Highlights
-
•
A novel peptidase Cj0511 in an important human bacterial pathogen Campylobacter jejuni has been characterized.
-
•
Proteolytic properties of Cj0511 protein were detected in whole cell lysates using zymography.
-
•
Enzymatic studies conducted with a purified protein confirmed the prediction of a serine peptidase.
-
•
The cj0511 mutant was severely attenuated in a chicken colonisation model, suggesting a role in infection.
Keywords: Peptidases, Proteases, Proteolysis, Protease inhibitors, Campylobacter, Colonisation
Abstract
According to MEROPS peptidase database, Campylobacter species encode 64 predicted peptidases. However, proteolytic properties of only a few of these proteins have been confirmed experimentally. In this study we identified and characterised a Campylobacter jejuni gene cj0511 encoding a novel peptidase. The proteolytic activity associated with this enzyme was demonstrated in cell lysates. Moreover, enzymatic studies conducted with a purified protein confirmed a prediction of it being a serine peptidase. Furthermore, cj0511 mutant was found to be severely attenuated in chicken colonisation model, suggesting a role of the Cj0511 protein in infection.
1. Introduction
Campylobacter jejuni is the most frequent causative agent of bacterial gastrointestinal diseases worldwide [1]. In particular, in England and Wales these bacteria cause the highest number of food borne infections and hospitalisations than any other bacterial pathogen, with over 8% annual increase in the number of cases reported in 2010 [2].
Bacterial peptidases may be involved in a variety of biological functions, such as degradation of unfolded proteins, protein secretion and processing, etc (reviewed in: [3]). A number of genes encoding putative peptidases have been identified in the genomes of C. jejuni. Some of these genes were found to be involved in stress response contributing to increased survival of bacteria in adverse conditions [4–7]. For example, a C. jejuni peptidase-related gene htrA is required for heat-shock resistance, oxygen tolerance and invasion of INT407 human epithelial cells [6]. Expression of another peptidase, ClpP, is associated with a biofilm formation [8]. The most recent review on Campylobacter peptidases was focused on just four enzymes including Lon, Clp, HtrA and FtsH [9]. Our previous studies demonstrated that a knock-out of a putative peptidase-encoding gene cj0511 resulted in attenuation of bacteria in Galleria mellonella model of infection [10]. Transposon inactivation of peptidase-related genes cj1068 and cj1215 resulted in reduction of invasive properties of C. jejuni strain 81-176 [11], and mutation in cj1228 (htrA) gene reduced bacterial attachment to host epithelial cells [12]. In this report we demonstrate a novel proteolytic activity associated with protein Cj0511.
2. Materials and methods
2.1. Bacterial strains and growth conditions
C. jejuni 11168H, a hypermotile derivative of strain NCTC 11168 [13], was maintained on blood agar plates. Bacteria were grown for two days at 37 °C in a VAIN cabinet (Don Whitely) with 85% Nitrogen, 5% Oxygen and 10% Carbon Dioxide. The Escherichia coli XL2 Blue MRF’ and XL1 Blue MRF’ strains (Stratagene), used in cloning experiments, were grown overnight at 37 °C on LB agar plates. The plates were supplemented with kanamycin (50 μg/ml) and/or chloramphenicol (10 μg/ml) when required.
2.2. Mutant construction
C. jejuni 11168H mutants were constructed via site-directed insertional mutagenesis. The BamHI fragment containing the kanamycin resistance (kanr) cassette from pJMK30 [14] was inserted into unique restriction sites within target gene-containing fragments from a 2 kb sequencing library [15] (Table 1). Blunt-end cloning was used where appropriate. The plasmid constructs were verified using restriction analysis and sequencing, transformed into C. jejuni via electroporation as described [16] and Kanr clones were selected. The mutants were verified by PCR using kanr cassette and gene specific primers. The kanr cassette was inserted in orientation excluding a possible polar effect on the downstream genes. This cassette does not contain a transcription terminator, and the lack of a polar effect, when this cassette was inserted in the same transcription polarity with the downstream genes, was previously confirmed experimentally using qPCR [17].
Table 1.
Mutants constructed in this study.
| Mutant | pUC18 construct | Insertion site | Position of the kanr cassette (% from gene start) |
|---|---|---|---|
| cj0511 | cam88g9 | StyI | 75 |
| cj1228 | cam67e8 | PsiI | 67 |
2.3. Complementation of mutant 11168H/cj0511::kanr
Complementation of the cj0511 mutant was performed using a previously developed system for heterogonous gene expression in Campylobacter [18]. The complementation system is based on integration of a gene of interest into a bacterial chromosome, and a constitutative gene expression under the control of the camr promoter. A previously described plasmid pRED [18] was used for the introduction of an intact copy of gene cj0511 into C. jejuni 11168H/cj0511:kanr mutant. Gene cj0511 was PCR amplified using the following primers
ak358 (ctcaatttaaatATGATGGAGCTTATTTTGAAAACAAAA) and
ak359 (gtcatttctagaTTATTGTCCTTGTTTGATATTTAAA).
The priming regions are shown in upper case, whilst lower case letters denote additional sequences containing SwaI and XbaI restriction sites used for cloning. The 1358 nt PCR product generated with these primers was inserted between SwaI and XbaI sites of the delivery vector pRED . The derived recombinant plasmids was used for transformation of 11168H/cj0511::kanr mutant via electroporation. Integration of the cj0511 gene into the bacterial chromosome was confirmed by PCR with primer pairs ak233/ak237, ak234/ak237 and ak235/ak237 as described previously [18].
2.4. Expression of a 6x His tagged protein Cj0511 (Cj0511His) of C. jejuni in E. coli
Gene cj0511 was PCR amplified using Hi-fidelity Taq polymerase Pwo (Roche) and the following primers:
cj0511F AGTCGGATCCAAGTTGATCAAAAAGAAGAGCAGGTTC
cj0511R CATTCTGCAGTTATTGTCCTTGTTTGATATTTAAAATTTTAATA.
The PCR product was inserted into PstI/BamHI site of pQE-32 plasmid (Qiagen). The recombinant plasmids were verified by restriction analysis and sequencing.
2.5. Protein expression and purification
Overnight E. coli cultures grown at 37 °C were diluted 1:50 with LB medium, grown to OD600 of 0.5–0.8, induced by adding isopropyl-β-D-thiogalactoside (IPTG) to 1 mM and incubated for further 2 h. Large scale preparation was carried out using 100 ml of induced cultures. Bacteria were pelleted by centrifugation and resuspended in CellLytic B reagent (Sigma). After centrifugation the soluble fraction of the lysate was used for protein purification using HisSelect™ Spin Columns (Sigma) according to manufacturer’s protocol. The fractions were analyzed on 12 % NuPAGE™ Novex Bis-Tris SDS–PAGE gels.
The gels were run using NuPAGE™ MOPS running buffer (Invitrogen) at 200 V for 1 h and stained with Coomassie Blue.
2.6. Protease assay
Concentration of the purified protein was determined using the BCA™ Protein Assay Kit (Pierce). Proteolytic activity was monitored on a 96-well microtiter plate using Protease Screening™ kit (Geno-Technolgy Inc) according to manufacturer’s protocol. Each well contained 50 μl of reaction containing 0.25 μg of purified protein and 2.5 μl peptidase substrate. Where appropriate, the reaction mixtures contained 0.5 μl of protease inhibitors (Geno Technology Inc) supplied at 100x concentrations. The final concentrations of the inhibitors were as follows: Antipain, 74 μM; Aprotinin, 0.3 μM; Bestatin, 130 μM; AEBSF, 1 mM; Phosphoramidon, 10 μg/ml; PMSF, 100 mM. The plate was incubated for 3 hours at 37 °C followed by addition of 50 μl of precipitation agent and incubated for further 10 min. The plate was then centrifuged at 4000g for 15 min, supernatant (80 μl) was transferred to a clean plate and 120 μl of assay buffer was added and absorbance was read at 550 nm. Statistical data analysis (P–value estimation) was performed using GraphPad Prism version 4.02 statistical analysis software package.
2.7. Zymography
Proteolytic activity testing of bacterial lysates and protein samples by zymography was conducted as previously described [19]. One loopful of bacteria from two-day 37 °C blood agar culture was suspended in 500 μl of PBS. The cells were spun down at 13,000 rpm for 5 min and washed twice in the PBS. The bacterial pellet was resuspended in 200 μl of 4% SDS and mixed with a pipette tip and stored at −80 °C until required. Samples diluted 1:1 with loading buffer were run on a 12% SDS casein gels for 30–45 minutes at 125 V. The gel was washed in 2.5% Triton X100 in water for 30 min with gentle agitation, followed by incubation in developing buffer (50 mM Tris-HCl pH 7.6, 0.2 M NaCl, 5 mM CaCl2, 0.2 % (v/v) Brij 35) for 3–5 h or overnight at 37 °C. The gel was stained with Coomassie Blue and visualized for zones of clearing. Inhibitor screening was performed as previously described [20].
2.8. Chicken colonization studies
Chicken colonisation studies were performed as described previously [21]. The animal work was conducted under the license approved by the UK Home Office within the Animal Scientific Procedures Act (ASPA). The work was reviewed by the local Ethical Committee of the Institute of Animal Health, Compton, United Kingdom, and meets UK legal requirements for studies involving animals. The work was done following ASPA guidelines and animals were condition scored to ensure no adverse effects were observed during the study. This work was classed as mild with no expected observable health effects, none were observed. All animals were euthanized using Schedule-1 procedures as outlined in Home Office Guidance.
The Campylobacter strains were grown individually for 48 h at 37 °C in Mueller Hinton broth under microaerophilic conditions. Specific-pathogen-free (SPF) Light Sussex chickens were inoculated orally on the day of hatch with 0.1 ml of Campylobacter-free adult gut-flora preparations. For this, one gram of cecal contents were taken from a 50-week-old SPF chicken, immediately after the bird had been killed and used to inoculate 10 ml of Luria-Bertani broth which was incubated for 24 h at 37 °C. Birds were housed in separate rooms in high-biosecurity accommodation until two weeks of age when they were used in colonization trials. Birds were fed a vegetable-based diet (Special Diet Services, Manea, Cambridgeshire, UK) ad libitum. Colonization trails were done in groups of three birds. Birds were orally inoculated with 0.1 ml of Mueller-Hinton broth culture containing 108 c.f.u. of the desired Campylobacter strain. Seven days post-infection birds were euthanized. Decimal dilutions of cecal contents were made in phosphate buffered saline (PBS) and plated onto Campylobacter blood-free agar for colony counting. Plates were incubated microaerobically for 48 h at 37 °C.
3. Results
3.1. Peptidases in C. jejuni NCTC 11168
According to the annotation of C. jejuni NCTC 11168 genome at Sanger Institute (www.sanger.ac.uk/Projects/C_jejuni/) and a comprehensive peptidase database MEROPS (merops.sanger.ac.uk/), the genome of this strain contains at least 45 putative peptidase-related genes. As predicted by PSORT (psort.nibb.ac.jp/form.html), most of these enzymes are either cytoplasmic or inner-membrane located (data not shown), with only six of them predicted to be periplasmic or outer membrane located, with none predicted to be extracellular (Table 1S).
Using zymography and casein as a substrate and a total cell lysate of C. jejuni 11168H we detected several bands associated with peptidase activities (Fig. 1A, lane 2), namely 42 kDa, 49 kDa and 51 kDa, as well as some high molecular weight bands in the range of 170 kDa.
Fig. 1.
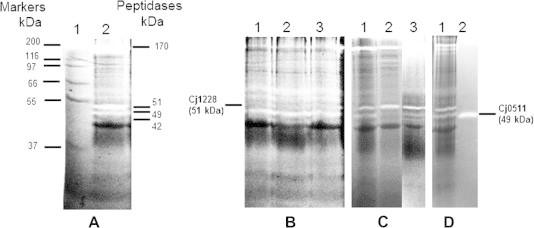
Identification of peptidase activities of C. jejuni strain 11168H using zymography. (A) 1, size markers (Mark 12, Invitrogen); 2, 11168H. (B) 1–3, 11168H/cj1228::kanr mutant (three clonal isolates). (C) 1, 11168H; 2, 11168H/cj0511::kanr; 3, 11168H/cj0511::kanr/cj0511+ (complement). (D) 1, 11168H; 2, purified Cj0511His protein.
As expected, the most prominent lower molecular weight band at 51 kDa corresponded to HtrA peptidase, since it was affected by a cj1228 (htrA) mutation (Fig. 1B, lanes 1–3). The results confirm proteolytic activity of a native form of protein Cj1228 (HtrA), which was previously detected with a fusion HtrA-His protein after expression and purification from E. coli [22]. The cj1228 mutation also resulted in disappearance of high molecular weight bands in the range of 170 kDa (Fig. 1B, lanes 1–3), representing oligomeric forms of HtrA [22,23]. Inactivation of gene cj0511 resulted in disappearance of a 49 kDa band corresponding to a predicted size of this protein (Fig. 1C, lane 2). This result was confirmed by the restoration of activity after complementation of this mutation in cis (Fig. 1C, lane 3). The intensity of the band in complementation derivative was lower than in the wild type strain, which was not unexpected as full restoration of activity requires fine tuning of gene expression, whilst the complementation system we were using employs a constitutative (unregulated) promoter.
3.2. Expression in E. coli, purification, stability and enzymatic activity of 6x His-tagged derivative of protein Cj0511 (Cj0511His)
Expression of protein Cj0511His in E. coli using a tightly regulated expression system pQE-32 allowed its purification as a highly stable protein (Fig. 1S). Using zymography assay and casein as a substrate we were able to detect proteolytic activity of the purified Cj0511His (Fig. 1D).
Proteolytic activity of Cj0511His in the presence of various inhibitors was tested using a peptidase assay kit (Fig. 2 A, B). Whilst statistically valid reduction in activity in the presence of antipain, aprotin, AEBSF and PMSF inhibitors specific for serine peptidases was detected, no effect on activity in the presence of metallo-peptidase-specific inhibitors bestatin and phosphoramidon was found. The results confirmed a prediction that Cj0511 is a serine peptidase.
Fig. 2.
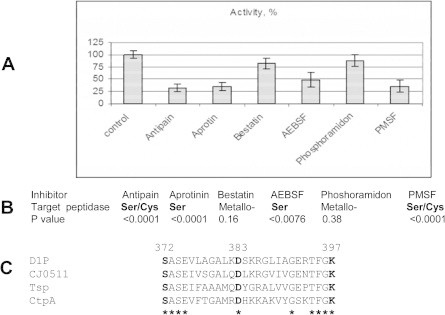
Cj0511His activity is affected by serine and serine/cystein but not metallo peptidase specific inhibitors. (A) Effect of various inhibitors on enzymatic activity of Cj0511His. The error bars denote standard deviations. (B) Statistical analysis of changes using one-way Anova. The experiments were repeated three times, with three technical replicates in each test. Text in bold denotes statically valid difference (P < 0.01) compared to control (no inhibitors). (C) Conservation of amino acid residues required for enzymatic activity of serine peptidase D1P (highlighted in bold) in Cj0511 and related proteins (see text). The identical amino acid residues are denoted by asterisks. The Swissprot amino acid sequence database accession numbers are: spO04073.1 (D1P), spP23865 (TspA) and spQ4L6D0.2 (CtpA).
3.3. Peptidase Cj0511 is required for colonisation
Inactivation of gene cj0511 in C. jejuni strain 11168H resulted in severe attenuation of colonisation in chickens (Fig. 3). The cj0511 mutation affected the ability of C. jejuni to colonise chickens reducing c.f.u. per gram of cecal content by over six logs. Importantly, the cj0511 mutant retained full motility of the wild type strain (Fig. 2S) suggesting that attenuation in chicken model of infection was not a result of changes in flagella expression, which can occur in C. jejuni due to phase variation [13].
Fig. 3.
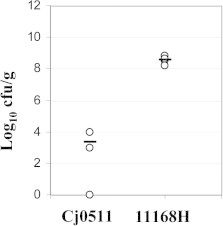
Attenuation of mutant 11168H/cj0511::kanr in chicken colonisation model of infection. Post-mortem cecal contents counts of Campylobacter strains used in this study. Counts were determined at 7 days post-infection and are displayed as the Log 10 of the cfu/g. Values of 0 represent undetectable levels of Campylobacter <100 cfu/g. Open circles indicate each individual bird. The bars represent average values. Cj0511 denotes 11168H/cj0511::kanr mutant.
4. Discussion
In this study we used mutagenesis, complementation, zymography and enzymatic studies to confirm that cj0511 gene encodes a serine peptidase. Cj0511 was found to be glycosylated [24]. Our findings, demonstrating enzymatic activity of this protein after expression in E. coli, lacking protein glycosylation apparatus, suggests that glycosylation of Cj0511 is dispensable for its proteolytic activity.
The role of Cj0511 protein in Campylobacter lifestyle remains to be elucidated. According to MEROPS database (merops.sanger.ac.uk/), Cj0511 belongs to S41 family of C-terminal processing peptidases, which recognize a C-terminal tripeptide, Xaa-Yaa-Zaa, in which Xaa is preferably Ala or Leu, Yaa is preferably Ala or Tyr and Zaa is preferably Ala. In other bacteria (e.g. in E. coli) these peptidases may be involved in degradation of incorrectly synthesized proteins, which, after being tagged with Leu-Ala-Ala tripeptide, become a target for the recognition and degradation by these enzymes (merops.sanger.ac.uk/).
In other bacteria, extracytoplasmic peptidases were shown to play a role in pathogenesis. For example, Chlamydial peptidase CPAF is translocated into the host cell cytosol and degrades transcription factors RFX5 and USF-1 impairing host cell immune response [25]. Interestingly, C. jejuni was found to be able to disrupt tight junctions in colonic epithelial cells [26]. Subsequently, a role of Campylobacter HtrA peptidase in degradation of E-cadherin junctions between epithelial cells was demonstrated [27]. It would be interesting to explore a possible role of other cell surface and/or secreted peptidases in this process. Along with MOMP, PEB2 and PEB3, both Cj1228 and Cj0511 peptidases have recently been identified as cell surface proteins with a potential as vaccine candidates [28]. Such cell-surface located proteins often play a role as virulence factors and are involved in direct interaction of pathogenic bacteria with host cells [29]. The number of secreted, outer-membrane located and periplasmic enzymes is likely to be underestimated when using common bioinformatics software such as PSORT. In particular, Cj0511 protein is predicted to be inner membrane located due to the presence of ‘uncleavable leader peptide’ despite being found on the cell surface [28].
Some peptidases with sequence similarity to Cj0511 were found to play a role in virulence. For example, a CPAF peptidase homologue from Chlamydia is involved in degradation of host transcription factors [25]. The amino acid sequence of Cj0511 has the highest level of similarity to Bartonella bacilliformis peptidase CtpA. A similar protein, Prc of E. coli, is involved in processing of penicillin-binding protein 3 [30]. Another homologue of Cj0511 is a CtpA protein of Rhizobium leguminosarum, which is important for bacterial viability on complex semi-solid media and for resistance to desiccation and detergents, indicating a role in cell envelope formation [31]. A Ser-Asp-Lys triad in the active site of Tsp peptidase similar to Cj0511 was found to be essential for peptidase activity [32]. The respective residues are conserved in Cj0511 and other members of the Tsp family of peptidases, even including a distantly related D1P peptidase from Scenedesmus obliquus (algae) required for processing of photosystem II D1 protein (Fig. 2C) [33].
Our results confirm prediction that Cj0511 is a serine peptidase. However, the active centre for this peptidase is unusual due to the lack of inhibitory effect by chymostatin, which is specific for chymotripsin-like Ser peptidases. Concordant with our data is a finding that another member of this family of enzymes, CtpA, despite belonging to Ser peptidases and the presence of Ser/Lys catalytic diad, is insensitive to certain serine peptidase inhibitors [34].
The investigation of bacterial peptidases as potential drug targets is an emerging area of research. The mechanism of action of an anti-ulcer drug ebrotidine used for the treatment of Helicobacter pylori (closely related to C. jejuni) infection was in part attributed to its strong inhibitory action of Helicobacter peptidases degrading gastric mucin [35]. Furthermore, the studies of peptidases in Porphyromonas gingivalis, allowed the development of specific bacterial peptidase inhibitors with potential therapeutic applications against periodontal disease [36]. More recently, inhibitors of a peptidase toxin produced by B. anthracis [37] were found to have a protective effect against mice infection with this pathogen [38,39]. A new class of antibiotics, acyldepsipeptides, which targets ClpP, is known to reduce S. aureus and Enterococcus faecalis load in liver, spleen and lung upon infection in mice [40].
Our data on the identification and characterisation of C. jejuni Cj0511 peptidase are important for better understanding of biology of this major bacterial pathogen. Further investigation of Campylobacter peptidases and their inhibitors may assist in the development of novel antibacterial strategies.
Appendix A. Supplementary data
Expression and purification of Cj0511His protein after overexpression in E. coli. 1, cell lysate; 2, soluble fraction; 3, pellet; 4, flow through; 5 purified protein; M, size markers (SeeBlue, Invitrogen).
Motility test. Three clonal isolates of strain 11168H/cj0511::kanr (clones 1–3) were stabbed into semi-solid (0.3%) Mueller Hinton agar plate and grown for two days at 37 °C. Compared to recipient strain 11168H, no difference in motility was observed.
Putative peptidases in C. jejuni NCTC 11168 (excluding non-peptidase homologues Cj0190, Cj0463, Cj1005. Cj1205 and Cj1275).
References
- 1.Silva J., Leite D., Fernandes M., Mena C., Gibbs P.A., Teixeira P. Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2011;2:200. doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. (2011). Campylobacter infections per year in England and Wales, 2000-2010. Health Protection Agency (HPA). http://www.hpa.org.uk/2011.
- 3.Raivio T. Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 2005;56:1119–1128. doi: 10.1111/j.1365-2958.2005.04625.x. [DOI] [PubMed] [Google Scholar]
- 4.Konkel M.E., Kim B.J., Klena J.D., Young C.R., Ziprin R. Characterization of the thermal stress response of Campylobacter jejuni. Infect. Immun. 1998;66:3666–3672. doi: 10.1128/iai.66.8.3666-3672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park S.F. Campylobacter jejuni stress responses during survival in the food chain and colonization. In: Ketley J.M., Konkel M.E., editors. Campylobacter. Molecular and Cellular Biology. Horizon Bioscience; Norfolk: 2005. pp. 311–330. [Google Scholar]
- 6.Brondsted L., Andersen M.T., Parker M., Jorgensen K., Ingmer H. The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl. Environ. Microbiol. 2005;71:3205–3212. doi: 10.1128/AEM.71.6.3205-3212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohn M.T., Ingmer H., Mulholland F., Jorgensen K., Wells J.M., Brondsted L. Contribution of conserved ATP-dependent proteases of Campylobacter jejuni to stress tolerance and virulence. Appl. Environ. Microbiol. 2007;73:7803–7813. doi: 10.1128/AEM.00698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalmokoff M., Lanthier P., Tremblay T.L., Foss M., Lau P.C., Sanders G., Austin J., Kelly J., Szymanski C.M. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 2006;188:4312–4320. doi: 10.1128/JB.01975-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingmer H., Brondsted L. Proteases in bacterial pathogenesis. Res. Microbiol. 2009;160:704–710. doi: 10.1016/j.resmic.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Champion O.L., Karlyshev A.V., Senior N.J., Woodward M., La Ragione R., Howard S.L., Wren B.W., Titball R.W. Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity RID D-2797-2011 RID E-8955-2010 RID F-7204-2010. J. Infect. Dis. 2010;201:776–782. doi: 10.1086/650494. [DOI] [PubMed] [Google Scholar]
- 11.Novik V., Hofreuter D., Galan J.E. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 2010;78:3540–3553. doi: 10.1128/IAI.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baek K.T., Vegge C.S., Brondsted L. HtrA chaperone activity contributes to host cell binding in Campylobacter jejuni. Gut Pathog. 2011;3:13. doi: 10.1186/1757-4749-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlyshev A.V., Linton D., Gregson N.A., Wren B.W. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology. 2002;148:473–480. doi: 10.1099/00221287-148-2-473. [DOI] [PubMed] [Google Scholar]
- 14.van Vliet A.H., Wooldridge K.G., Ketley J.M. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 1998;180:5291–5298. doi: 10.1128/jb.180.20.5291-5298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkhill J., Wren B.W., Mungall K., Ketley J.M., Churcher C., Basham D., Chillingworth T., Davies R.M., Feltwell T., Holroyd S., Jagels K., Karlyshev A.V., Moule S., Pallen M.J., Penn C.W., Quail M.A., Rajandream M.A., Rutherford K.M., van Vliet A.H., Whitehead S., Barrell B.G. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 16.Wassenaar T.M., Fry B.N., van der Zeijst B.A. Genetic manipulation of Campylobacter: evaluation of natural transformation and electro-transformation. Gene. 1993;132:131–135. doi: 10.1016/0378-1119(93)90525-8. [DOI] [PubMed] [Google Scholar]
- 17.Hichey T.E., Mcveigh A.L., Scott D.A., Michielutti R.E., Bixby A., Carroll S.A., Bourgeois A.L., Guerry P. Campylobacter jejuni cytolethal distending toxin mediates release of interleukin-8 from intestinal epithelial cells. Infect. Immun. 2000;68:6535–6541. doi: 10.1128/iai.68.12.6535-6541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karlyshev A.V., Wren B. Development and application of an insertional system for gene delivery and expression in Campylobacter. Appl. Environ. Microbiol. 2005;71:4004–4013. doi: 10.1128/AEM.71.7.4004-4013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Patron C., Radomski M.W., Davidge S.T. Vascular matrix metalloproteinase-2 cleaves big endothelin-1 yielding a novel vasoconstrictor. Circ. Res. 1999;85:906–911. doi: 10.1161/01.res.85.10.906. [DOI] [PubMed] [Google Scholar]
- 20.Michaud D. Gel electrophoresis of proteolytic enzymes. Anal. Chim. Acta. 1998;372:173–185. [Google Scholar]
- 21.Jones M.A., Marston K.L., Woodall C.A., Maskell D.J., Linton D., Karlyshev A.V., Dorrell N., Wren B.W., Barrow P.A. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 2004;72:3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baek K.T., Vegge C.S., Skorko-Glonek J., Brondsted L. Different contributions of HtrA protease and chaperone activities to Campylobacter jejuni stress tolerance and physiology. Appl. Environ. Microbiol. 2011;77:57–66. doi: 10.1128/AEM.01603-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoy B., Geppert T., Boehm M., Reisen F., Plattner P., Gadermaier G., Sewald N., Ferreira F., Briza P., Schneider G., Backert S., Wessler S. Distinct roles of secreted HtrA proteases from Gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J. Biol. Chem. 2012;287 doi: 10.1074/jbc.C111.333419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young N.M., Brisson J.R., Kelly J., Watson D.C., Tessier L., Lanthier P.H., Jarrell H.C., Cadotte N., St Michael F., Aberg E., Szymanski C.M. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 25.Zhong G., Fan P., Ji H., Dong F., Huang Y. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 2001;193:935–942. doi: 10.1084/jem.193.8.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M.L., Ge Z., Fox J.G., Schauer D.B. Disruption of tight junctions and induction of proinflammatory cytokine responses in colonic epithelial cells by Campylobacter jejuni. Infect. Immun. 2006;74:6581–6589. doi: 10.1128/IAI.00958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehm M., Hoy B., Rohde M., Tegtmeyer N., Baek K.T., Oyarzabal O.A., Brondsted L., Wessler S., Backert S. Rapid paracellular transmigration of Campylobacter jejuni across polarized epithelial cells without affecting TER: role of proteolytic-active HtrA cleaving E-cadherin but not fibronectin. Gut Pathogen. 2012;4:3. doi: 10.1186/1757-4749-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prokhorova T.A., Nielsen P.N., Petersen J., Kofoed T., Crawford J.S., Morsczeck C., Boysen A., Schrotz-King P. Novel surface polypeptides of Campylobacter jejuni as traveller’s diarrhoea vaccine candidates discovered by proteomics. Vaccine. 2006;24:6446–6455. doi: 10.1016/j.vaccine.2006.05.085. [DOI] [PubMed] [Google Scholar]
- 29.Wells T.J., Tree J.J., Ulett G.C., Schembri M.A. Autotransporter proteins: novel targets at the bacterial cell surface. FEMS Microbiol. Lett. 2007;274:163–172. doi: 10.1111/j.1574-6968.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- 30.Hara H., Yamamoto Y., Higashitani A., Suzuki H., Nishimura Y. Cloning, mapping, and characterization of the Escherichia-Coli Prc Gene, which is involved in C-terminal processing of penicillin-binding protein-3. J. Bacteriol. 1991;173:4799–4813. doi: 10.1128/jb.173.15.4799-4813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gilbert K.B., Vanderlinde E.M., Yost C.K. Mutagenesis of the carboxy terminal protease CtpA decreases desiccation tolerance in Rhizobium leguminosarum. FEMS Microbiol. Lett. 2007;272:65–74. doi: 10.1111/j.1574-6968.2007.00735.x. [DOI] [PubMed] [Google Scholar]
- 32.Keiler K.C., Sauer R.T. Identification of active site residues of the Tsp protease. J. Biol. Chem. 1995;270:28864–28868. doi: 10.1074/jbc.270.48.28864. [DOI] [PubMed] [Google Scholar]
- 33.Liao D.I., Qian J., Chisholm D.A., Jordan D.B., Diner B.A. Crystal structures of the photosystem II D1 C-terminal processing protease. Nat. Struct. Biol. 2000;7:749–753. doi: 10.1038/78973. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto Y., Inagaki N., Satoh K. Overexpression and characterization of carboxyl-terminal processing protease for precursor D1 protein: regulation of enzyme-substrate interaction by molecular environments. J. Biol. Chem. 2001;276:7518–7525. doi: 10.1074/jbc.M008877200. [DOI] [PubMed] [Google Scholar]
- 35.Slomiany B.L., Piotrowski J., Mojtahed H., Slomiany A. Ebrotidine effect on the proteolytic and lipolytic activities of Helicobacter pylori. Gen. Pharmacol. 1992;23:203–206. doi: 10.1016/0306-3623(92)90010-h. [DOI] [PubMed] [Google Scholar]
- 36.Curtis M.A., Aduse Opoku J., Rangarajan M., Gallagher A., Sterne J.A., Reid C.R., Evans H.E., Samuelsson B. Attenuation of the virulence of Porphyromonas gingivalis by using a specific synthetic Kgp protease inhibitor. Infect. Immun. 2002;70:6968–6975. doi: 10.1128/IAI.70.12.6968-6975.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turk B.E., Wong T.Y., Schwarzenbacher R., Jarrell E.T., Leppla S.H., Collier R.J., Liddington R.C., Cantley L.C. The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor. Nat. Struct. Mol. Biol. 2004;11:60–66. doi: 10.1038/nsmb708. [DOI] [PubMed] [Google Scholar]
- 38.Popov S.G., Popova T.G., Hopkins S., Weinstein R.S., MacAfee R., Fryxell K.J., Chandhoke V., Bailey C., Alibek K. Effective antiprotease-antibiotic treatment of experimental anthrax. BMC Infect. Dis. 2005;5:25. doi: 10.1186/1471-2334-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shoop W.L., Xiong Y., Wiltsie J., Woods A., Guo J., Pivnichny J.V., Felcetto T., Michael B.F., Bansal A., Cummings R.T., Cunningham B.R., Friedlander A.M., Douglas C.M., Patel S.B., Wisniewski D., Scapin G., Salowe S.P., Zaller D.M., Chapman K.T., Scolnick E.M., Schmatz D.M., Bartizal K., Maccoss M., Hermes J.D. Anthrax lethal factor inhibition. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7958–7963. doi: 10.1073/pnas.0502159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brotz-Oesterhelt H., Beyer D., Kroll H.P., Endermann R., Ladel C., Schroeder W., Hinzen B., Raddatz S., Paulsen H., Henninger K., Bandow J.E., Sahl H.G., Labischinski H. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression and purification of Cj0511His protein after overexpression in E. coli. 1, cell lysate; 2, soluble fraction; 3, pellet; 4, flow through; 5 purified protein; M, size markers (SeeBlue, Invitrogen).
Motility test. Three clonal isolates of strain 11168H/cj0511::kanr (clones 1–3) were stabbed into semi-solid (0.3%) Mueller Hinton agar plate and grown for two days at 37 °C. Compared to recipient strain 11168H, no difference in motility was observed.
Putative peptidases in C. jejuni NCTC 11168 (excluding non-peptidase homologues Cj0190, Cj0463, Cj1005. Cj1205 and Cj1275).


