH and subsequent histological analysis. In embryos at around E7.5 (head fold) stage, we found that ISM1 is expressed in the anterior mesendoderm (Fig. 3B–E). ISM1 expression seems to broaden as the embryo develops and it includes new domains. In embryos at E8.5 (4–5 somites) stage, ISM1 transcripts are found in the MHB, somites, anterior presomitic mesoderm, and lateral plate mesoderm (Fig. 3F–I). Expression of ISM1 seems to be absent from the last formed somite. Weaker staining can also be observed in the foregut endoderm. In embryos at E9.5 (28–29 somites) stage, ISM1 transcripts are found in the MHB, otic vesicle, developing somites, and neural tube (Fig. 3J–M). ISM1 expression in the developing somites is limited to the dermomyotome, where it seems to follow a gradient along the AP (anterior–posterior) axis: stronger expression is observed in more posterior regions than at more anterior positions. Interestingly, the expression in the dorsal aspect of the neural tube seems to follow an opposite gradient along the AP axis, and stronger staining is found in more anterior levels. Altogether, our spatiotemporal analysis shows that ISM1 is dynamically expressed in several structures of the mouse embryo.
Figure 3.ISM1 expression during early mouse embryonic development. (A) RT-PCR for full-length ISM1 in mouse embryos at distinct developmental stages, from E6.75 to E12.5. Strong levels of ISM1 mRNA are detected from E7.75. GADPH is used as an internal control. (B–M) ISH for ISM1 of E7.5, E8.5 and E9.5 mouse embryos. Whole-mount anterior (B) and lateral (C) views and cross-sections (D and E) of E7.5 (head fold) embryos. ISM1 is expressed in the anterior mesendoderm. Whole-mount lateral (F) and posterior (G) views and cross-sections (H and I) of E8.5 (4–5 somites) embryos. ISM1 is observed in the MHB, somites, presomitic mesoderm, and lateral plate mesoderm. Whole-mount lateral (J) view and cross-sections (K–M) of E9.5 (38–29 somites) embryos. ISM1 is found in the MHB, ear vesicle, dermomyotome, and neural tube. Orientation of the embryos along the AP axis is indicated. A, anterior; da, dorsal aorta; der, dermomyotome; dm, dorsal mesentery; ee, extraembryonic endoderm; fe, foregut endoderm; fl, forelimb; he, heart; hf, head fold; lpm, lateral plate mesoderm; nd, node; ne, neuroepithelium; nt, neural tube; oc, optic cup; ov, otic vesicle; P, posterior; psm, presomitic mesoderm; scl, sclerotome; so, somite. Scale bar = 20 μm in (B, C, F, G and J); scale bar = 50 μm in (D, E, H, and I); scale bar = 100 μm in (K–M).
Tissue and organ specificity of ISM1 expression during mouse postnatal development
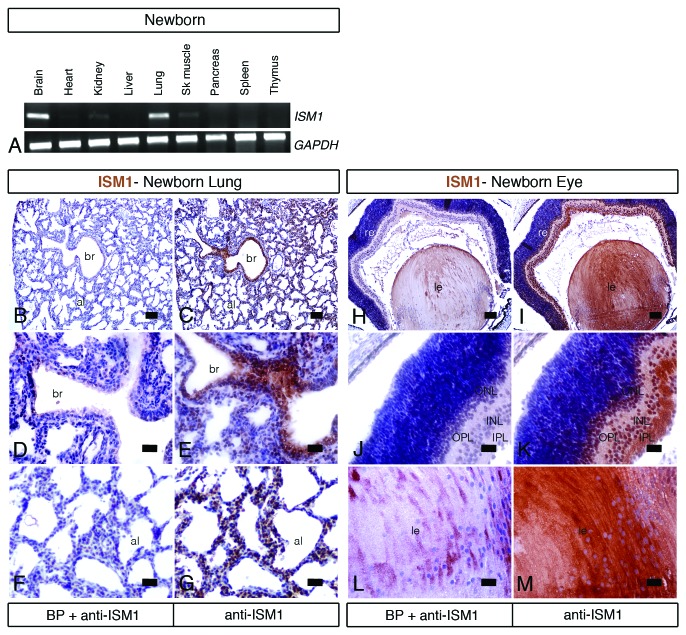
In addition to the embryonic stages, we have further examined ISM1 expression during mouse postnatal development. We have first examined ISM1 expression by RT-PCR analysis of several tissues collected from the newborn (postnatal day 0) mouse (Fig. 4A). We found high levels of ISM1 transcripts in the brain and lung tissues. Moderate to low levels are detected in the kidney, skeletal muscle, and heart. Other organs, such as liver, pancreas, spleen, and thymus, seem negative for ISM1 expression. Since the lung is one of the organs that expresses the highest levels of ISM1, we have further characterized its spatial distribution by IHC (immunohistochemistry) (Fig. 4B–G). We have used a custom produced polyclonal rabbit antibody against the synthetic peptide FPRQRFPPETGHPSC that corresponds to amino acids 81 to 94 of the mouse ISM1 protein. We found that ISM1 protein is detected in the epithelium of both the bronchi (Fig. 4C and E) and alveoli (Fig. 4C and G). In addition to the lung, we have also performed IHC analysis in the eye of the newborn (postnatal day 0) mouse. ISM1 is strongly expressed in distinct structures of the eye, such as the lens and the retina. Within the retina, ISM1 is observed in the inner plexiform layer as well as in both the inner and outer nuclear layers. As a control for the specificity of our staining, pre-absorption of the anti-ISM1 antibody with a BP (blocking peptide) reveals no staining either in the lung tissue (Fig. 4B, D, and E) or in the eye (Fig. 4H, J, and L). Taken together, our results show that ISM1 has a tissue- and organ-specific distribution in the newborn mouse: ISM1 is highly expressed in the lung, brain, and eye.
Figure 4.ISM1 expression in the newborn mouse. (A) RT-PCR for full-length ISM1 in different tissues of the newborn (postnatal day 0) mouse. GADPH is used as an internal control. Brain and lung tissues show the highest level of ISM1 mRNA. Moderate to low levels are detected in kidney, heart, and skeletal muscle. (B–M) Immunohistochemistry for ISM1 in adjacent transversal sections of the lung (B–G) and eye (H–M) of the newborn mouse counterstained with hematoxylin. Pre-absorption of the anti-ISM1 antibody with a BP reveals no staining (B, D, and E). ISM1 protein is detected in the epithelium of both the bronchi (C and E) and alveoli (C and F) of the lung. In the eye, ISM1 immunostaining is observed in the retina cell layer (K) and in the lens (M). al, alveoli; br, bronchi; INL, inner nuclear layer; IPL, inner plexiform layer; le, lens; ONL, outer nuclear layer; OPL, outer plexiform layer. Scale bar = 200 μm in (B, C, H, and I); scale bar = 50 μm in (D–G, J, K, L, and M).
Tissue and organ specificity of ISM1 expression in the adult mouse
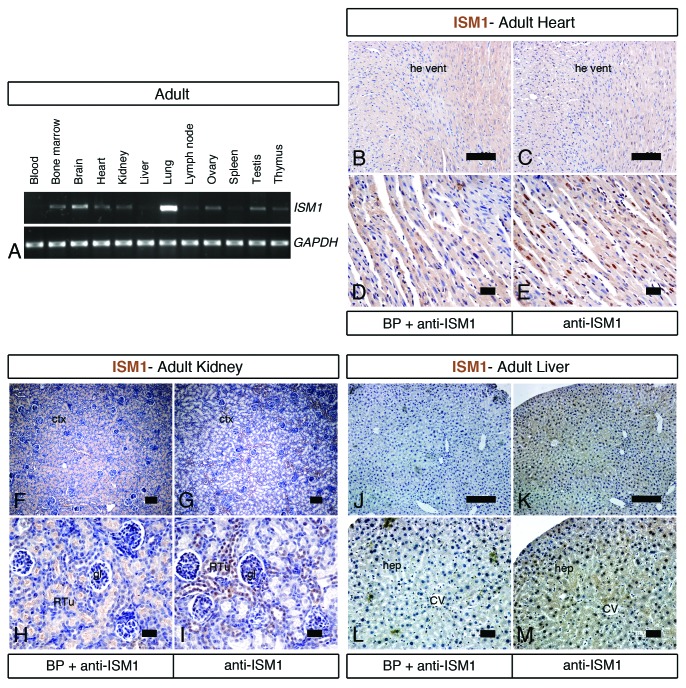
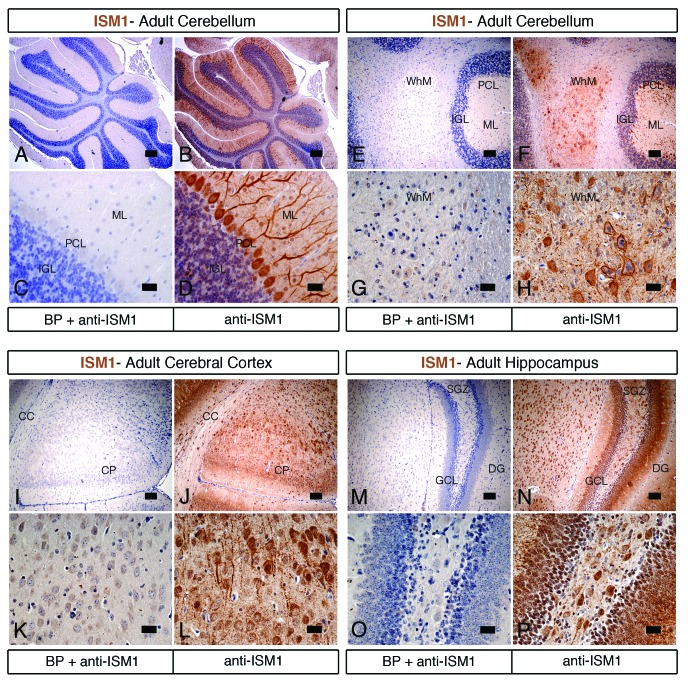
In addition to the embryonic and postnatal stages, we have further extended our study of ISM1 expression to the adult mouse. We have performed RT-PCR analysis of diverse tissues and organs collected from the adult (3-mo-old) mouse (Fig. 5A). Consistent with the results from the newborn stage, we found that the lung and the brain are the organs that express the highest levels of ISM1 transcripts. Other organs, such as the heart, kidney, ovary, testis, as well as the bone marrow, express moderate levels of ISM1. Lower levels are detected in the liver, lymph node, spleen, and thymus. No ISM1 transcripts are found in the blood. We have then examined the spatial distribution of ISM1 protein by IHC in some of these organs. In the lung and eye, IHC analysis for ISM1 reveals the same pattern as in the newborn mouse (data not shown). In the heart, ISM1-positive cells are observed in the myocardium of the ventricle (Fig. 5B–E). As only some cells are positive for ISM1, it remains to be determined whether they correspond to cardiac myocytes and/or Purkinje fibers. In the cortex of the kidney, ISM1-positive staining occurs in a subset of renal tubules (Fig. 5F–I). In the liver, ISM1 staining is weak, diffuse, and ubiquitous (Fig. 5J–M). This is consistent with the data from RT-PCR analysis showing low levels of ISM1 in the liver. Pre-adsorption of the anti-ISM1 antibody with a BP reveals no staining in the heart (Fig. 5B and D), kidney (Fig. 5F and H), and liver (Fig. 5J and K). In both newborn and adult mice, the brain is one of the organs that express the highest levels of ISM1. From IHC analysis, we found that ISM1 is strongly expressed in the cerebellum (Fig. 6A–H), cerebral cortex (Fig. 6I–L), and hippocampus (Fig. 6M–P) of the adult mouse. Within the cerebellum, ISM1-positive cells are found in the molecular, Purkinje cell, and internal granular layers (Fig. 6A–D). In addition, positive staining for ISM1 is observed in the white matter (Fig. 6E–H). Within the cerebral cortex, ISM1 staining is detected in the cortical plate cells (Fig. 6I–L), while in the hippocampus, ISM1-positive cells are found in the dentate gyrus (Fig. 6M–P). Pre-adsorption of the anti-ISM1 antibody with a BP reveals no staining in the cerebellum (Fig. 6B, D, E, and G), cerebral cortex (Fig. 6I and K), and hippocampus (Fig. 6M and O). These results suggest that ISM1 activity may be particularly required in organs such as the lung, brain, eye, or kidney.
Figure 5.ISM1 expression in the adult mouse. (A) RT-PCR for full-length ISM1 in different tissues of the adult (3-mo-old) mouse. GADPH is used as an internal control. The highest levels of ISM1 mRNA are observed in lung and brain tissues. Lower ISM1 expression levels are found in bone marrow, heart, kidney, ovary, and testis. (B–M) Immunohistochemistry for ISM1 in adjacent sections of the heart (B–E), kidney (F–I), and liver (J–M) of the adult mouse counterstained with hematoxylin. Pre-absorption of the anti-ISM1 antibody with a BP reveals no staining in the heart (B and D), kidney (F and H), and liver (J and L). ISM1-positive cells are found in the myocardium of the heart ventricle (C and E), renal tubules of cortex of the kidney (G and I), and hepatocytes of the liver (K and M). ctx, cortex; CV, central vein; gl, glomerulus; hep, hepatocyte; he vent, heart ventricle; RTu, renal tubules. Scale bar = 200 μm in (F and G); scale bar = 100 μm in (B, C, J, and K); scale bar = 50 μm in (D, E, H, I, L, and M).
Figure 6.ISM1 expression in the adult mouse brain. (A–P) Immunohistochemistry for ISM1 in adjacent sections of the brain counterstained with hematoxylin. Pre-absorption of the anti-ISM1 antibody with a BP reveals no staining in the cerebellum (A, C, E, and G), cerebral cortex (I and K), and hippocampus (M and O). In sagittal sections of the cerebellum, ISM1-positive cells are present in the molecular, Purkinje cell and inner granular layers (B and D). In coronal sections of the cerebellum, ISM1 immunostaining is observed in the white matter cells (F and H). In coronal sections of the cerebral cortex, cells positive for ISM1 are present in the cortical plate (J and L). In coronal sections of the hippocampus, ISM1-positive cells are found in dentate gyrus (N and O). CC, corpus collosum; CP, cortical plate; DG, dentate gyrus; GCL, granular cell layer; IGL, internal granular layer; ML, molecular layer; PCL, Purkinje cell layer; SGZ, subgranular zone; WhM, white matter. Scale bar = 200 μm in (A, B, E, F, I, J, M, and and N); scale bar = 50 μm in (C, D, G, H, K, L, O, and M).
Dynamic ISM1 expression during early chick development
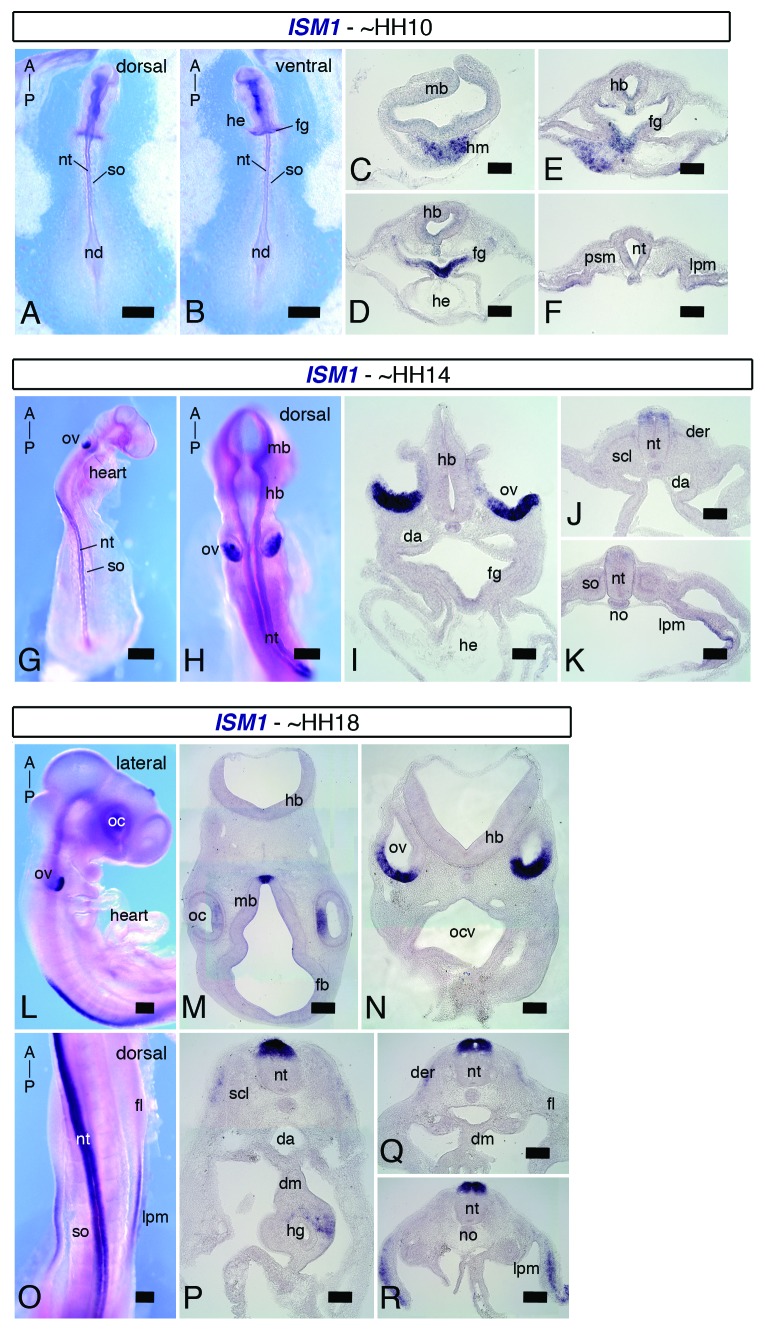
The absence of data relative to ISM1 in the mouse had led us to examine its spatiotemporal expression from early embryonic stages to adulthood. In the chick embryo, as for the mouse, almost no information relative to ISM1 is available. In fact, only 1 paper has described ISM1 as among the top 20 genes that are expressed in the anterior primitive streak of embryos at stage HH (Hamburger and Hamilton) 4.13 This prompts us to investigate ISM1 expression pattern during early chick embryonic development. We have performed whole-mount ISH and subsequent histological analysis (Fig. 7A–R). In embryos at stage HH10 (10 somites), ISM1 expression is spatially restricted to the anterior regions, such as the head mesoderm and foregut endoderm (Fig. 7A–F). In embryos at stage HH14 (21–22 somites), ISM1 transcripts are strongly detected in the epithelium of the ventral otic vesicle, and a weak expression starts to be observed along the neural tube in the anterior trunk region (Fig. 7G–K). As the embryo continues to develop, additional domains of ISM1 expression can be identified. In embryos at stage HH18 (31–33 somites), in addition to the otic vesicle, ISM1 is expressed in the epithelium of the optic cup and ventral midbrain. ISM1 is also found in the neural tube, dermomyotome, and lateral plate mesoderm (Fig. 7L–R). Interestingly, ISM1 expression in the dorsal aspect of the neural tube starts at level of somite 5–6, and it seems to follow an AP gradient, where stronger staining is observed at more anterior than posterior levels. As for the mouse, chick ISM1 is dynamically expressed in distinct structures during early embryonic development. ISM1 is particularly prominent in the ear, eye, and spinal cord primordia, and thus it may play an important role for proper generation of these organs.
Figure 7.ISM1 expression during early chick embryonic development. ISH for ISM1 in chick embryos at stages HH10, HH14 and HH18. Whole mount dorsal (A) and ventral (B) views and transversal sections (C–F) of HH10 (10 somites) embryos. ISM1 is expressed in the head mesoderm and foregut endoderm. Whole-mount dorso-lateral (G) and dorsal (H) views and cross-sections (I–K) of HH14 (21–22 somites) embryos. ISM1 is strongly detected in the otic vesicles and moderately in the dorsal aspect of the neural tube. Whole-mount lateral (L) and dorsal (O) views and transversal sections (M–R) of HH18 (28–29 somites) embryos. ISM1 mRNA is observed in the optic cups, otic vesicles, neural tube, and lateral plate mesoderm. Orientation of the embryos along the AP axis is indicated. A, anterior; da, dorsal aorta; der, dermomyotome; dm, dorsal mesentery; fb, forebrain; fg, foregut; fl, forelimb; hb, hindbrain; he, heart; hm, head mesoderm; lpm, lateral plate mesoderm; nd, node; nt, neural tube; oc, optic cup; ov, otic vesicle; P, posterior; scl, sclerotome; so, somite. Scale bar = 50 μm in (A, B and, G); scale bar = 25 μm in (H); scale bar = 20 μm in (L and O); scale bar = 100 μm in (C–F, I–K, M, N, and P–R).
Discussion
In the present work, we have focused on the study of ISM1, the founder member of a new family of proteins, the function of which remains largely unknown. We have first isolated and characterized ISM1 in mouse and chick and found that their protein sequence shows a high degree of homology (Figs. 1 and 2). We have then examined the spatiotemporal distribution of ISM1 in the mouse, from early embryonic stages to adulthood, as well as in the chick embryo. Our results show that ISM1 is dynamically expressed in several structures of the developing embryo, and, thus, it may be important for their proper development. In the mouse embryo, ISM1 transcripts are found in tissues such as the anterior mesendoderm, paraxial and lateral plate mesoderm (Fig. 3). ISM1 is also found in discrete regions of the nervous system, namely in the MHB and trunk neural tube. Additionally, ISM1 is present in the somites and its derivatives, such as dermomyotome. In the newborn and adult mouse, ISM1 is prominently expressed in the lung and brain (Figs. 4–6). In fact, it has been previously reported that ISM1 is expressed at a significantly higher level in lung comparing to other tissues.8 Our IHC analysis showed that ISM1 is widely distributed in the respiratory epithelial cells of the bronchi and alveoli and does not seem to be expressed by a particular cell type (Fig. 4B–G). Altogether, ISM1 may be important for organ homeostasis in the adult, in addition to its putative role during embryonic and postnatal development. In the chick embryo, ISM1 transcripts are mainly detected in tissues such as the anterior head mesoderm, otic and optic vesicles, lateral plate mesoderm, trunk neural tube, and dermomyotome (Fig. 7). Although there are similarities in the expression pattern of ISM1 in the chick and mouse embryo, they are distinct and present remarkable differences.
An interesting finding of our study is that ISM1 seems to have a conserved expression in the MHB in the mouse (this work), frog,1 and zebrafish,4 but not in the chick (this work) embryos. The MHB acts as an organizing center for the surrounding tissues, and it is crucial for formation of the tectum and cerebellum.14,15 Members of the FGF family of proteins, together with WNT, SHH, and TGFβ, mediate most of the organizer functions of the MHB. As initially postulated by Pera et al.,1 ISM1 may act in concert with these signaling molecules and orchestrate the elaborated cellular diversity that characterizes these brain regions. ISM1 may be particularly important for the formation of the cerebellum. The cerebellum is fundamental for motor control and cognitive functions such as attention and language.16 These complex activities depend on the action of 3 main neuronal subclasses: granule, Purkinje, and deep cerebellar neurons. We found that ISM1 is expressed in both the granule and Purkinje neurons (Fig. 6A–H). ISM1 may thus be important for development of the cerebellum as well as for its proper motor and cognitive functions. As described, we were unable to detect ISM1 transcripts in the chick MHB (Fig. 7). Several reasons may account for this striking observation. One simple explanation could be that ISM1 is expressed at low levels, and the ISH method is not sensitive enough for its detection. However, taking into account the prominent MHB expression described in frog,1 zebrafish,4 and mouse (this work), this hypothesis seems unlikely to us. Other possibility is that it could reflect species-specific differences in the function of the MHB and/or tectum and cerebellum development, yet the current understanding is that the MHB function in organizing brain development is generally conserved among species.15 Another possibility is that the putative function of ISM family of proteins in brain development may be played by ISM2 at the expense of ISM1 in the chick embryo. The existence of the paralog ISM2 is predicted in many species, such as mouse, chick, frog, and zebrafish. Indeed, ISM2 has been identified in the human as TAIL1 but its function remains unknown.17 Identification, cloning and analysis of the ISM2 would provide relevant information.
Another consistent difference in ISM1 expression pattern between mouse and chick is related to its expression in paraxial mesoderm and its derivatives, the somites and dermomyotome. In the mouse, ISM1 is strongly expressed in the anterior presomitc mesoderm that gives rise to the somites that also express high levels of ISM1 (Fig. 3F–I). As the somite differentiates, ISM1 expression becomes localized to the dermomyotome (Fig. 3J–M). In the chick embryo, ISM1 transcripts were not detected in the presomitic mesoderm or somites but were observed in the dermomyotome (Fig. 7O and P). The dermomyotome will form the dermis of the skin, the tendons and ligaments, as well as the skeletal muscle of the body.18 The differences observed between mouse and chick embryo may be related to the different mechanisms controlling dermomyotome specification and/or differentiation in these species. Our results suggest that ISM1 may be involved in the development of a particular paraxial mesoderm-derived cell lineage(s), such as the skeletal muscle. In fact, we found that ISM1 transcripts in the skeletal muscle of the newborn mouse (Fig. 4A) as well as in the dermis of the skin (data not shown).
In both mouse and chick embryo, ISM1 is expressed in dorsal aspect of the trunk neural tube (Fig. 3J–M and 7G–R). Interestingly, this expression starts at the level of somite 5–6 and follows a gradient along the AP axis of the embryo, where stronger staining is found in more anterior levels. These results suggest that ISM1 may contribute to spinal cord formation. The spinal cord is composed of distinct classes of neurons at precise locations along its dorsal–ventral axis as a result of the combinatorial activity of several secreted molecules.19 SHH and BMP/WNT are the key players for patterning the ventral and dorsal neural tube, respectively. As ISM1 has a regionalized distribution in the neural tube, it may be required for the generation of a particular type(s) of neurons. Such role of ISM1 may be analogous in both mouse and chick.
We believe that our detailed and thorough study on the spatiotemporal expression of ISM1 in the mouse from embryo to adulthood as well as in the chick embryo provide the foundation for a better understanding of the function(s) of ISM family of proteins, in particular of ISM1, which remains largely unexplored. Our results strongly suggest that ISM1 is likely to play other functions than its already described role as an angiogenesis inhibitor.
Materials and Methods
Mouse
Wild-type C57BL/6 mice were used in this study. All the experiments were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR) and conform to all guidelines of the Laboratory Animal Unit of the University of Hong Kong. For embryo collection, E0.5 corresponds to the morning at which the vaginal plug was observed. Pregnant females were sacrificed, and embryos were dissected in ice-cold PBS (phosphate buffered saline) and immediately transferred to Trizol (15596–026, Invitrogen) (for RNA isolation) or fixed by immersion in 4% PFA (paraformaldehyde) (for whole-mount ISH analysis). A total of 4–5 and 8–10 embryos per stage were used for RNA extraction and ISH analysis, respectively. For newborn and adult tissue collection, the mice were sacrificed and the subsequently processed as follows: for RNA isolation, excess blood was drained from the dissected organs with the help of filter paper, and the tissues were then transferred to Trizol (15596–026, Invitrogen); for IHC analysis, the mice were subjected to transcardial perfusion. Briefly, after opening of the thoracic cavity and exposure of the heart, a needle was carefully inserted into the left ventricle while a cut was made to the right atrium of the heart; PBS followed by 4% PFA were perfused, until a pale color was observed in the liver. After dissection, the organs were post-fixed in 4% PFA for 2 h (for newborn) or overnight (for adult) at 4 °C. The tissues were then processed for paraffin sectioning. A total of 2 newborn mice at postnatal day 0 and 2 adult 3 mo-old mice were used.
Chick embryos
Fertilized chick eggs were obtained from Jinan Poultry Co. (Tin Hang Technology) and incubated in a humidified atmosphere. Embryos were staged accordingly to HH table.20 For RNA isolation, embryos at different stages (from HH4 to HH24) were mixed and transferred to Trizol (15596–026, Invitrogen). For whole-mount ISH, the embryos were fixed in 4% PFA overnight at 4 °C. A total of 4–5 embryos and 8–10 embryos per stage were used for RNA extraction and ISH analysis, respectively.
Cloning of mouse and chick ISM1 cDNA
Total RNA was isolated using Trizol (15596–026, Invitrogen) according to the manufacturer’s instruction. RQ1 DNase I (M6101, Promega) was used to eliminate genomic DNA contamination from the RNA samples before synthesis of first strand cDNA. Briefly, 2 μg of total RNA were reverse transcribed using Oligo(dT)15 primer (C1101, Promega) and M-MLV reverse transcriptase (M170, Promega). The first strand cDNA was then used as template for PCR with the high-fidelity Platinum Pfx DNA Polymerase (11708–013, Invitrogen). Mouse full-length ISM1 cDNA was amplified from adult lung tissue using the following set of primers: forward 5′-CGCGAAGCTT GACTCAAGAG GATGGTGCGC C-3′ and reverse 5′-CGCGGAATTC GTGCAGTGCA TTTGTGTCTC C-3′. Chick ISM1 partial cDNA (corresponding to exons 2 to 6) was amplified from a mixed pool of embryo samples at different stages using the following set of primers: forward 5′-CGCGAAGCTT GTGTGTTTTT CCCAGAACAC GCTCA-3′ and reverse 5′-CCGCTCGAGC CCAAGTCAGT TCTTTGGCAT CCAG-3′. Both mouse and chick ISM1 cDNA were cloned into pcDNA3, and positive clones were identified by restriction enzyme digestion and sequencing analyses.
ISM1 protein sequence analysis
The complete amino acid sequence of ISM1 from human (H. sapiens, NP_543016.1), mouse (M. musculus, NP_001263418.1), chick (G. gallus, XP_415036.4), zebrafish (D. ranio, NM_001012376.1), anole (A. carolinensis, XM_003215294.1), turtle (C. pitca bellii, XP_005287456.1), and frog (X. laevis, NP_001082228.1) were aligned using Clutal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). The alignment was then converted into a phylogenetic tree using Jalview. Other bioinformatics analysis of the mouse ISM1 protein included identification of its signal peptide using SignalIP3.0 (http://www.cbs.dtu.dk/services/SignalP-3.0/) ant of its putative N-glycosylation sites using NetNGly1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/).
RT-PCR analysis
RNA extraction and reverse transcription was performed as described above. The first strand cDNA was then used as template for PCR using DyNAzyme II DNA Polymerase (F-501, Thermo Scientific). For amplification of mouse full-length ISM1, the primers used were: forward 5′-GCCTGGCTGC GGAACTGCTC-3′ and reverse 5′-TTAGTACTCT CTGGCTTCTT GGA-3′. GADPH was used as an internal control: forward 5′-CATGGCCTTC CGTGTTCCTA-3′ and reverse 5′-CCTGCTTCAC CACCTTCTTG AT-3′.
In vitro transcription for generation of RNA probes
DIG (digoxigenin)-labeled antisense RNA probes against ISM1 transcripts were prepared from the plasmid constructs containing the cloned mouse or chick cDNA that were linearized by Hind III (R0104, New England Biolabs) and purified by phenol-chloroform extraction. The DIG RNA Labeling Mix (11277073910, Roche) was used with the RNA Polymerase Sp6 (P1085, Promega) in the in vitro transcription reaction following manufacturer’s instructions. RQ1 DNase I (M6101, Promega) was used to remove excess of template plasmid DNA. DIG-labeled RNA probes were then purified by ProbeQuant G-50 Micro Columns (28-9034-08, GE Healthcare) and diluted in an equal volume of formamide (F7508, Sigma Aldrich).
Whole-mount ISH and subsequent histological analysis
Whole-mount ISH using DIG-labeled antisense RNA probes was performed as previously described (Henrique et al., 1995). Briefly, the fixed embryos were treated with methanol, hydrogen peroxide, and proteinase K accordingly to their stages. After post-fixation, the embryos were pre-hybridized for 3 h prior to incubation with DIG-labeled probe overnight at 68 °C. The embryos were then washed first with pre-warmed post-hybridization solution for 2 h at 68 °C, followed by several washes with 0.1% Tween-20 in PBS at room temperature. The embryos were next blocked in 20% normal sheep serum in 0.1% Tween-20 in PBS prior to incubation with sheep anti-DIG APh (Alkaline Phosphatase)-conjugated antibody (11093274910, Roche) overnight at 4 °C. After being extensively washed for 1–2 d, APh activity was detected using 0.35 mg/mL of NBT (nitro blue tetrazolium) and 0.175 mg/mL of BCIP (5-bromo-4-chloro-3-indolyl phosphate). Once color has developed as desired, the embryos were washed several times and fixed in 4% PFA. Whole-mount embryos were photographed in a Leica MZ10F stereomicroscope coupled with a Leica DFC310FX camera and processed for histological analysis. For cryostat tissue sections, the fixed embryos were cryo-protected in 15% sucrose overnight at 4 °C, embedded in gelatin, frozen and kept at −80 °C. Serial sections of 20-μm thickness were performed in a Leica CM3050S cryostat and photographed using an Olympus BX51 microscope equipped with a Spot RT3 CCD camera. Images and figures were processed with Adobe Photoshop CS5.
IHC in paraffin sections
After post-fixation, the newborn and adult tissues were washed with PBS and prepared for paraffin embedding. Briefly, after dehydration through an ethanol series, the samples were cleared with xylene and incubated in paraffin wax for 5 h at 60 °C. After incubation overnight at room temperature, the wax solution was discarded and the samples were then incubated twice in paraffin wax 60 °C for 4 h in the vacuum. The paraffin blocks were sectioned at 6-μm thickness using a rotary microtome (Microm HM340E, Thermo Scientific). The paraffin sections were de-waxed and cleared with xylene. After being rehydrated with an ethanol series and washed with water, sections were subjected to heat induced antigen retrieval. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide. Sections were then incubated with blocking solution (5% normal goat serum and 2% bovine serum albumin in PBS) followed by incubation with a rabbit polyclonal anti-ISM1 antibody overnight at 4 °C. This antibody was custom produced by GenScript Corp against the synthetic peptide FPRQRFPPETGHPSC of the mouse ISM1 protein, corresponding to amino acids 81 to 94. As a control, adjacent sections were incubated with anti-ISM1 antibody that has been pre-adsorbed with a BP that corresponds to the antigen sequence. Both experimental and control sections were processed simultaneously. Detection of positive staining was performed with Dako Envision+ Dual Link System HRP (DAB+) (K4065, Dako) following manufacturer’s instructions and until the brown color has developed as desired. The sections were counterstained with Mayer’s hematoxylin (MHS1, Sigma Aldrich) and photographed using an Olympus BX51 microscope equipped with a Spot RT3 CCD camera. Images and figures were processed using Adobe Photoshop CS5.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work is supported by General Research Fund HKU 787011M and Small Funding Project CRCG HKU 201001176085.
Glossary
Abbreviations:
- AMOP
adhesion-associated domain in MUC4 and other proteins
- AP
anterior-posterior
- BP
blocking peptide
- E
embryonic day
- HH
Hamburger and Hamilton
- IHC
immunohistochemistry
- ISH
in situ hybridization
- MHB
midbrain-hindbrain boundary
- PBS
phosphate buffered saline
- PFA
paraformaldehyde
- RT-PCR
reverse transcriptase-polymerase chain reaction
- TSR1
thrombospondin type 1 repeat
References
- 1.Pera EM, Kim JI, Martinez SL, Brechner M, Li SY, Wessely O, De Robertis EM. Isthmin is a novel secreted protein expressed as part of the Fgf-8 synexpression group in the Xenopus midbrain-hindbrain organizer. Mech Dev. 2002;116:169–72. doi: 10.1016/S0925-4773(02)00123-5. [DOI] [PubMed] [Google Scholar]
- 2.Weidinger G, Thorpe CJ, Wuennenberg-Stapleton K, Ngai J, Moon RT. The Sp1-related transcription factors sp5 and sp5-like act downstream of Wnt/β-catenin signaling in mesoderm and neuroectoderm patterning. Curr Biol. 2005;15:489–500. doi: 10.1016/j.cub.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 3.Bennett JT, Joubin K, Cheng S, Aanstad P, Herwig R, Clark M, Lehrach H, Schier AF. Nodal signaling activates differentiation genes during zebrafish gastrulation. Dev Biol. 2007;304:525–40. doi: 10.1016/j.ydbio.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiang W, Ke Z, Zhang Y, Cheng GH, Irwan ID, Sulochana KN, Potturi P, Wang Z, Yang H, Wang J, et al. Isthmin is a novel secreted angiogenesis inhibitor that inhibits tumour growth in mice. J Cell Mol Med. 2011;15:359–74. doi: 10.1111/j.1582-4934.2009.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawler J, Hynes RO. The structure of human thrombospondin, an adhesive glycoprotein with multiple calcium-binding sites and homologies with several different proteins. J Cell Biol. 1986;103:1635–48. doi: 10.1083/jcb.103.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker RP. The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol. 2004;36:969–74. doi: 10.1016/j.biocel.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Ciccarelli FD, Doerks T, Bork P. AMOP, a protein module alternatively spliced in cancer cells. Trends Biochem Sci. 2002;27:113–5. doi: 10.1016/S0968-0004(01)02049-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Chen M, Venugopal S, Zhou Y, Xiang W, Li Y-H, Lin Q, Kini RM, Chong Y-S, Ge R. Isthmin exerts pro-survival and death-promoting effect on endothelial cells through alphavbeta5 integrin depending on its physical state. Cell Death Dis. 2011;2:e153–10. doi: 10.1038/cddis.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 12. Du J, Takeuchi H, Leonhard-Melief C, Shroyer KR, Dlugosz M, Haltiwanger RS, Holdener BC. O-fucosylation of thrombospondin type 1 repeats restricts epithelial to mesenchymal transition (EMT) and maintains epiblast pluripotency during mouse gastrulation. Dev Biol. 2010;346:25–38. doi: 10.1016/j.ydbio.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alev C, Wu Y, Kasukawa T, Jakt LM, Ueda HR, Sheng G. Transcriptomic landscape of the primitive streak. Development. 2010;137:2863–74. doi: 10.1242/dev.053462. [DOI] [PubMed] [Google Scholar]
- 14.Raible F, Brand M. Divide et Impera--the midbrain-hindbrain boundary and its organizer. Trends Neurosci. 2004;27:727–34. doi: 10.1016/j.tins.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Martinez S, Andreu A, Mecklenburg N, Echevarria D. Cellular and molecular basis of cerebellar development. Front Neuroanat. 2013;7:18. doi: 10.3389/fnana.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White JJ, Sillitoe RV. Development of the cerebellum: from gene expression patterns to circuit maps. Wiley Interdiscip Rev Dev Biol. 2013;2:149–64. doi: 10.1002/wdev.65. [DOI] [PubMed] [Google Scholar]
- 17.Rossi V, Beffagna G, Rampazzo A, Bauce B, Danieli GA. TAIL1: an isthmin-like gene, containing type 1 thrombospondin-repeat and AMOP domain, mapped to ARVD1 critical region. Gene. 2004;335:101–8. doi: 10.1016/j.gene.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Buckingham M, Vincent SD. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr Opin Genet Dev. 2009;19:444–53. doi: 10.1016/j.gde.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–22. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 20.Hamburger V, Hamilton HL, Normal HH. Table. J Morphol. 2004;88:1–42. [PubMed] [Google Scholar]