Abstract
The transformation/transcription domain-associated protein (TRRAP) is a common component of many histone acetyltransferase (HAT) complexes. Targeted-deletion of the Trrap gene led to early embryonic lethality and revealed a critical function of TRRAP in cell proliferation. Here, we investigate the function of TRRAP in murine B cells. To this end, we ablated Trrap gene in a B cell-restricted manner and studied its impact on B-cell development and proliferation, a pre-requisite for class switch recombination (CSR), the process that allows IgM-expressing B lymphocytes to switch to the expression of IgG, IgE, or IgA isotypes. We show that TRRAP deficiency impairs B-cell development but does not directly affect CSR. Instead, cells induced to proliferate undergo apoptosis. Our findings demonstrate a central and general role of TRRAP in cell proliferation.
Keywords: B lymphocyte, immunoglobulin gene, TRRAP, class switch recombination, proliferation
Introduction
Upon antigen challenge, activated B cells proliferate and undergo CSR, which plays an important role in antibody diversification. CSR targets specifically the immunoglobulin heavy chain locus and occurs between highly repetitive S regions located upstream of the constant genes. The process requires GL transcription of S sequences that initiates from their upstream promoters. In addition, CSR absolutely requires AID to initiate DNA cleavages, which culminate in DSBs at partner S sequences.1 DNA ends are formed and repaired by components of the non-homologous end-joining DNA repair pathway2 in the G1 phase of the cell cycle.3,4
TRRAP is a highly conserved member of the PI3KK/ATM (phosphatidylinositol-3-OH-kinase-like kinases/ataxia-telangiectasia mutated) family and a cofactor of many HAT complexes in yeast and mammalian cells.5 Previous work suggests that one function of TRRAP is to recruit HAT complexes to transcription factors bound to their target promoters.6-10 Other studies showed that TRRAP was recruited to chromatin surrounding sites of DNA DSBs and was involved in DNA repair, by promoting chromatin relaxation through histone acetylation, thus facilitating the recruitment of a subset of DNA repair proteins.11
Targeted deletion of Trrap gene in the mouse indicated an essential function in early embryonic development, mitotic checkpoint, and cell cycle progression.10,12 The early embryonic lethality hampered the study of potential tissue-specific functions of TRRAP and/or its involvement in tissue-specific processes. In the present study, we sought to investigate the in vivo role of the murine TRRAP during B-cell development, CSR and proliferation upon B cell-specific ablation of Trrap gene.
Results and Discussion
B cell-specific deletion of TRRAP by CD19–Cre system
In order to obtain a B cell-specific deletion of Trrap gene, mutant mice with a floxed Trrap gene12 (hereafter Tf/f mice) were crossed with CD19–Cre mice that express Cre sequence under the transcriptional control of CD19-regulatory elements.13 The homozygous mutant mice harboring Cre transgene (CD19cre/creTf/f mice) were chosen for further analysis in order to increase the deletion efficiency of Trrap.14
Early B-cell development in TRRAP-deficient mice
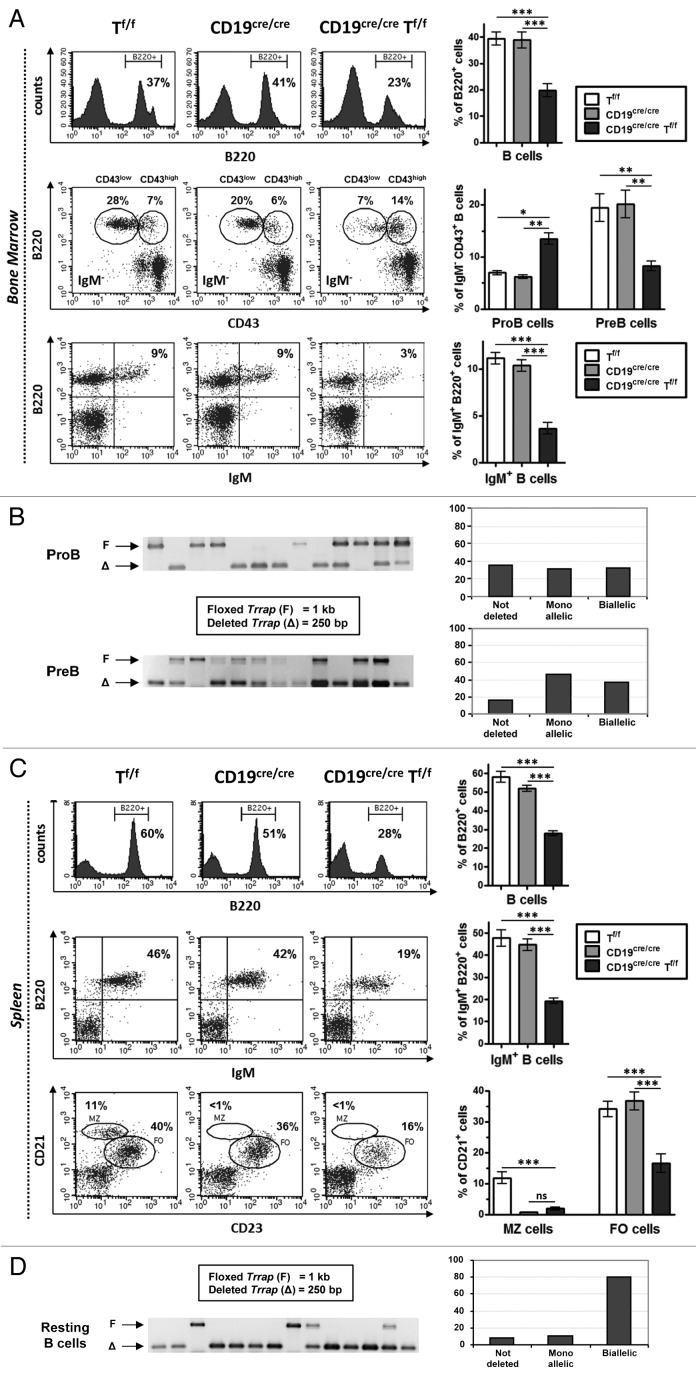
To assess the impact of Trrap deletion on B-cell development, bone marrow cells were stained with specific antibodies and analyzed by flow cytometry. In agreement with previous findings,15 we found no significant difference between CD19cre/cre mice compared with Tf/f and WT controls (Fig. 1A and data not shown). In contrast, a ~2-fold decrease was found for total B220+ cells in CD19cre/creTf/f mice (Fig. 1A, top panels). Analysis of B-cell populations revealed an ~2-fold accumulation of pro-B (B220+CD43highIgM−) cells, an ~3-fold decrease of pre-B (B220+CD43lowIgM−) cells (Fig. 1A, middle panels), and an ~3-fold decrease in the B220+IgM+ population in CD19cre/creTf/f mice (Fig. 1A, bottom panels).
Figure 1. Effect of TRRAP deficiency on B-cell development. (A) Top: Bone marrow cells with the indicated genotypes were stained with anti-B220-APC. Representative plots are shown (n = 6). Middle: For pro-B and pre-B cell populations, bone marrow cells were stained with anti-B220-APC, anti-IgM-FITC, and anti-CD43-PE and gated on IgM− population (n = 6). Bottom: For immature (and circulating) B cells, bone marrow cells were stained with B220-APC and anti-IgM-PE (n = 5). Comparative histograms are shown on the right panels. (B) Single pro-B or pre-B cells were sorted in 96-well plates and subjected to PCR using primer pairs that distinguish between the floxed (1 kb) and the deleted (250 bp) alleles of Trrap. An example of positive wells (with a detectable PCR product on agarose gels) is shown. The histograms in the right panels depict the percentage of deleted alleles among 92 positive wells (pro-B cells) and 62 positive wells (pre-B cells). (C) Spleen cells were stained with anti-B220-APC (top) (n = 6), with anti-B220-APC and anti-IgM-PE (middle) (n = 5), or with CD21-APC and anti-CD23-FITC (bottom) (n = 5). The right panels show the statistical data for each staining. (D) Single resting B cells were sorted and analyzed as in (B). The histograms show the percentage of deleted alleles among 95 positive wells.
In order to check the deletion efficiency of Trrap in the bone marrow of CD19cre/creTf/f mice, we resorted to single-cell PCR on genomic DNAs purified from sorted B-cell populations. In pro-B cells, the majority of cells (~67%) either deleted a single allele of Trrap (~31%) or had no deletion at all (36%), whereas 33% deleted both alleles (Fig. 1B). In pre-B cells, the monoallelically deleted fraction represented 47% of the total population, while its biallelically deleted counterpart represented 37% (Fig. 1B).
These data reveal that B-cell development in the bone marrow is compromised in TRRAP-deficient mice with an accumulation of pro-B cells and a decrease of pre-B and immature B-cell populations. While these results suggest that TRRAP is important for early B-cell development, the fact that some B cells still develop could be due to a lack of or monoallelic deletion of Trrap, or to the persistence of sufficient levels of TRRAP in the case of biallelic deletion. It is also possible that the living cells that we detect are non-proliferating (see below). In this case, doubly deleted pre-B cells, which undergo a burst of proliferation, would disappear from the population, which is manifest in the reduction of immature B-cell population.
In unstimulated spleen, there was an ~2-fold decrease in the total B220+ population and the B220+IgM+ population in CD19cre/creTf/f mice (Fig. 1C, top and middle panels). The marginal zone B cells were totally absent in CD19cre/creTf/f mice, as a consequence of the loss of CD19 (Fig. 1C), and follicular B cells were reduced (~2.5-fold) in CD19cre/creTf/f mice (Fig. 1C, bottom panels).
Single-cell PCR analysis of sorted unstimulated B cells from CD19cre/creTf/f spleen showed that the majority (~80%) has lost both Trrap alleles (Fig. 1D). This finding, together with the relatively mild reduction of IgM-expressing splenic B cells (Fig. 1C), suggests that although TRRAP is important for late B-cell development, the majority of resting B cells that have deleted both alleles may not die immediately, but possibly upon induction of cell proliferation (see below).
CSR in the absence of TRRAP
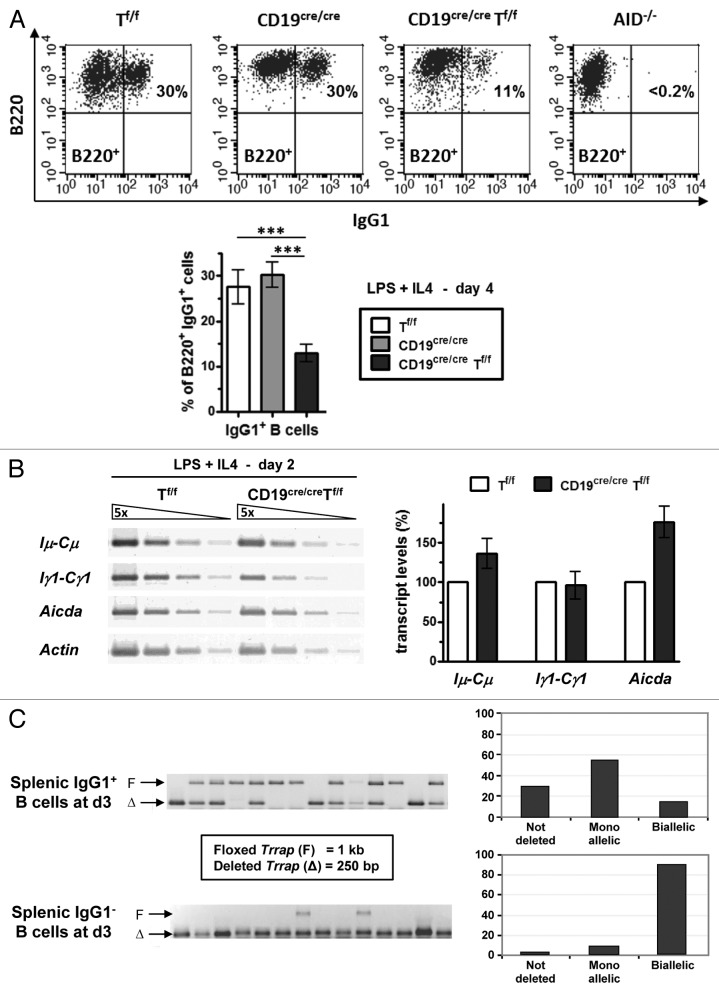
As a first step in the analysis of the role of TRRAP in CSR, sorted CD43− resting B cells from spleens were induced to switch to IgG1 by culturing B cells in the presence of LPS+IL4. At day 4 post-stimulation, the percentage of IgG1+ was reduced in CD19cre/creTf/f mice (Fig. 2A). These results suggest a possible defect in CSR in activated CD19cre/creTf/f B-cell populations, which is unlikely to result from decreased Iμ-, Iγ1-derived GL transcripts or AID transcripts, as no significant alteration of these transcripts was seen at day 2 post-stimulation (Fig. 2B).
Figure 2. Reduced CSR in the absence of TRRAP. (A) CD43− sorted splenic B cells with the indicated genotypes were induced to switch to IgG1 with LPS+IL4. At day 4 post-stimulation, the cells were stained with anti-B220-APC and anti-IgG1-FITC (top). The statistical data are shown in the bottom panel (n = 7). (B) The levels of GL and Aicda transcripts were estimated by semi-quantitative RT-PCR. Total RNA was prepared at day 2 from LPS+IL4-activated B cells and reverse transcribed. Five-fold serial dilutions of single-strand cDNAs were amplified by using appropriate primers. Actin transcripts were used for normalization (n = 5). (C) Resting B cells were induced to switch for 3 d and stained with anti-IgG1-FITC. The IgG1+ fraction was sorted and analyzed as described in Figure 1B. The histograms show the percentage of deleted alleles among 95 positive wells. The IgG1− fraction was sorted and analyzed as described in Figure 1B. The histograms show the percentage of deleted alleles among 46 positive wells.
To check the status of Trrap deficiency in switched B cells, purified CD19cre/creTf/f B cells were induced to switch to IgG1 with LPS+IL4, then IgG1+ and IgG1− cells were sorted for single-cell PCR analysis. The results clearly show that the majority (~85%) of IgG1+ B cells have at least one copy of Trrap (Fig. 2C). In contrast, the vast majority (~90%) of IgG1- B cells has deleted both alleles, suggesting that this TRRAP-deficient population may represent a population that could not lead CSR to completion or that is undergoing early apoptosis (see below).
Combined, the data suggest that biallelic deletion of Trrap does not lead immediately to cell death, but does so upon a proliferative burst. This would imply a major effect of TRRAP deficiency on the proliferative ability of B cells. If so, then inducing resting B cells to switch, which requires prior proliferation, would lead to massive death, and the switched B cells would most likely originate from few TRRAP-proficient cells. Indeed, single-cell PCR revealed that only 15% of IgG1+ cells had a biallelic deletion of Trrap, strongly suggesting that TRRAP is required for proliferation of B cells, so that they can engage in the CSR pathway. In stark contrast, the vast majority of IgG1- B cells had a biallelic deletion of Trrap, and it is conceivable that this TRRAP-deficient population mainly represents the population undergoing early apoptosis. Thus, the defect in CSR seen in the bulk double-mutant population is likely an indirect consequence, due to a failure of activated B cells to proliferate, culminating in cell death.
B-cell proliferation in the absence of TRRAP
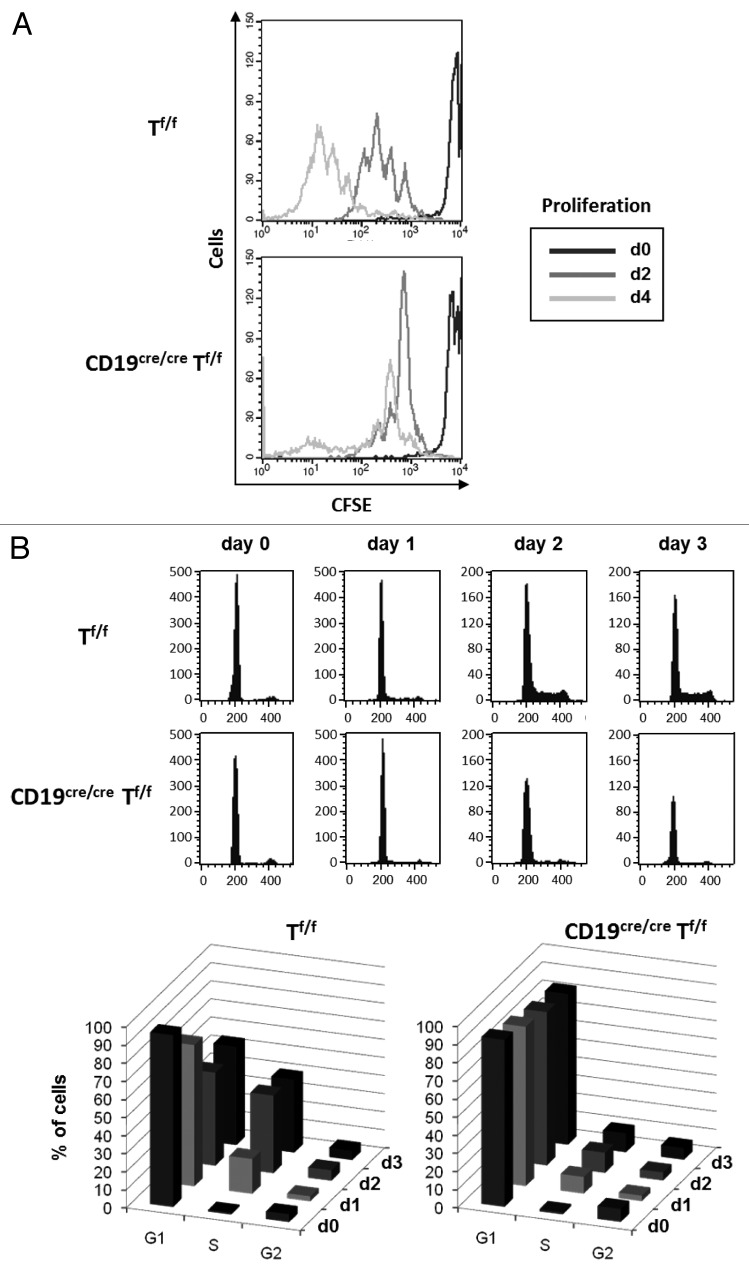
In order to check that the diminished CSR in TRRAP-deficient B cells is due to an impaired proliferation, we performed a proliferation assay and cell cycle analysis on CD19cre/creTf/f and Tf/f B cells. The CD43- sorted splenic B cells were labeled with CFSE, induced to switch with LPS+IL4, and their proliferation ability was tracked by FACS. We found that proliferation of CD19cre/creTf/f B cells was markedly reduced after the first division (Fig. 3A).
Figure 3. Role of TRRAP in B-cell proliferation and cell cycle progression. (A) Negatively (CD43−) sorted B cells were stained with CFSE and induced to switch with LPS+IL4, and their proliferation ability tracked by FACS for the indicated days (n = 3). (B) Resting B cells were induced to switch with LPS+IL4 for 3 d. Aliquots were labeled on a daily basis with PI and cell distribution at the cell cycle phases analyzed by FACS using ModFit software. The bottom panels are a compilation of the data shown in the top panels; n = 2 with 3 mice of each genotype in each experiment.
To further strengthen this finding, we performed a cell cycle analysis. CD43- sorted CD19cre/creTf/f and Tf/f B cells were induced to switch with LPS+IL4, and the distribution of cell populations at the cell cycle phases was tracked on a daily basis by FACS. The percentage of cells having undergone DNA synthesis was already ~2 times larger at day 1 post-stimulation in Tf/f compared with CD19cre/creTf/f B-cell populations, and while the percentage of cells in S phase increased in Tf/f B-cell population at day 2, it remained virtually constant in CD19cre/creTf/f B-cell population, where the majority of cells were blocked at G1 (Fig. 3B, top and bottom panels). Thus B cell-restricted deletion of Trrap leads to a major defect in cell cycle progression.
B-cell death in the absence of TRRAP
We next asked whether the defect in proliferation was associated with cell death. To this end, purified CD19cre/creTf/f and Tf/f B cells were activated with LPS+IL4, and the percentage of dead cells was monitored by FACS by using PI. The percentage of dead cells increased with time for both populations; however, the percentage was clearly higher for CD19cre/creTf/f B-cell population (~2-times) (Fig. 4A). This was confirmed by analyzing the percentage of early apoptotic B cells (B220+ Annexin5+ 7-AAD−) among the B cells that have been induced to proliferate. This percentage steadily increased with time for CD19cre/creTf/f B cells (Fig. 4B). These data strongly suggest that CD19cre/creTf/f B cells that have been induced to proliferate trigger an enhanced programmed cell death in the absence of TRRAP.
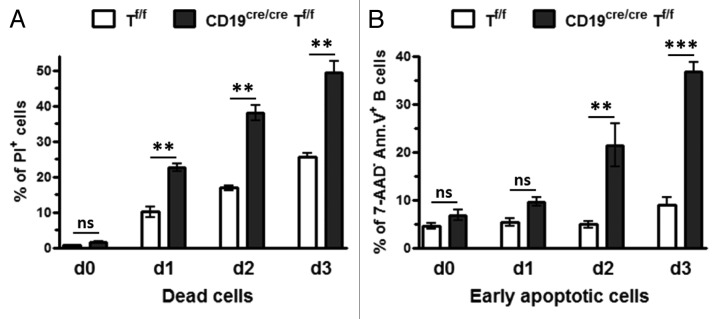
Figure 4. Cell death in TRRAP-deficient B cells. (A) The percentage of dead cells was monitored by labeling the cells that have been induced to proliferate with PI. Standard variations are shown, n = 2 with 3 mice of each genotype in each experiment. (B) To quantify the percentage of early apoptotic cells (B220+7-AAD− Annexin V+), resting B cells were induced to proliferate with LPS+IL4 for 3 d. Every day, an aliquot was removed, labeled with anti-B220-APC, anti-Annexin V-PE, and 7-AAD and gated on 7-AAD− population; n = 2 with 3 mice of each genotype in each experiment.
Our findings hint to a general role of TRRAP, probably more critically in proliferating cell populations. The present study provides evidence that it is indeed the case in B cells. But the requirement for TRRAP in proliferation10,12,16 does obviously not preclude a role in the repair of DNA DSBs during CSR. Given the evolutionary conservation of TRRAP function,5 it is conceivable that TRRAP may be recruited to sites of DSBs at S regions, where it could promote chromatin remodeling and DNA repair. In the absence of TRRAP, DSBs generated at G1 phase may not be repaired, leading to the triggering of DNA damage checkpoint and cell cycle arrest. If the DSBs are not processed properly after a prolonged period, the cell will undergo apoptosis.
In conclusion, our study demonstrates that TRRAP is necessary for normal B-cell development and B-cell proliferation, supporting a ubiquitous and critical role for TRRAP in proliferation.
Materials and Methods
Mice
Tf/f and CD19–Cre mice have been described.12,13 The experiments on mice have been performed according to the CNRS Ethical Committee guidelines.
FACS analyses
Bones from 6- to 8-wk-old mice were flushed with 10% FCS-containing RPMI 1640. Single-cell suspensions were stained with APC-conjugated anti-B220 (Biolegend), PE-conjugated anti-CD43 (BD PharMingen), and PE- or FITC-conjugated anti-IgM (Biolegend). For FACS acquisition, the data were obtained on 3 × 104 viable cells. Pro-B (B220+IgM−CD43high) and pre-B (B220+IgM−CD43low) populations were sorted using a Becton Dickinson apparatus (FACSAria II-Sorp). Single-cell suspensions from spleens of 6- to 8-wk-old mice (5 × 105 cells/assay) were labeled by using anti-B220-APC, anti-IgM-PE, anti-CD21-APC, and anti-CD23-FITC (Biolegend). Controls to exclude non-specific binding of anti-sera to dead cells included isotype controls, PI, and gating on live cells. CSR assays were performed as described17 starting with 750 000 cells/ml, and adjusted to this concentration at days 2 and 3. CD43− B cells were activated with 20 μg/ml of LPS (S. typhimurium, Sigma) and 10 ng/ml of IL-4 (R&D Systems). At day 4, cells were stained with anti-B220-APC and anti-IgG1-FITC (Biolegend). In all FACS acquisitions, dead cells were excluded by labeling with PI.
RT-PCR
At day 2 post-stimulation, aliquots of cells were removed. Total RNA preparation, the primers used to amplify Sμ, Sγ1 GL, Aicda, and actin transcripts, the RT-PCR conditions, and the expected sizes of the PCR products have been described.17 For quantification, agarose gels were dried for 1 h at 80 °C using a gel dryer (BioRad), stained with SYBR green I (Fisher Bioblock) for 1 h, and scanned by using a phosphorimager and Image Quant software (Molecular dynamics). After substracting background levels, the signals in the diluted lanes were normalized against actin signals.
Proliferation assay
CD43- B cells were labeled with CFSE (Molecular Probes) according to manufacturer’s instructions. The cells were induced to switch as described above. The number of cell divisions was measured by FACS at days 2 and 4.
Cell cycle analysis
CD43− B cells were induced to switch as described above. Every 24 h, aliquots were removed and washed with ice-cold PBS. Cells were then labeled with ice-cold PI staining solution (25 μg/ml PI, 0.1% W/V tri-sodium citrate, 1 μg/ml RNase-A, 0.1% triton X-100) and analyzed by FACS (Becton Dickinson FACScan) by adjusting the flow below 200 events/second.
Cell death and early apoptosis
CD43- B cells were induced to switch as described above. Every 24 h, aliquots were removed, labeled with PI, or with anti-B220-APC, 7-amino-actinomycin D (7-AAD) and anti-annexin V-PE as per manufacturer’s instructions (BD PharMingen) and analyzed by FACS (Becton Dickinson FACScalibur).
Single-cell PCR
Pro-B and pre-B cells were stained as described above. CD43− splenic B cells were either stained with anti-B220-APC and sorted, or induced to switch as described above. Cells were labeled with anti-IgG1-FITC, and the IgG1+ and IgG1− fractions were sorted. Two sorts were performed in parallel for each population, a single-cell sort in 96-well plates and a standard bulk sort to check the purity of the sorted cells (>95%). Fifteen μl of lysis buffer (1×PCR buffer, 100 μg/ml proteinase K, 0.2 mM dNTPs, I2.1 [AGAGTAGAGC GTCATTGTC] and G7.0 [GACAAAACCA ACGACAGAGC] primers at 0.5 µM, in H20) were immediately added per well, incubated at 56 °C for 20 min, followed by incubation at 95 °C for 5 min in a thermocycler (BioRad). After the samples were cooled down, a PCR was performed as follows: 0.15 μl of GoTaq (Promega) were added, then heated at 95 °C for 2 min; 30 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 2 min; 5 min of final elongation at 72 °C. Three microliters were then subjected to a new round of PCR in the same conditions.
Statistical analysis
Results are expressed as mean ± SEM (GraphPad Prism), and overall differences between values from control and mutant mice were evaluated by a ANOVA parametric test with Newman–Keuls post test or the Kruskal–Wallis non-parametric test with Dunn post test, or by a 2-tailed unpaired Student t test. The difference between means is significant if P value < 0.05 (*), very significant if P value < 0.01 (**), and extremely significant if P value < 0.001 (***).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank K Rajewsky for providing CD19-Cre mice and D Trouche for critical reading of the manuscript. We also thank P Mercier and F L’Faqihi/V Duplan-Eche, at the IPBS transgenesis and Purpan CPTP plate-forms respectively, for their excellent work. C.L. was supported by an MRES and G.C. was supported by the INCa. This work was funded by the INCa (PL-96-52, INCa_4513), ANR (ANR-07-BLAN-0080-01/0080-03), ARC (Grant SFI20101201441), LCC-Comité de Haute-Garonne, and Cancéropôle GSO.
Author Contributions
C.L., G.C., N.P., and C.S. performed experiments, C.L., G.C., N.P., and A.A.K. analyzed the data, M.M. managed the mouse lines, Z.H. and A.A.K. designed the project, and A.A.K. wrote the manuscript.
Glossary
Abbreviations:
- AID
activation-induced cytidine deaminase
- CSR
class switch recombination
- DSBs
double-strand breaks
- GL
germ-line
- HAT
histone acetyltransferase
- PI
propidium iodide
- S
switch
- TRRAP
transformation/transcription domain-associated protein
References
- 1.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boboila C, Alt FW, Schwer B. Classical and alternative end-joining pathways for repair of lymphocyte-specific and general DNA double-strand breaks. Adv Immunol. 2012;116:1–49. doi: 10.1016/B978-0-12-394300-2.00001-6. [DOI] [PubMed] [Google Scholar]
- 3.Rush JS, Liu M, Odegard VH, Unniraman S, Schatz DG. Expression of activation-induced cytidine deaminase is regulated by cell division, providing a mechanistic basis for division-linked class switch recombination. Proc Natl Acad Sci U S A. 2005;102:13242–7. doi: 10.1073/pnas.0502779102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrader CE, Guikema JEJ, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–71. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- 5.Murr R, Vaissière T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–72. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, Lüscher B. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–7. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–7. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 8.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–82. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herceg Z, Li H, Cuenin C, Shukla V, Radolf M, Steinlein P, Wang ZQ. Genome-wide analysis of gene expression regulated by the HAT cofactor Trrap in conditional knockout cells. Nucleic Acids Res. 2003;31:7011–23. doi: 10.1093/nar/gkg902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Cuenin C, Murr R, Wang ZQ, Herceg Z. HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J. 2004;23:4824–34. doi: 10.1038/sj.emboj.7600479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–9. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 12.Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet. 2001;29:206–11. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- 13.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–8. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwenk F, Sauer B, Kukoc N, Hoess R, Müller W, Kocks C, Kühn R, Rajewsky K. Generation of Cre recombinase-specific monoclonal antibodies, able to characterize the pattern of Cre expression in cre-transgenic mouse strains. J Immunol Methods. 1997;207:203–12. doi: 10.1016/S0022-1759(97)00116-6. [DOI] [PubMed] [Google Scholar]
- 15.Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 1995;376:352–5. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- 16.Ichim G, Mola M, Finkbeiner MG, Cros MP, Herceg Z, Hernandez-Vargas H. The histone acetyltransferase component TRRAP is targeted for destruction during the cell cycle. Oncogene. 2014;33:181–92. doi: 10.1038/onc.2012.570. [DOI] [PubMed] [Google Scholar]
- 17.Haddad D, Oruc Z, Puget N, Laviolette-Malirat N, Philippe M, Carrion C, Le Bert M, Khamlichi AA. Sense transcription through the S region is essential for immunoglobulin class switch recombination. EMBO J. 2011;30:1608–20. doi: 10.1038/emboj.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]