Abstract
Renal cell carcinoma of clear-cell type (ccRCC) is an enigmatic tumor type, characterized by frequent inactivation of the VHL gene (infrequently mutated in other tumor types), responsiveness to angiogenesis inhibitors, and resistance to both chemotherapy and conventional radiation therapy. ccRCC tumors exhibit substantial mutation heterogeneity. Recent studies using massively parallel sequencing technologies have implicated several novel driver genes. In VHL wild-type tumors, mutations were discovered in TCEB1, which encodes Elongin C, a protein that binds to VHL and is required for its function. Several additional tumor suppressor genes have been identified near the VHL gene, within a region that is frequently deleted in ccRCC on chromosome 3p: SETD2, BAP1, and PBRM1. Mutations in BAP1 and PBRM1 are largely mutually exclusive and are associated with different tumor biology and patient outcomes. In addition, the mTORC1 pathway is deregulated by mutations in MTOR, TSC1, PIK3CA, and PTEN in approximately 20% of ccRCCs. Mutations in TSC1, and possibly other genes, may predict for sensitivity to mTORC1 inhibitors. These discoveries provide insight into ccRCC development and set the foundation for the first molecular genetic classification of the disease, paving the way for subtype-specific therapies.
INTRODUCTION
Kidney tumors are estimated to be diagnosed in more than 270,000 individuals every year worldwide.1 More than 65,000 new diagnoses and approximately 13,680 patient deaths as a result of tumors of the kidney and renal pelvis were projected in the United States for 2013.2 In the United States, 15% to 20% of individuals present with lymph node metastases and a similar percentage have distant involvement at the time of diagnosis.3 In the metastatic setting, renal cancer remains largely incurable. The majority of malignant kidney tumors are renal cell carcinomas (RCC) and approximately 70% are RCCs of clear-cell type (ccRCC).4
MOLECULAR GENETICS OF ccRCC
The Cancer Genome Atlas consortium analyzed over 400 tumor/normal pairs.5 On average, ccRCCs exhibit less than 20 DNA copy-number alterations, fewer changes than in colon and breast cancers.5 Proportionally, however, there is an overrepresentation of copy-number alterations involving whole chromosome arms.5 RNA fusions (resulting from translocation events) were observed in 10% to 20% of ccRCCs and the vast majority of them were unique.5 A second study by Sato et al6 evaluated more than 100 ccRCCs using whole genome or exome sequencing. Overall, ccRCCs are characterized by one to two somatically acquired single nucleotide variants or small insertions and deletions (indels) per megabase pair (approximately 3,000 to 6,000 mutations per tumor).5,6 Most of these mutations occur outside coding regions. Protein-coding regions account for approximately 1% of the genome and are subject to approximately 1% of the mutations, suggesting that mutations occur randomly.6 Table 1 lists genes mutated in ccRCC in both studies.5,6
Table 1.
ccRCC-Mutated Genes
| Genes | TCGA Cohort |
Japanese Cohort* |
||
|---|---|---|---|---|
| Tumors With Mutation (%) | Passenger Probability (q value) | Tumors With Mutation (%) | Passenger Probability (q value) | |
| VHL | 52.3 | < .0001 | 39.6† | < .0001 |
| PBRM1 | 32.9 | < .0001 | 26.4 | < .0001 |
| SETD2 | 11.5 | < .0001 | 11.3 | < .0001 |
| BAP1 | 10.1 | < .0001 | 7.5 | < .0001 |
| MTOR | 6 | < .0001 | 5.7 | .0431 |
| TCEB1 | 0.7 | .0566 | 4.7‡ | < .0001 |
| PIK3CA | 2.9 | < .0001 | 4.7 | .0268 |
| KDM5C | 6.7 | < .0001 | 3.8 | .12 |
| TP53 | 2.2 | < .0001 | 2.8 | .0176 |
| PTEN | 4.3 | < .0001 | 1.9 | .116 |
Abbreviations: ccRCC, clear-cell renal cell carcinoma; TCGA, The Cancer Genome Atlas.
Mutations found by whole exome sequencing.
Including complementary approaches overall VHL mutation rate, 66%.
There is significant mutation heterogeneity within ccRCC tumors.7 According to their prevalence, somatic mutations are classified into ubiquitous, shared, and private mutations.7 Ubiquitous mutations are present in every tumor cell. Shared and private mutations are found in progressively smaller subclones. Overall, mutation prevalence reflects the time of mutation acquisition, with ubiquitous mutations representing early, truncal events and shared and private representing progressively more distant subclones or branches. However, this timeline may be distorted by later mutations with a disproportionate proliferative advantage or other factors.
According to their significance, mutations are classified into drivers and passengers.8 Driver mutations include those implicated in tumor initiation and progression. Ubiquitous mutations are not necessarily driver mutations. Indeed, unselected mutations acquired during the normal process of DNA replication in the cell lineage that ultimately results in the initial tumor clone represent ubiquitous passengers.9 Only a subset of mutations (possibly fewer than 10 protein-coding gene mutations) are drivers. In addition, driver mutations may be found among shared and private mutations.
Mutation heterogeneity may be advantageously exploited. The best therapeutic targets may be found in pathways deregulated by ubiquitous driver mutations present in every tumor cell. These mutations may be more easily identified by exploiting mutation heterogeneity. Furthermore, tumors likely develop as a set of conditional dependencies in which new mutations build on the confines imposed by pre-existing mutations,10 and the degree of dependency of a tumor on a pathway may be related to how early the corresponding mutation occurred. This conditional or contextual nature of oncogenic mutations fits well with the empiric observation that mutations exert their protumorigenic effect in a tissue-dependent manner.11 For example, in dominantly inherited familial cancer-prone syndromes, tumors develop in a subset of tissues despite the presence of the mutation in every diploid cell.
Experimentally, whether a mutation is ubiquitous can be inferred from sampling multiple areas of the tumor.7 In addition, mutant-allele ratios (MAR), referring to the fraction of mutant over mutant plus wild-type alleles for each mutation, may also help determine the prevalence of a mutation. Ubiquitous heterozygous mutations have MARs of approximately 0.5. However, if the mutation arose later and is only present in 50% of the tumor cells, the MAR would be 0.25. Similar MARs may be found in mutations arising around the same time, and this approach was used by Sato et al6 to define subclonal populations. However, MARs are confounded by DNA copy-number alterations as well as by contamination with normal DNA (from stroma or inflammatory cells). While cumbersome, the problem of contamination may be resolved by implanting the tumors in mice, which results in the selective expansion of tumor cells while the stroma is replaced by the host.12 Although the focus of this article is on genetic events, epigenetic alterations most likely contribute to cancer development.13
VHL COMPLEX IS BROADLY INACTIVATED IN ccRCC
The von Hippel-Lindau (VHL) gene is inactivated by either mutation or methylation in over 80% of ccRCC.6,14–16 VHL was originally identified as the gene responsible for the ccRCC-predisposing syndrome, von Hippel-Lindau.17 VHL is a two-hit tumor suppressor gene and, typically, one allele is inactivated through an intragenic mutation and the second is deleted as part of large deletion. The VHL gene is on chromosome 3p25.3 and deletions in this region, which often involve the whole short arm of chromosome 3, are observed in approximately 90% of ccRCC.18–20 At times, a VHL mutation is found without a 3p deletion. However, a deletion may have occurred, accompanied by duplication of the remaining chromosomal region, resulting in copy-neutral loss of heterozygosity (LOH). Consistent with this, ccRCCs with VHL mutations and copy-neutral LOH exhibited VHL MARs that were higher than for control genes (mutated genes in diploid regions without LOH).6 In this setting, both alleles of VHL would be inactivated by the same mutation.
The VHL protein forms a complex with Elongin B, Elongin C, Cul2, and Rbx1 that functions as an E3 ubiquitin ligase toward, most prominently, the α subunits of HIF (hypoxia-inducible factor) transcription factors (Fig 1).21 Many mutations in VHL disrupt protein expression, but missense mutations often cluster in the interface between VHL and Elongin C.22 Interestingly, the Elongin C gene (called TCEB1) is mutated in 0.5% to 5% of ccRCCs (Table 1).6 TCEB1 mutations are uniformly associated with LOH of 8q21, where TCEB1 is located.6 As expected, these mutations are exclusive with VHL mutations (P < .0001).6 This is consistent with the notion that mutations in either VHL or TCEB1 are sufficient to inactivate the function of the complex.
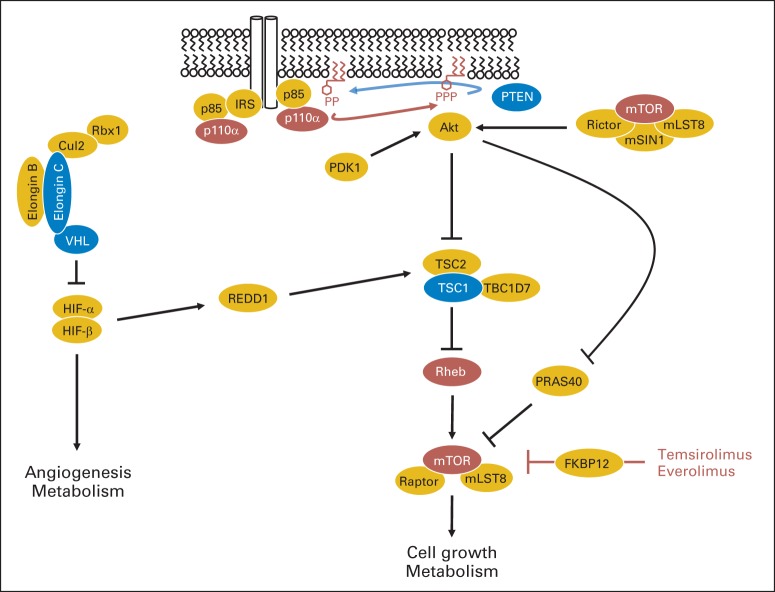
Fig 1.
Interplay between VHL and mTORC1 pathways. In the presence of growth factors, transphosphorylation by the intracellular domains of receptor tyrosine kinases leads to recruitment of the regulatory subunit of class IA PI3K, p85 (either directly or through adaptor proteins like IRS) and releases its inhibition of the catalytic subunit (p110α; encoded by the PIK3CA gene), which phosphorylates phosphatidylinositol-4,5-trisphosphate to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3). PIP3 recruits interacting proteins to the plasma membrane, such as Akt, which is phosphorylated and activated by PDK1 and mTORC2 (mTOR complex 2). Akt phosphorylates TSC2, which is in a complex with TSC1 and TBC1D7, releasing its inhibition on Rheb. Activated Rheb binds to and activates mTORC1. mTORC1 is inhibited by PRAS40, and this inhibition is also released by AKT activation. mTORC1 is also inhibited by REDD1 in a manner that requires TSC1/TSC2. REDD1 is transcriptionally induced by both HIF-1 and HIF-2, which are activated following the inactivation of the VHL complex through mutations in either VHL or TCEB1 (encoding Elongin C). mTORC1 is inhibited by temsirolimus and everolimus, which interact with FKBP12 and subsequently bind to mTORC1. Brown ovals, oncoproteins activated by mutation in ccRCC; blue ovals, tumor-suppressor proteins inactivated by mutation in ccRCC.
Enigmatically, TCEB1 mutations are not typical loss-of-function mutations. All mutations reported by Sato et al6 were missense mutations at two conserved residues, Tyr79 (n = 7) and Ala100 (n = 1). These mutations seemingly interfere with VHL binding to Elongin C and lead to the stabilization of HIF-α subunits. However, the pattern of mutation suggests that the situation is more complex. Perhaps other functions of elongin C need to be preserved.
Overall, Sato et al6 found evidence of VHL complex inactivation in 92% of ccRCC (97 of 106 tumors). In this cohort, VHL mutations were found in 66% (70 of 106 tumors),VHL methylation in 21% (22 of 106 tumors), and TCEB1 mutation in 5% (five of 106 tumors). Whether the VHL complex is inactivated in the remaining tumors is unclear. In the nine remaining ccRCCs, no mutations were found in other complex components.6 However, immunohistochemistry (IHC) analyses showed that seven tumors expressed HIF-α (HIF-1α, HIF-2α, or both) at levels comparable to VHL-deficient tumors and several of these had low mutation numbers, raising the possibility that some mutations may have been missed. In addition, VHL may have been inactivated through mutations outside sequenced regions. The remaining two ccRCCs had no detectable HIF-α expression. One of these was a translocation carcinoma involving the TFE3 gene, and translocation carcinomas may lack VHL mutations.5 Thus, most, if not all, ccRCC may have deregulation of the VHL pathway.
PBRM1 IS THE SECOND MOST FREQUENTLY MUTATED GENE IN ccRCC
Polybromo 1 (PBRM1) is mutated in approximately 45% of ccRCC.23 Lower mutation frequencies in recent studies5,6 may reflect reduced sensitivity, and other studies have shown comparable mutation frequencies.12 The majority of mutations are truncating, and PBRM1 functions as a two-hit tumor suppressor gene.23 Furthermore, PBRM1 is on the same chromosome arm as VHL and the second allele is frequently codeleted with VHL.23 As expected, most PBRM1 mutations are accompanied by loss of the protein.12 Analyses of MARs (as well as tumor sampling studies) show that PBRM1 mutations may be ubiquitous.6,7
PBRM1 encodes BAF180, a component of a nucleosome-remodeling complex. Nucleosomes are histone octamers composed, typically, of two copies of each of four canonical histone proteins (H2A, H2B, H3, and H4), around which 147 bp of DNA are wrapped.24 DNA binding to histones limits its accessibility to transcription factors and RNA polymerases. DNA accessibility is regulated by remodelers, which unwrap, reposition, and eject nucleosomes.25
There are currently four different families of remodeler complexes, including the switching defective/sucrose nonfermenting (SWI/SNF) family.25 These families differ in subunit composition and biologic function.25 SWI/SNF complexes are organized around an ATPase that provides energy to break DNA/histone contacts, brahma homolog (BRM), and brahma-related gene 1 (BRG1).26,27 According to the subunit composition, SWI/SNF complexes are subdivided into BRG1-associated factor (BAF) and polybromo BRG1-associated factor (PBAF) complexes (Table 2). Both contain approximately 15 subunits, but whereas BAF complexes contain either a BRM or BRG1 subunit, only a BRG1 is in PBAF complexes.27,28 BAF complexes are thought to be targeted to chromatin by BAF250 proteins, whereas targeting of PBAF complexes involves BAF180 and BAF200.27,29
Table 2.
SWI/SNF Genes and Proteins
| Location | Gene | Subunit | Complex | Mutated in ccRCC |
|---|---|---|---|---|
| ATPase | ||||
| 9p22.3 | SMARCA2 | BRM | BAF | + |
| 19p13.2 | SMARCA4 | BRG1 | BAF/PBAF | ++ |
| Targeting | ||||
| 1p35.3 | ARID1A | BAF250A | BAF | +++ |
| 6q25.1 | ARID1B | BAF250B | BAF | ++ |
| 12q12 | ARID2 | BAF200 | PBAF | |
| 3p21 | PBRM1 | BAF180 | PBAF | (+++++++++++++++)2 |
| Other | ||||
| 12q13.2 | SMARCC2 | BAF170 | BAF/PBAF | ++ |
| 3p21.31 | SMARCC1 | BAF155 | BAF/PBAF | |
| 12q13-q14 | SMARCD1 | BAF60A | BAF/PBAF | ++ |
| 17q23-q24 | SMARCD2 | BAF60B | BAF/PBAF | |
| 7q35-q36 | SMARCD3 | BAF60C | BAF/PBAF | |
| 17q21.2 | SMARCE1 | BAF57 | BAF/PBAF | |
| 3q26.33 | ACTL6A | BAF53A | BAF/PBAF | |
| 7q22 | ACTL6B | BAF53B | BAF/PBAF | |
| 22q11 | SMARCB1 | BAF47 | BAF/PBAF | + |
BAF180 (encoded by the PBRM1 gene) contains six tandem bromodomains, two bromo-adjacent homology (BAH) domains and a high-mobility group (HMG) box.29 Bromodomains bind acetylated lysine residues in histone tails and may target PBAF to chromatin (Fig 2A). Different BAF180 bromodomains show different affinities for acetylated lysine residues in vitro, and BAF180 may target PBAF to a specific pattern of acetylated lysines.29,31 Disruption of a single bromodomain may suffice to abrogate tumor suppressor function.12,31
Fig 2.

PBRM1- and BAP1-mutant tumors are associated with different biology, pathologic features, and outcomes, setting the foundation for a molecular genetic classification of clear-cell renal cell carcinoma (ccRCC). (A) BAF180 (encoded by the PBRM1 gene) contains six tandem bromodomains that bind to acetylated lysine residues in histone tails, thereby localizing the PBAF chromatin remodeling complex to specific chromatin regions and regulating gene expression. (B) BAP1 interacts with HCF-1 and functions to deubiquitinate proteins, including histone H2AK119ub1. By deubiquitinating its substrates, BAP1 may inhibit protein degradation or, in the case of H2A, alter gene expression. PBRM1- and BAP1-mutant tumors are associated with different gene expression signatures, pathologic features, mTORC1 activation, and outcomes (Kapur et al30). (C) Pie chart representation of ccRCC subtypes and their approximate frequencies. HR, hazard ratio; other, tumors without detectable mutations in PBRM1 and BAP1.
Other genes encoding subunits of the SWI/SNF complex are mutated in ccRCC but at much lower frequencies (Table 2).5,6,32 However, these mutations are not exclusive with PBRM1,6,23 and how they cooperate in tumor development is unclear.
How PBRM1 loss promotes tumorigenesis is poorly understood. In keeping with its role in nucleosome remodeling, ccRCCs deficient in PBRM1 are associated with a distinct gene-expression signature.30 PBRM1-mutant ccRCCs are enriched for genes in pathways implicated in the cytoskeleton and cell motility.30 In addition, reintroduction of PBRM1 into PBRM1-deficient cells induces the expression of the cyclin-dependent kinase inhibitor p21.33 This is accompanied by a reduction in cell proliferation.33 Finally, PBRM1 was identified in a small-hairpin RNA (shRNA) screen for genes whose inactivation would extend the proliferative capacity of primary fibroblasts in culture.34 Thus, PBRM1 appears to regulate cell proliferation. Studies in insect cells and mice suggest that SWI/SNF complexes are in a functionally antagonistic relationship with polycomb group proteins.26 However, whether this will offer opportunities for therapeutic intervention remains to be determined.35
SETD2 GENE
The gene encoding SET domain containing protein 2 (SETD2) is somatically mutated in approximately 10% to 15% of ccRCCs (Table 1).5,6,36,37 Like VHL and PBRM1, SETD2 is a two-hit tumor suppressor gene and is located on chromosome 3p. SETD2 mutations tend to be in the shared group.6,7 Analyses of data provided by Sato et al6 show that SETD2 MARs are lower than VHL MARs in one third of ccRCCs, suggesting that in these tumors SETD2 mutations are subclonal. In addition, sampling studies have shown different SETD2 mutations in different samples of the same tumor.7 This mutation convergence suggests a high selective pressure to mutate SETD2 in some contexts; a meta-analysis suggests that SETD2 mutations cooperate with mutations in PBRM1.11 Though the molecular basis remains unclear, both BAF180 and SETD2 converge on histones, one as a reader (BAF180) and the other as a writer (SETD2).
How biallelic SETD2 inactivation leads to ccRCC is unclear. The SETD2 protein is a nonredundant histone H3 lysine 36 trimethylating (H3K36me3) enzyme.38 Though H3K36 methylation is generally linked to active transcription, it is also associated with alternative splicing and transcriptional repression.39 Interestingly, a recent study has linked SETD2 and H3K36me3 to DNA mismatch repair,40 and microsatellite instability was found in a subset of ccRCC.41 In addition, a link has been reported in ccRCC between SETD2 mutation and DNA methylation.5
BAP1 IS A DRIVER OF TUMOR AGGRESSIVENESS
The BRCA1 associated protein-1 (BAP1) gene is mutated in 10% to 15% of patients with ccRCC.12,42 BAP1 was originally identified in a yeast two-hybrid screen for BRCA1-interacting proteins,43 but endogenous BAP1 seems not to bind BRCA1 in mammalian cells. Guo et al42 performed exome sequencing in a small number of ccRCCs with targeted sequencing of selected genes in an expansion cohort. They reported a list of 12 genes that mutated in ccRCC at frequencies higher than expected by chance alone. This list included the TSC1 gene, which was reported previously to be mutated in ccRCC,44 BAP1, and several other genes not identified in other studies.5,6,42 A similar approach focusing on high-grade ccRCC led to the identification of BAP1 mutations in ccRCC by our group.12
BAP1 is a two-hit tumor suppressor gene located on chromosome 3p between the VHL and PBRM1 genes. The BAP1 protein interacts with host cell factor-1 (HCF-1), a protein that serves as a scaffold for several chromatin remodeling complexes.45–47 BAP1 mutations are typically associated with loss of the protein.12 As determined by analyses of results provided by Sato et al,6 MARs for BAP1 are similar to VHL in 70% to 80% of ccRCCs and substantially lower in the rest. These data suggest that in a subset of tumors, BAP1 mutations are subclonal. Recent studies using a validated IHC test in approximately 1,400 ccRCCs identified focal loss of BAP1 in 2% to 3% of ccRCCs.48 However, the number of tumors with focal loss of BAP1 is likely to be much larger, as a single section per tumor was examined.
Frequent somatic mutation of BAP1 was first described in metastasizing uveal melanoma and subsequently in malignant pleural mesothelioma.49,50 Notably, BAP1 is also mutated in the germline.51,52 Germline BAP1 mutations are associated with a syndrome characterized by uveal and cutaneous melanoma, mesothelioma, and RCC.51–54 The presence of different tumors in individual families51,53 suggest that the specific mutation alone does not dictate the tumor spectrum. In two families, ccRCC was the dominant feature.53,54 The finding that germline mutations in BAP1 predispose to ccRCC suggest that BAP1 loss can initiate RCC development.
BAP1 is a deubiquitinase of the ubiquitin C-terminal hydrolase (UCH) family.43 BAP1 localizes to the nucleus, and nuclear localization is required for BAP1 tumor-suppressor function.55 BAP1 contains an N-terminal catalytic domain, an HCF-1 binding motif (HBM), and a C-terminal UCH37-like domain (ULD).56 The catalytic domain is often targeted by missense mutations in ccRCC.12
In Drosophila, BAP1 (Calypso) functions as an H2A deubiquitinase.57 Similarly, mammalian BAP1 is able to deubiquitinate H2AK119ub1.57 Drosophila Calypso is a polycomb repressive deubiquitinase that silences genes implicated in body planning and patterning.57 Polycomb complexes regulate different gene expression programs in different lineages.58,59 This cell-context dependency also characterizes BAP1. Furthermore, BAP1 deubiquitinates different proteins in different cell types,60 and BAP1 can both promote and suppress cell proliferation in a cell-type dependent manner.61
An important difference between Calypso and mammalian BAP1 is that Calypso lacks the HBM motif implicated in binding to HCF-1 (Fig 2B). This motif may be important as most BAP1 in cells seems to be bound to HCF-1.46 In addition, mutation of the motif impairs the growth suppressive function of BAP1 in renal cancer cells.12 HCF-1 serves as a chromatin scaffold protein for multiple histone modifying enzymes.62 HCF-1 is also a substrate for BAP1,45,56,60 but this seems to be cell-type specific and its relevance in renal cancer is unclear.12
BAP1-deficient ccRCCs are characterized by a specific gene-expression signature.30 This signature is enriched for pathways implicated in growth factor and phosphatidylinositol 3-kinase (PI3K) signaling.30 Consistent with this, BAP1-deficient tumors exhibit increased mammalian (or mechanistic) target of rapamycin (mTOR) complex 1 (mTORC1) activation.12 In addition, BAP1 mutations in tumors seemingly correlate with methylation changes of polycomb repressive complex 2 (PRC2) target genes.6
Interestingly, BAP1 and PBRM1 mutations in ccRCC are largely mutually exclusive (Fig 2C).12 As determined by a meta-analysis, the odds of having mutations in BAP1 are reduced by 70% in PBRM1-mutated tumors.11 The molecular basis of this relationship is unknown. Mutation exclusivity is often interpreted to mean that genes are in the same pathway (such as VHL and TCEB1). However, BAP1- and PBRM1-mutant tumors are associated with different histologic features, biology, and outcomes (Fig 2). Whereas BAP1-mutant tumors tend to be of high grade and may show coagulative necrosis, PBRM1-mutant tumors may be of high or low grade and less frequently exhibit necrosis.12,30,37 PBRM1- and BAP1-mutant tumors are associated with characteristic, but independent, gene-expression signatures.30 In addition, BAP1-mutant tumors tend to be associated with mTORC1 activation, but PBRM1-mutant tumors are not.12,30 Finally, BAP1- and PBRM1-mutant tumors are associated with markedly different outcomes in patients. In the localized or locoregional setting, patients with BAP1-mutant tumors have a 2.5- to three-fold higher hazard ratio for death than those with PBRM1-mutant tumors.30 Similar results have been obtained in several other studies.5,6,37,63,64 An IHC test has been developed with high sensitivity and specificity for BAP1 loss.12,48,64 Using this test, BAP1 loss seems to be an independent predictor of outcome.48 In a retrospective analysis of approximately 1,400 patients with nonmetastatic ccRCC, loss of BAP1 in the tumor was associated with an increased risk of ccRCC-associated death (hazard ratio, 3.06; 95% CI, 2.28 to 4.10; P = 6.77 × 10−14). BAP1 loss remained an independent predictor after adjusting for University of California, Los Angeles–integrated staging system (UISS) variables and in patients with low stage, size, grade, and necrosis (SSIGN) scores.48 The higher aggressiveness of BAP1-mutant tumors is reminiscent of its role in uveal melanoma, in which BAP1 mutation correlated with metastasizing potential.65 However, whether BAP1 and PBRM1 predict for outcomes in patients with metastatic disease remains to be determined. The largely exclusive nature of BAP1 and PBRM1 mutations in ccRCC coupled with associated differences in tumor biology and outcomes establishes a foundation for a molecular genetic classification of ccRCC (Fig 2C).
A small percentage of tumors harbor mutations in both BAP1 and PBRM1, and these tumors seem to be the most aggressive.30 Though mutation heterogeneity in tumors is well documented, evidence from analyses of tumorgrafts and IHC studies suggests that these mutations co-occur in the same tumor cells.12 However, double-mutant tumors should be distinguished from tumors harboring subclones individually mutated for one or the other gene, and their outcomes may be different.
Though BAP1 mutations are not always ubiquitous,6,48 because of their association with tumor aggressiveness and poor outcomes, BAP1-regulated pathways may be appropriate therapeutic targets. As a tumor suppressor, BAP1 itself is not a suitable target. There is a correlation between BAP1 loss and mTORC1 activation, but this effect does not seem to be direct.12 However, whether BAP1 loss sensitizes to mTORC1 inhibitors remains to be determined. In addition, loss of BAP1 may sensitize ccRCCs to radiation, but this effect is modest.12 Therapeutic targets may arise from a greater understanding of the mechanism of BAP1 action and, in particular, identifying the enzyme responsible for ubiquitinating relevant BAP1 substrates.
MODEL OF ccRCC DEVELOPMENT
VHL, PBRM1, SETD2, and BAP1 are within a 50 Mb stretch on chromosome 3p, in a region that is lost in approximately 90% of sporadic ccRCCs.11 Deletion of this region simultaneously inactivates one allele of four ccRCC tumor-suppressor genes, leaving cells vulnerable to the loss of the remaining allele.11
The available data support the following model of ccRCC development (Fig 3). ccRCC may be initiated by an intragenic mutation of VHL, followed by the loss of chromosome 3p. VHL mutations are an initiating event and VHL inactivation has been observed in isolated cells lining tubules and in single-layered cysts.66,67 Mutations in the remaining PBRM1 allele would contribute to transformation and may synergize with subsequent mutations in SETD2. A second path involves mutation of the remaining BAP1 allele, which may confer greater aggressiveness. The frequency of tumors simultaneously mutated for BAP1 and PBRM1 is lower than expected11,12; simultaneous inactivation of these two tumor-suppressor genes in the same tumor cell may reduce fitness. However, because simultaneous mutations do occur in some tumors, there may be a context-dependent advantage.
Fig 3.
Model for clear-cell renal cell carcinoma (ccRCC) development. The genes VHL, BAP1, and PBRM1 are all located on chromosome (Chr) 3p (SETD2 is also in this region; not shown). Following an intragenic mutation in VHL, loss of 3p, which is observed in the majority of ccRCCs, inactivates the remaining VHL allele along with one allele of BAP1 and PBRM1. Subsequent mutation in the remaining PBRM1 or BAP1 allele results in ccRCC with different pathologic features and outcomes.
In a fraction of ccRCCs, there are no deletions of 3p; instead, there is copy-neutral LOH.6 Analyses of data provided6 reveal that these tumors also exhibit mutations in PBRM1, SETD2, and BAP1. Overall, MARs for these genes are similar to those observed in VHL, suggesting that, as for VHL, mutations in these genes preceded the chromosome 3p duplication event.
It is noteworthy that SMARCC1 (encoding BAF155, a subunit of both BAF and PBAF complexes) is located on 3p21.31, between the VHL and PBRM1 genes (Table 2). Because of its location, one copy of SMARCC1 is lost in most ccRCCs. This would make inactivating the second allele as accessible to the tumor cell as the inactivation of PBRM1. However, whereas PBRM1 is mutated in 45% of ccRCCs, mutations in SMARCC1 have not been detected among 459 kidney tumors with information in COSMIC.32 This difference may be biologically significant and suggests that, in contrast to BAF180, BAF155 is not a ccRCC-tumor suppressor.35 Furthermore, BAF155 function may be required for cell fitness.68–70 Because of the selective loss of one allele in ccRCC, these tumors may be particularly sensitive to strategies inhibiting BAF155-dependent BAF/PBAF complexes.
The evolution of ccRCCs with mutations in TCEB1 may be different from those with mutations in VHL, as TCEB1 is on chromosome 8. Sato et al6 provided extensive data on five tumors with TCEB1 mutations. Mutations in PBRM1, SETD2, and BAP1 were found in only one tumor (which had a BAP1 mutation). The absence of PBRM1 mutations potentially highlights the importance of the physical location of tumor-suppressor genes in tumor evolution.
MUTATIONS IN mTORC1 PATHWAY GENES
Growth factor signaling pathways are frequently deregulated in cancer.71 In ccRCC, however, receptor tyrosine kinases are rarely mutated.5,6 Receptor activation leads to the recruitment of adaptor proteins, as well as class IA PI3K, to the plasma membrane.72 Class IA PI3Ks are made up of a catalytic subunit (p110) and a regulatory subunit (p85; Fig 1). Among the different catalytic subunits, p110α (encoded by the PIK3CA gene) is the most frequently mutated in tumors.72 PIK3CA is mutated in 2% to 5% of ccRCCs.5,6 PIK3CA mutations tend to be missense mutations5,6 and include mutations reported previously in other tumor types to increase PI3K activity in vitro. PI3K catalyzes the formation of the second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3) at the plasma membrane, which is downregulated by the tumor suppressor phosphatase, phosphatase and tensin homolog (PTEN). PTEN was previously shown to be mutated in ccRCC.73 PTEN mutations are loss-of-function mutations and occur in 1% to 5% of ccRCCs (Table 1).5,6 Activating mutations in PIK3CA or inactivating mutations in PTEN should increase PIP3 levels, leading to recruitment to the plasma membrane of proteins with PIP3-binding domains, such as AKT isoforms. AKT phosphorylates multiple substrates, including tuberous sclerosis complex 2 (TSC2), which forms a complex with TSC1.74 The TSC1/TSC2 complex functions as a tumor suppressor. TSC1 stabilizes TSC2,75 and TSC1 was previously reported to be mutated in approximately 4% of ccRCCs.44 Although our group did not find mutations in TSC2,76 TSC2 may also be mutated in ccRCC.5,32
The TSC1/TSC2 complex functions as a GTPase-activating protein (GAP) leading to reduced levels of the active GTP-bound form of Ras homolog enriched in brain (Rheb). RHEB mutations were identified in four ccRCCs from the Cancer Genome Atlas.5,32 Three of these mutations affected the same amino acid (Tyr35). Tyr35 assists in TSC2-mediated GTP hydrolysis,77 and by reducing the activity of TSC2, mutations at Tyr35 may increase Rheb-GTP levels (L. Kinch and J. Brugarolas, unpublished data, July 2013). GTP-bound Rheb binds to and activates mTORC1.78 Thus, mutations in RHEB represent another potential mechanism to active mTORC1.
MTOR is mutated in approximately 5% of ccRCCs (Table 1).5,6 mTOR nucleates two different complexes (mTORC1 and mTORC2). mTORC1 is implicated in cell growth control and may be the relevant target in tumorigenesis. mTOR is a serine/threonine kinase composed of HEAT repeats (approximately 40 amino acids each), which make up the N-terminal half of the protein, and a kinase domain that is flanked by two domains: FAT and FATC.79 Recent structural studies revealed that the kinase domain adopts a bilobed structure with a central cleft that binds ATP. The kinase domain contains several insertions, including an approximately 100 amino acid insertion corresponding to the FKBP12/rapamycin-binding (FRB) domain, a domain that binds to rapamycin (also called sirolimus). The majority of MTOR mutations found in renal cancer are missense mutations.5,6,32 However, unlike activating mutations in other oncogenes, MTOR mutations affect an extensive number of residues. Seventy percent of MTOR mutations in renal cancer converge on two domains, the kinase and FAT domains. Several mutations map to regions implicated in restricting substrate accessibility.80 A few mutations (p.L1460P, p.S2215Y, and p.R2505P)32 have been evaluated in vitro and increase mTORC1 activity.81 These mutations did not appear to increase mTORC2 activity, suggesting that mTORC1 is the relevant oncogenic complex.81 Importantly, in two mutations examined (p.L1460P and p.S2215Y), sensitivity to sirolimus was preserved.81 MTOR mutations have been hypothesized to sensitize to sirolimus analogs such as temsirolimus and everolimus.82 However, mutations mapping to the FRB domain may affect binding to sirolimus (as well as temsirolimus and everolimus) and could confer resistance.
Though mutation frequencies in many genes that encode components of this pathway fail to reach statistical significance, as a whole, the mTORC1 pathway seems to be activated by somatic mutation in approximately 20% of ccRCCs.5,6 Mutations in proximal mTORC1 regulators may predict responsiveness to mTORC1 inhibitors clinically. The first TSC1 mutation reported in a ccRCC was found in a patient that remained on everolimus in the second line for 13 months after progressing on sunitinib after 3 months of treatment.44 This led us to hypothesize that TSC1 mutations clinically predicted for responsiveness to mTORC1 inhibitors.44,83 This concept is supported by emerging data in renal cancer and other tumor types.82,84 As for TSC1, mutations in TSC2 and RHEB may predict for responsiveness to mTORC1 inhibitors clinically. However, whether mutations in genes encoding proteins more distant to mTORC1, such as PIK3CA and PTEN, predict for responsiveness to mTORC1 inhibitors is less certain.
A negative feedback loop links VHL and mTORC1 pathways (Fig 1).83 mTORC1 is downregulated in response to a variety of stresses including hypoxia85,86 and this is mediated, at least in part,87 by regulated in development and DNA damage response 1 (REDD1).88 REDD1 expression is directly induced by both HIF-1 and HIF-2 in ccRCC,44 and REDD1 induction is sufficient to inhibit mTORC1.88 Like many other HIF-target genes, REDD1 is consistently upregulated in most ccRCCs.44 However, mTORC1 is often activated in ccRCC.89 This may be accomplished by mutations inactivating TSC1 (which is required for REDD1 signaling)44,88 or PTEN.44 However, this accounts for only a small percentage of tumors and how mTORC1 is reactivated in the rest despite REDD1 induction, remains unknown.
CONCLUSION AND FUTURE DIRECTIONS
Discoveries about the molecular genetics of ccRCC have shed light on tumor development, have led to the identification of previously unknown subtypes with different biology and outcomes, and may help with more accurate prognostication. These discoveries set the foundation for the next generation of molecularly targeted therapies.
Acknowledgment
I thank Payal Kapur, MD, for discussions on this topic. I apologize to colleagues whose work was not cited owing to space limitations or oversight, and I would appreciate being notified about any omissions.
Footnotes
Supported by Grants No. RP101075 and RP130603 from Cancer Prevention Research Institute of Texas and Grants No. R01CA129387 and R01CA175754 from the National Institutes of Health.
Author's disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHOR'S DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author indicated no potential conflicts of interest.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975-2010. Bethesda, MD: National Cancer Institute; 2013. [Google Scholar]
- 4.Baldewijns MM, van Vlodrop IJ, Schouten LJ, et al. Genetics and epigenetics of renal cell cancer. Biochim Biochim Biophys Acta. 2008;1785:133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Creighton CJ, Morgan M, Gunaratne PH, et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 7.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomasetti C, Vogelstein B, Parmigiani G. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci U S A. 2013;110:1999–2004. doi: 10.1073/pnas.1221068110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peña-Llopis S, Christie A, Xie XJ, et al. Cooperation and antagonism among cancer genes: The renal cancer paradigm. Cancer Res. 2013;73:4173–4179. doi: 10.1158/0008-5472.CAN-13-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peña-Llopis S, Vega-Rubín-de-Celis S, Liao A, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyn H, Esteller M. DNA methylation profiling in the clinic: Applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 14.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14:4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latif F, Tory K, Gnarra J, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 18.Zbar B, Brauch H, Talmadge C, et al. Loss of alleles of loci on the short arm of chromosome 3 in renal cell carcinoma. Nature. 1987;327:721–724. doi: 10.1038/327721a0. [DOI] [PubMed] [Google Scholar]
- 19.Beroukhim R, Brunet JP, Di Napoli A, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Ye Y, Yang H, et al. Genome-wide profiling of chromosomal alterations in renal cell carcinoma using high-density single nucleotide polymorphism arrays. Int J Cancer. 2009;125:2342–2348. doi: 10.1002/ijc.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen C, Kaelin WG., Jr The VHL/HIF axis in clear cell renal carcinoma. Semin Cancer Biol. 2013;23:18–25. doi: 10.1016/j.semcancer.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: Implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- 23.Varela I, Tarpey P, Raine K, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson MA, Kouzarides T, Huntly BJ. Targeting epigenetic readers in cancer. N Engl J Med. 2012;367:647–657. doi: 10.1056/NEJMra1112635. [DOI] [PubMed] [Google Scholar]
- 25.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 26.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 27.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 28.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson M. Polybromo-1: The chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91:309–319. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapur P, Peña-Llopis S, Christie A, et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: A retrospective analysis with independent validation. Lancet Oncol. 2013;14:159–167. doi: 10.1016/S1470-2045(12)70584-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brownlee PM, Chambers AL, Oliver AW, et al. Cancer and the bromodomains of BAF180. Biochem Soc Trans. 2012;40:364–369. doi: 10.1042/BST20110754. [DOI] [PubMed] [Google Scholar]
- 32.Wellcome Trust Sanger Institute. Catalog of Somatic Mutations in Cancer. http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/
- 33.Xia W, Nagase S, Montia AG, et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107:14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugarolas J. PBRM1 and BAP1 as novel targets for renal cell carcinoma. Cancer J. 2013;19:324–332. doi: 10.1097/PPO.0b013e3182a102d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hakimi AA, Ostrovnaya I, Reva B, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A report by MSKCC and the KIRC TCGA research network. Clin Cancer Res. 2013;19:3259–3267. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F, Mao G, Tong D, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo G, Gui Y, Gao S, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2012;44:17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 43.Jensen DE, Proctor M, Marquis ST, et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097–1112. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 44.Kucejova B, Peña-Llopis S, Yamasaki T, et al. Interplay between pVHL and mTORC1 pathways in clear-cell renal cell carcinoma. Mol Cancer Res. 2011;9:1255–1265. doi: 10.1158/1541-7786.MCR-11-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machida YJ, Machida Y, Vashisht AA, et al. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179–34188. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu H, Mashtalir N, Daou S, et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol. 2010;30:5071–5085. doi: 10.1128/MCB.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sowa ME, Bennett EJ, Gygi SP, et al. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joseph RW, Kapur P, Serie DJ, et al. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low risk clear cell renal cell carcinoma. Cancer. doi: 10.1002/cncr.28521. doi: 10.1002/cncr.28521 [epub ahead of print on December 30, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bott M, Brevet M, Taylor BS, et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat Genet. 2011;43:668–672. doi: 10.1038/ng.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022–1025. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiesner T, Obenauf AC, Murali R, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popova T, Hebert L, Jacquemin V, et al. Germline BAP1 mutations predispose to renal cell carcinomas. Am J Hum Genet. 2013;92:974–980. doi: 10.1016/j.ajhg.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farley MN, Schmidt LS, Mester JL, et al. A novel germline mutation in BAP1 predisposes to familial clear-cell renal cell carcinoma. Mol Cancer Res. 2013;11:1061–1071. doi: 10.1158/1541-7786.MCR-13-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventii KH, Devi NS, Friedrich KL, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res. 2008;68:6953–6962. doi: 10.1158/0008-5472.CAN-08-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misaghi S, Ottosen S, Izrael-Tomasevic A, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181–2192. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheuermann JC, de Ayala Alonso AG, Oktaba K, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sauvageau M, Sauvageau G. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mills AA. Throwing the cancer switch: Reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dey A, Seshasayee D, Noubade R, et al. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science. 2012;337:1541–1546. doi: 10.1126/science.1221711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eletr ZM, Wilkinson KD. An emerging model for BAP1's role in regulating cell cycle progression. Cell Biochem Biophys. 2011;60:3–11. doi: 10.1007/s12013-011-9184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kristie TM, Liang Y, Vogel JL. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim Biochim Biophys Acta. 2010;1799:257–265. doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gossage L, Murtaza M, Slatter AF, et al. Clinical and pathological impact of VHL, PBRM1, BAP1, SETD2, KDM6A, and JARID1c in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2014;53:38–51. doi: 10.1002/gcc.22116. [DOI] [PubMed] [Google Scholar]
- 64.Kapur P, Christie A, Raman JD, et al. BAP1 immunohistochemistry predicts outcomes in a multi-institutional cohort with clear cell renal cell carcinoma. J Urol. 2014;191:603–610. doi: 10.1016/j.juro.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 65.Harbour JW. The genetics of uveal melanoma: An emerging framework for targeted therapy. Pigment Cell Melanoma Res. 2012;25:171–181. doi: 10.1111/j.1755-148X.2012.00979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mandriota SJ, Turner KJ, Davies DR, et al. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 67.Montani M, Heinimann K, von Teichman A, et al. VHL-gene deletion in single renal tubular epithelial cells and renal tubular cysts: Further evidence for a cyst-dependent progression pathway of clear cell renal carcinoma in von Hippel-Lindau disease. Am J Surg Pathol. 2010;34:806–815. doi: 10.1097/PAS.0b013e3181ddf54d. [DOI] [PubMed] [Google Scholar]
- 68.Ho L, Ronan JL, Wu J, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sohn DH, Lee KY, Lee C, et al. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J Biol Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- 70.Kim JK, Huh SO, Choi H, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sellers WR. A blueprint for advancing genetics-based cancer therapy. Cell. 2011;147:26–31. doi: 10.1016/j.cell.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 72.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, et al. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 73.Kondo K, Yao M, Kobayashi K, et al. PTEN/MMAC1/TEP1 mutations in human primary renal-cell carcinomas and renal carcinoma cell lines. Int J Cancer. 2001;91:219–224. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1034>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 74.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benvenuto G, Li S, Brown SJ, et al. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 2000;19:6306–6316. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- 76.Kucejova B, Sunny NE, Nguyen AD, et al. Uncoupling hypoxia signaling from oxygen sensing in the liver results in hypoketotic hypoglycemic death. Oncogene. 2011;30:2147–2160. doi: 10.1038/onc.2010.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mazhab-Jafari MT, Marshall CB, Ishiyama N, et al. An autoinhibited noncanonical mechanism of GTP hydrolysis by Rheb maintains mTORC1 homeostasis. Structure. 2012;20:1528–1539. doi: 10.1016/j.str.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 78.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fingar DC, Blenis J. Target of rapamycin (TOR): An integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 80.Yang H, Rudge DG, Koos JD, et al. mTOR kinase structure, mechanism and regulation. Nature. 2013;497:217–223. doi: 10.1038/nature12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sato T, Nakashima A, Guo L, et al. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Voss MH, Hakimi AA, Chen Y, et al. Effective monotherapy despite intratumor heterogeneity: Clonal convergence within the PI3K pathway and sensitivity to mTOR inhibitors in patients with advanced renal cell carcinoma (RCC) J Clin Oncol. 2013;31(suppl):302s. abstr 4573. [Google Scholar]
- 83.Brugarolas J. Research translation and personalized medicine. In: Figlin RA, Rathmell WK, Rini BI, editors. Renal Cell Carcinoma: Translational Biology, Personalized Medicine, and Novel Therapeutic Targets. New York, NY: Springer US; 2012. pp. 161–191. [Google Scholar]
- 84.Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338:221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tinton SA, Buc-Calderon PM. Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett. 1999;446:55–59. doi: 10.1016/s0014-5793(99)00185-4. [DOI] [PubMed] [Google Scholar]
- 86.Brugarolas J. MTORC1 signaling and hypoxia. In: Polunovsky VAA, Houghton PJJ, editors. mTOR Pathway and mTOR Inhibitors in Cancer Therapy. Cancer Drug Discovery and Development, Humana Press; 2010. pp. 75–97. [Google Scholar]
- 87.Wolff NC, Vega-Rubin-de-Celis S, Xie XJ, et al. Cell-type-dependent regulation of mTORC1 by REDD1 and the tumor suppressors TSC1/TSC2 and LKB1 in response to hypoxia. Mol Cell Biol. 2011;31:1870–1884. doi: 10.1128/MCB.01393-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brugarolas J, Lei K, Hurley RL, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pantuck AJ, Seligson DB, Klatte T, et al. Prognostic relevance of the mTOR pathway in renal cell carcinoma: Implications for molecular patient selection for targeted therapy. Cancer. 2007;109:2257–2267. doi: 10.1002/cncr.22677. [DOI] [PubMed] [Google Scholar]