Abstract
Background
In the present study we have assessed whether the Carpathian Mountains represent a genetic barrier in East Europe. Therefore, we have analyzed the mtDNA of 128 native individuals of Romania: 62 of them from the North of Romania, and 66 from South Romania.
Results
We have analyzed their mtDNA variability in the context of other European and Near Eastern populations through multivariate analyses. The results show that regarding the mtDNA haplogroup and haplotype distributions the Romanian groups living outside the Carpathian range (South Romania) displayed some degree of genetic differentiation compared to those living within the Carpahian range (North Romania).
Conclusion
The main differentiation between the mtDNA variability of the groups from North and South Romania can be attributed to the demographic movements from East to West (prehistoric or historic) that differently affected in these regions, suggesting that the Carpathian mountain range represents a weak genetic barrier in South-East Europe.
Keywords: Mitochondrial DNA, Carpathian Mountains, Romanian population, Genetic barrier
Background
Assessing the genetic structure of different populations is important for understanding the population history of a certain geographic area from a genetic point of view. It is also important from the epidemiological point of view to avoid spurious association between genetic markers and certain diseases [1,2]. The genetic structure of European populations has been the focus of several recent studies which have identified that geographical distance is more important than cultural and linguistic affinities when explaining the genetic distance between populations [3,4].
While most of the genetic studies of European populations have focused on the Western part of the continent, few studies have assessed the genetic structure of Eastern Europe. Among these, one study assessing the variability of the Y-chromosome has proposed that, although the population structure of the Carpathian basin is relatively homogenous, the Carpathian range from the present territory of Romania represents a genetic barrier between populations living in the East and West of this barrier and mountain populations encompassed within the Carpathian arch [5]. In this study, those populations outside the Carpathian range seemed to show more genetic similarities with East Europe and the Mediterranean regions, while the populations living within the Carpathian range in Transylvania were closer to Central and Western European populations [5]. Other Y-chromosome studies have proposed a homogenous structure of populations in the Balkans [6] or the Dnieper-Carpathian basin [7], but these studies have only analysed Romanian populations from outside the Carpathian range. To our knowledge, no studies have been performed to assess the mtDNA population structure in Romanians.
The present population of Romania is homogenous from a cultural and linguistic point of view, with an absence of language dialects within the borders of the country despite the relatively large and diverse geographic areas inhabited. Several historical events impacted the demography of Romanian populations inside and outside the Carpathian range. In this regard, although relatively little is known about potential differences in the Neolithic and the Bronze age, an impact of Celtic migration has been reported mainly in Transylvania (inside the Carpathian range), rather than in the planes East and South of the mountains [8]. During the Iron age and Antiquity the population was represented both inside and outside the Carpathian range by Geto-Dacian tribes, later incorporated in the Roman Empire after the conquest of Dacia by emperor Traianus in 106 AD. Several waves of migrations from both West (Gothic tribes) and East (Huns, Slavs, Magyars, Cumans) have differently impacted the country. While the Middle Ages were characterized by Hungarian and Germanic influences in the territory West of the Carpathians. The Greek and Turkish influences dominated in South and East of the country [9]. The unity of Romania was realized only during the 19th - 20th century, and was completed after the First World War. Therefore, while Romanians are seen in genetic and medical studies as one homogenous group, potential differences may have important epidemiological consequences. In order to test the hypothesis that the Carpathian mountains represents a genetic barrier differentiating populations within Romania, we have assessed mtDNA distributions in Romanian populations from outside (South Romania) and within (North Romania) the Carpathian range (Figure 1).
Figure 1.
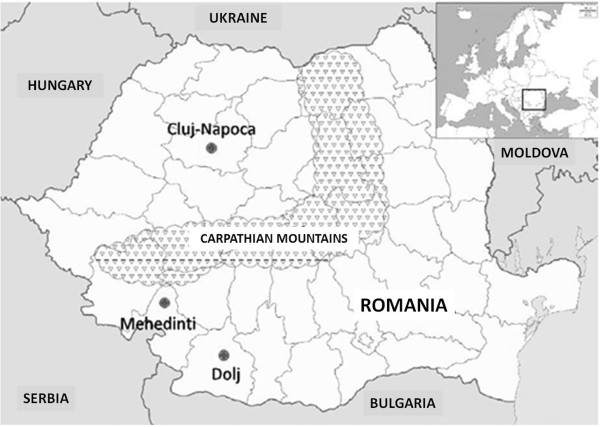
Geographical location of the two Romania populations analyzed in the present study. North Romania (Cluj-Napoca) and South Romania (Dolj and Mehedinti).
Results
The mitochondrial variability obtained in the population from the North of Romania (i.e., within the Carpathian range) (N = 62) was classified in 51 different sequences or haplotypes, which indicates a high degree of sequence diversity (0.9905 ± 0.0072) (Table 1 and Additional file 1). Seven out of the 51 mitochondrial sequences obtained in this population show up in more than one individual, being the rCRS sequence the most frequent one (Table 2). This mitochondrial haplotype variability was classified into ten mitochondrial haplogroups (H, U, K, T, J, HV, W, M, X and A). Haplogroup H was the most frequent (59.7%) while haplogroups M, X and A were the least frequent ones (1.61% each haplogroup). Besides, haplogroups K and HV, show up also at a low frequency (3.23% each haplogroup) (Table 2).
Table 1.
Sequence and haplogroup diversity indices for mtDNA lineages in North and South Romania populations
| North of Romania | South of Romania | |
|---|---|---|
| N. individuals |
62 |
66 |
| N. sequences (K) |
51 |
43 |
| Sequence diversity (Ĥ ± sd) |
0.9905 ± 0.0072 |
0.9804 ± 0.0059 |
| Nucleotide diversity (π ± sd) |
0.01180 ± 0.00089 |
0.01068 ± 0.00082 |
| N. polymorphic sites (s) |
58 |
51 |
| N. haplogroups |
10 |
7 |
| Haplogroup diversity | 0.7542 ± 0.0461 | 0.6256 ± 0.0622 |
Table 2.
Distribution of the frequencies (%) of the mtDNA sequences (HT) and haplogroups (HG) obtained in North Romania population
| HT | HT(N) | HT (%) | HG | HG (%) |
|---|---|---|---|---|
| rCRS |
5 |
8.06 |
H |
|
| 16070G |
3 |
4.84 |
H36 |
|
| 16189C 16356C 16362C |
2 |
3.23 |
H1b |
|
| 16311C |
2 |
3.23 |
H2b |
|
| 16092C 16293G 16311C |
2 |
3.23 |
H11 |
|
| 16129A |
1 |
1.61 |
H |
|
| 16093C |
1 |
1.61 |
H |
|
| 16153A |
1 |
1.61 |
H |
|
| 16354 T |
1 |
1.61 |
H2a1 |
|
| 16357C |
1 |
1.61 |
H |
|
| 16092C 16189C |
1 |
1.61 |
H |
|
| 16209C 16261 T |
1 |
1.61 |
H |
|
| 16304C 16311C |
1 |
1.61 |
H5 |
|
| 16093C 16129A 16207G |
1 |
1.61 |
H |
|
| 16241G 16311C |
1 |
1.61 |
H |
|
| 16093C 16249C 16362C |
1 |
1.61 |
H6 |
|
| 16278 T 16293G 16311C |
|
1.61 |
H11a |
|
| 16080G 16183C 16189C 16223 T 16356C |
1 |
1.61 |
H1b |
|
| 16162G |
1 |
1.61 |
H1a |
|
| 16304C 16311C 16391A |
1 |
1.61 |
H5 |
|
| 16093C 16223 T |
1 |
1.61 |
H |
|
| 16362C |
1 |
1.61 |
H6 |
|
| 16288C 16311C 16362C |
1 |
1.61 |
H8 |
|
| 16148 T 16256 T 16319A |
1 |
1.61 |
H13a2 |
|
| 16287 T |
1 |
1.61 |
H12 |
|
| 16189C 16304C |
1 |
1.61 |
H5 |
|
| 16093C 16311C |
1 |
1.61 |
H |
59.68 |
| 16075C 16256 T 16270 T 16399G |
1 |
1.61 |
U5a1 |
|
| 16189C 16270 T |
1 |
1.61 |
U5b1 |
|
| 16093C 16189C 16270 T 16301 T |
1 |
1.61 |
U5b1b |
|
| 16146G 16342C |
1 |
1.61 |
U8a1 |
|
| 16169 T 16356C |
1 |
1.61 |
U4 |
|
| 16051G 16092C 16129C 16183C 16189C 16362C |
1 |
1.61 |
U2 |
|
| 16189C 16192 T 16256 T 16270 T 16291 T 16399G |
1 |
1.61 |
U5a1 |
|
| 16256 T 16270 T 16399G |
1 |
1.61 |
U5a1 |
11.29 |
| 16224C 16311C |
2 |
3.23 |
K |
3.23 |
| 16126C 16294 T 16296 T 16304C |
2 |
3.23 |
T2b |
|
| 16126C 16163G 16186T 16189C 16294 T |
1 |
1.61 |
T1a |
|
| 16126C 16294 T 16296 T 16362C 16390A 16399G |
1 |
1.61 |
T |
6.45 |
| 16069 T 16126C 16261 T |
1 |
1.61 |
J1 |
|
| 16069 T 16126C |
1 |
1.61 |
J |
|
| 16069 T 16126C 16189C |
1 |
1.61 |
J |
4.84 |
| 16298C 16362C |
1 |
1.61 |
HV0 |
|
| 16298C 16311C |
1 |
1.61 |
HV0 |
3.23 |
| 16223 T 16292 T 16399G |
1 |
1.61 |
W |
|
| 16223 T 16292 T 16355 T 16362C |
1 |
1.61 |
W |
|
| 16147 T 16223 T 16286 T 16292 T |
1 |
1.61 |
W |
|
| 16172C 16223 T 16231C 16292 T |
1 |
1.61 |
W |
6.45 |
| 16129A 16223 T 16291 T 16298C |
1 |
1.61 |
M5 |
1.61 |
| 16183C 16189C 16223 T 16255A 16278 T |
1 |
1.61 |
X2c |
1.61 |
| 16039A 16188 T 16189C 16223 T 16290 T 16319A 16356C 16362C | 1 | 1.61 | A | 1.61 |
The mitochondrial variability obtained in the population from the South of Romania (N = 66), was classified in 43 different sequences or haplotypes, which indicates also a high sequence diversity (0.9804 ± 0.0059), but lower than that obtained in the sample from the North of Romania (Table 1 and Additional file 1). Thirteen out of the 43 mitochondrial haplotypes obtained in this population appear in more than one individual (Table 3). As in the sample from the North of Romania, the rCRS haplotype was also the most frequent one. This mitochondrial variability was classified within seven mitochondrial haplogroups (H, U, K, T, J, HV and W). Haplogroup H showed the highest frequency (47%), although it was lower than the frequency of haplogroup H in the sample from the North of Romania (59.7%). Haplogroup U showed a noticeable frequency (17%), higher than in the sample from North Romania (11.3%) (Table 2 and Table 3). As regards haplogroups M, X and A, they were not observed in the South Romanian sample (Table 3).
Table 3.
Distribution of the frequencies (%) of the mtDNA sequences (HT) and haplogroups (HG) obtained in the population from South Romania
| HT | HT (N) | HT (%) | HG | HG (%) |
|---|---|---|---|---|
| rCRS |
6 |
9.09 |
H |
|
| 16183C 16189C |
5 |
7.58 |
H |
|
| 16294 T 16304C |
3 |
4.55 |
H5 |
|
| 16070G |
2 |
3.03 |
H36 |
|
| 16189C 16384A |
1 |
1.52 |
H5 |
|
| 16124C 16304C |
1 |
1.52 |
H |
|
| 16209C 16304C |
1 |
1.52 |
H5 |
|
| 16182C 16183C 16189C |
1 |
1.52 |
H |
|
| 16188G |
1 |
1.52 |
H |
|
| 16291 T 16343G 16390A |
1 |
1.52 |
H |
|
| 16042A 16288C |
1 |
1.52 |
H1 |
|
| 16093C 16189C |
1 |
1.52 |
H1 |
|
| 16113C 16311C |
1 |
1.52 |
H |
|
| 16343G 16390A |
1 |
1.52 |
H |
|
| 16189C 16384A |
1 |
1.52 |
H5 |
|
| 16139 T |
1 |
1.52 |
H |
|
| 16093C 16311C |
1 |
1.52 |
H |
|
| 16183C 16189C 16356C 16362C |
1 |
1.52 |
H |
|
| 16189C 16311C |
1 |
1.52 |
H |
46.97 |
| 16356C |
3 |
4.55 |
U4 |
|
| 16256 T 16270 T 16399 |
2 |
3.03 |
U5a1 |
|
| 16051G 16179T 16356C 16362C |
2 |
3.03 |
U4c |
|
| 16197 T 16256 T 16270 T |
2 |
3.03 |
U5 |
|
| 16179 T 16356C |
1 |
1.52 |
U4c |
|
| 16192 T 16256 T 16270 T |
1 |
1.52 |
U5a |
16.67 |
| 16224C 16311C |
2 |
3.03 |
K |
|
| 16224C 16311C 16360 T |
2 |
3.03 |
K2a |
|
| 16224C 16311C 16362C |
1 |
1.52 |
K |
7.58 |
| 16182C 16183C 16189C 16294 T 16296 T |
1 |
1.52 |
T2 |
|
| 16126C 16294 T 16324C |
1 |
1.52 |
T |
|
| 16126C 16163G 16170G 16186 T 16189C 16323 T 16294 T 16316G |
1 |
1.52 |
T |
|
| 16126C 16193C 16294 T 16324C |
1 |
1.52 |
T |
6.06 |
| 16069 T 16126C |
2 |
3.03 |
J |
|
| 16066G 16069T 16126C 16145A 16172C 16261 T 16311C |
1 |
1.52 |
J1b |
|
| 16069 16126C 16261 T |
1 |
1.52 |
J |
|
| 16069 T 16126C 16224C |
1 |
1.52 |
J1c |
7.58 |
| 16298C |
3 |
4.55 |
HV0 |
|
| 16067 T |
1 |
1.52 |
HV0 |
|
| 16067 T 16126C |
1 |
1.52 |
HV0 |
|
| 16147 T 16221 T |
1 |
1.52 |
HV4 |
|
| 16298C 16311C |
1 |
1.52 |
HV0 |
10.61 |
| 16223 T 16286 T 16292 T 16311C |
2 |
3.03 |
W |
|
| 16051G 16189C 16192 T 16223 T 16292 T 16325G | 1 | 1.52 | W | 4.55 |
After performing the pairwise FST test (Additional file 2) based on mitochondrial haplogroup frequencies, we did not detect statistically significant differences between the Northern and Southern Romania population samples. When comparing these two samples with other population samples from Europe and the Near East, we observed that Southern Romanians did not show statistically significant differences with any other population. However, the Northern Romanians did show statistically significant differences with the Near Eastern populations (Additional file 2). Regarding the Romanian populations (North and South of Romania) statistically significant differences were detected when the comparison was based on haplotype frequencies (p = 0.00000 ± 0.0000, pairwise FST test). This could be due to the fact that North and South Romanian samples share only eight mitochondrial haplotypes. On the other hand, the analysis of the mitochondrial haplotype variability obtained in Romanian samples in the context of the Near Eastern and North East European populations, showed that 30 out of 51 haplotypes (58%) obtained in the North Romanian sample have been found only within the populations of North East Europe, and 21 out of 51 haplotypes (41%) have been found in both North East Europe and the Near Eastern populations. Regarding the Southern Romanian sample, 16 out of 43 haplotypes obtained (37%) have been found only in the Near Eastern populations, and 10 out of the 43 haplotypes obtained (23%) have been found in both the Near East and North East European populations.In the First Component (43% of the variance) of the Principal Component Analysis (PCA) (57% of total variance in two First Principal Components) (Figure 2), the European populations lie at one end of this axis, whereas the populations of the Near Eas tare located at the other end of this axis. The population of Northern Romania is within the range of the mitochondrial distribution observed for the other European populations, while the distribution of South Romanians was closer to that of the Near Eastern populations.
Figure 2.
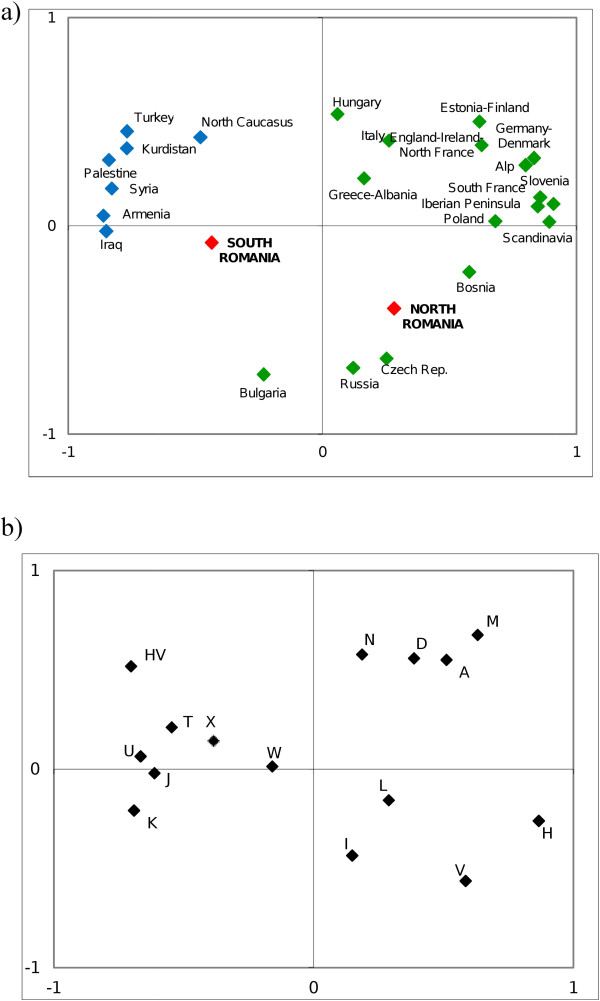
Principal Component Analysis (PCA) (57% of total variance in two First Principal Components). a) Distribution of the populations based on the frequencies of the mitochondrial haplogroups of North and South of Romania (in red), Europe (in green) and Near East (in blue). b) Correlation of the mtDNA haplogroups with the main axes of the PCA.
The mitochondrial haplogroups that show the higher value of correlation with the First Component are haplogroups H, HV and K (correlation coefficient 0,867, −0,703, −0,691 respectively) (Figure 2). Thus, the different position of North and South Romanian populations in the First Principal Component is explained by the distribution of the frequencies of haplogroups H, HV and K. In this regard, we observe that the North Romanian population is located alongside with other European populations because it has a high frequency for haplogroup H (59.7%) and low frequencies for the haplogroups HV and K (3.23% each haplogroup). On the contrary, the population of South Romania has a lower frequency of haplogroup H (46.9%) and higher frequencies of the haplogroups HV and K (10.61 and 7.58%, respectively); the frequency distribution of these haplogroups in South of Romanian is in the range of variability of some Near East populations [10].
The Bulgarian, Hungarian, Russian and Czech populations are located between the North and South Romanian populations in this First Component. Bulgaria shows more similarity with South Romania than with North Romania regarding the frequency distribution of haplogroups K (7.5%), HV (6%) and H (43%) [11]. However, the samples from the Czech Republic and Russia, show frequencies for haplogroups HV (4.2 and 1.6%, respectively) and K (2 and 3.9%, respectively) [12-14] that are closer to the frequency values described in the North Romanian sample (Figure 2).
The Second Component of the PCA, which explains 14% of the variance, did not show a clear clustering of populations. The haplogroup with a higher correlation with this Second Component is haplogroup M (correlation value 0.677). The populations from Northern Romania, Czech Republic, Russia and Bulgaria differ from the rest in their high frequency for haplogroup M (4.3-0.9%) [11-14], whereas haplogroup M is absent or very rare in other European and Near Eastern populations (Figure 2).
A Multidimensional Scaling (MDS) analysis, where all mitochondrial variability was taken into account, was also carried out to provide a two-dimensional view of the FST distance matrix. The analysis showed an RSQ of 0.98037 and a stress value of 0.07343, which indicates that the representation of the MDS obtained showed a good description of the real mitochondrial variability. In the MDS analysis, the Near Eastern and the European populations presented the greatest distance (Figure 3). Moreover, the analysis showed that the South Romanian population was located within the mitochondrial variability of the European populations, whereas North Romania is separated from the South Romanian population.In the MDS (Figure 3), the location of the Eastern European populations including North and South Romania, Bulgaria, Hungary, Czech Republic, Russia, Slovenia and Poland showed a heterogeneous distribution, without a clear clustering. In addition to that, the Czech and Russian populations were located closer to South Romania than to North Romania population, in contrast with the position showed in PCA analysis (Figure 2).The analysis of the genetic boundaries between the populations around Romania supported the four boundaries shown in Figure 4. The first boundary [(a), with 93% bootstrap support] was found between Hungary and both Romania (North and South) and Serbia. The second boundary [(b), with 83-92% bootstrap support] was found between Poland and both Hungary and Czech Republic. The third boundary [(c), with 82% bootstrap support] separated the populations of Russia from Poland. Finally, the fourth boundary [(d) with 80% bootstrap support] was found between North and South Romania.
Figure 3.
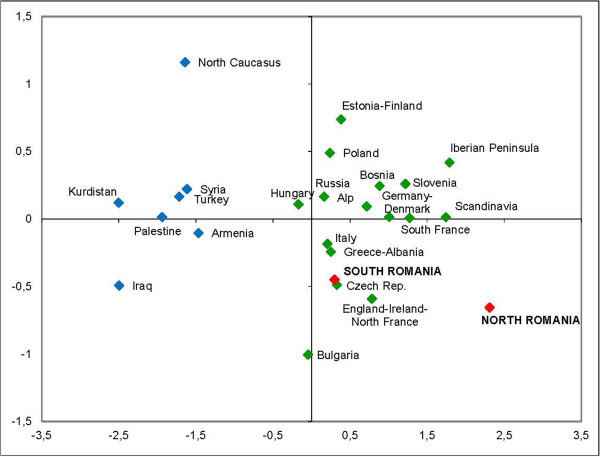
Multidimensional Scaling (MDS) based on the F ST genetic distances, calculated according to the distribution of the mtDNA haplogroup frequencies of different populations: North and South of Romania (in red), Europe (in green) and Near East (in blue).
Figure 4.
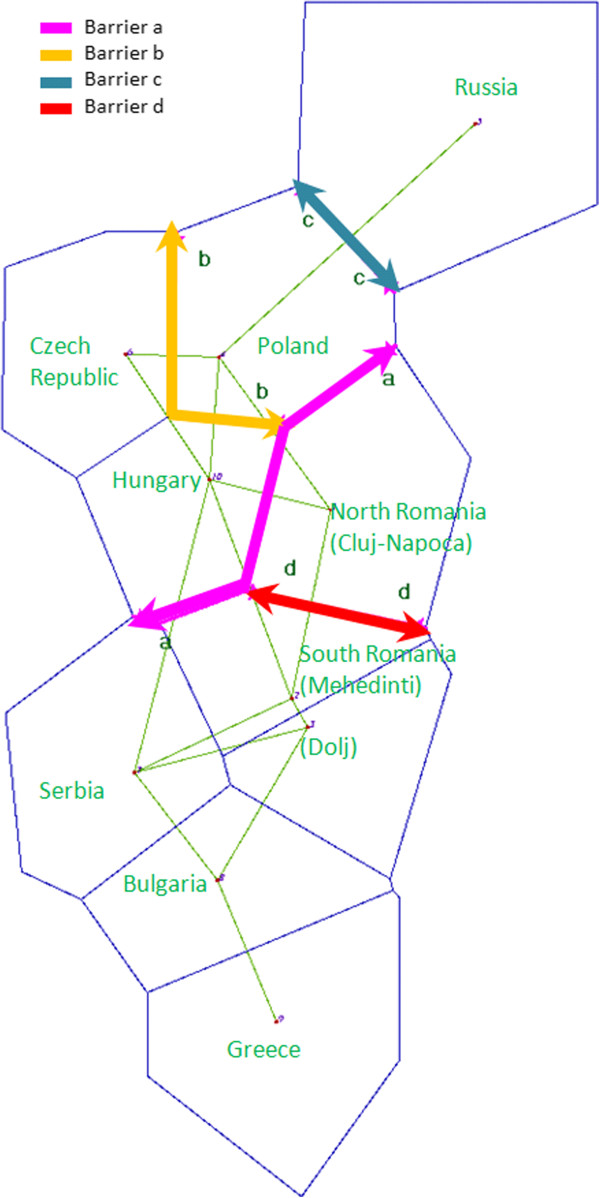
The first four genetic boundaries (lines: a, b, c and d) detected by BARRIER version 2.2 using genetic distance matrix based on the mtDNA haplogroup frequencies. The Romanian groups from the North and South (present study) have been considered together with the surrounding populations.
Next, we decided to obtain Median Joining Networks (MJN) for haplogroups J (Figure 5) and K (K2a) (Figure 6) because these haplogroups are considered as markers of the Neolithic expansion into Europe from the Near East [10,15,16]. For this MJN analysis, the HVS-I sequences of the North and South Romanian samples obtained in this study, have been considered together with a set of sequences of North East European and Near Eastern populations. The resulting MJN for haplogroup J and K (K2a) are shown in Figures 5 and 6, where the central node is represented by the polymorphisms which define each haplogroup, i.e. 16224–16311 in the case of haplogroup K and 16069–16126 for haplogroup J.In the case of haplogroup J (Figure 5), the MJN central node is defined by polymorphisms 16069–16126, which are shown by all of the samples. On the one hand, it can be observed the high sequence diversity within this haplogroup, and on the other, that the haplotypes of haplogroup J are more common in the Near East population than in North East European populations. With regard to the Romanian samples, it can be highlighted that those sequences found only in the South Romanian sample are also found within the Near Eastern populations.Regarding the MJN of haplogroup K (K2a), haplotype 16224–16311, located in the central node (Figure 6), has been described in both South and North Romanian groups, as well as in the North East European and Near Eastern populations. However, two haplotypes defined by polymorphisms 16224-16311-16360 and 16224-16311-16362 are shared only by one individual from the Near East population and a few individuals from the South Romanian sample.
Figure 5.
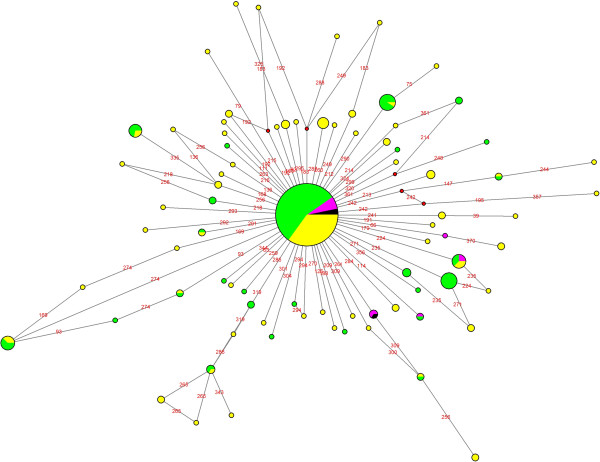
Median Joining Network of haplogroup J. Data encompass mtDNA HVS-I (nps 15999–16399). South Romanian sample in pink, North Romanian sample in black, Near East populations in green and North East Europe population in yellow.
Figure 6.
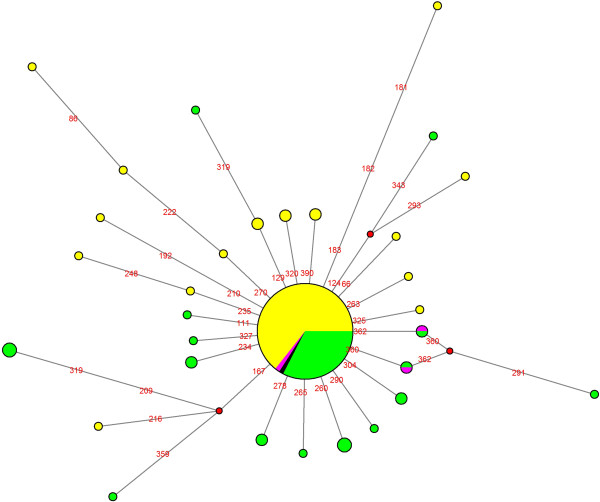
Median Joining Network of haplogroup K. Data encompass mtDNA HVS-I (nps 15999–16399). South Romanian sample in pink, North Romanian sample in black, Near East populations in green and North East Europe population in yellow.
Discussion
In the present study we carried out an assessment of the mtDNA variability of Romanian populations from outside (South Romanians) and within (North Romanians) the Carpathian mountain range, in order to assess its influence as a potential genetic barrier on the genetic variability of Romanians.
The North Romanian population exhibited several differences in the frequency distribution of certain mitochondrial haplogroups comparing with other populations. Haplogroup H showed a very high frequency (59.7%) (Table 2), while in Europe the frequency of haplogroup H reaches 42-55%, and in the Near East this is substantially lower, 23-33% [10]. Haplogroup U in North Romania showed a slightly lower frequency (11%) than in most European (13-19%) and Near Eastern populations (21-27%) [10]. Furthermore, in the North of Romania, a low frequency of haplogroup J was found (4.8%) compared with that observed in other populations of Europe and the Near East (Table 2, Figure 5).
The frequency of the remaining mitochondrial haplogroups in the North Romanian sample (K, T and HV) were within the European mitochondrial haplogroup frequency variation, except for haplogroups M, X and A, which are very rare in Europe. Haplogroups X and A are found mostly in Eastern Asia and America, and haplogroup M is the root of many of the haplogroups derived from the dispersion of Homo sapiens out of Africa, and it can be found mainly in Southern Asia, having a high frequency in Indian subcontinent [17]. The presence of these haplogroups in North Romanians, albeit at low frequencies, might be the result of the early medieval migrations of Asian populations of Huns, Avars, Magyars and Cumans that crossed the territory of Transylvania between the 5th and 11th century AD.
The mitochondrial haplogroup variability (H, U, J, K, T and W) of the sample from Southern Romania is, in general terms, within the range of variation of other European populations, except for haplogroups HV and K. The frequencies of haplogroup HV (10.6%) and K (7.6%) are closer to the range of variation described in the Near East (7-17% and 5–10.8%, respectively) compared with other European populations (0-7% and 2–6.2%, respectively) [10]. Regarding haplogroup K, the distribution of the haplotypes shown in the MJN is more similar to that of the Near East than to that of the European populations (Figure 6).
Therefore, the frequency distribution of mtDNA haplogroups in Romania indicates certain differences between the North and the South of the country (Table 2 and Table 3). Although the FST analysis based on haplogroup frequencies did not indicate statistically significant differences between North and South of Romania, the pairwise FST analysis based on mitochondrial sequence frequencies showed statistically significant differences between the two populations. The Northern Romanian haplogroup distribution is statistically significant different from that observed in the Near Eastern populations, while the haplogroup distribution from Southern Romania does not present such differences (Additional file 2). These results could be due to the fact that North Romanian sequences were closer to the sequences from North East Europe, but in the case of the South Romanian sequences, a better match to the Near East populations was observed.The distinction between North and South Romanian populations is also appreciated in the PCA where the South Romanian population is located closer to the range of variation of the Near Eastern populations, whereas the sample of Northern Romania is located within the range of European variation (Figure 2).
According to the First Component of the PCA (Figure 2), the distinction between the South and North of Romania populations can be explained by a different influence of Near Eastern populations. The South of Romania, as well as the Bulgarian population, presents a high frequency for haplogroup K; moreover subhaplogroup K2a, proposed as a possible marker of the dispersion of farming from the Near East [10,15,16], has been found in both samples (Tables 2 and 3). On the contrary, the North Romanian population (as well as Russia and the Czech Republic) did not show any individual belonging to haplogroup K2a, and the frequency of haplogroup K is in the range of variation of other European populations. Regarding the Second Component of the PCA, the samples of Bulgaria, Russia and Czech Republic can be clustered with the North Romanian sample due to the presence of haplogroup M (4.3-0.9%), which is absent or very rare in Europe and Near East, having higher frequency in Western and Southern Asia.
Therefore, the distribution of mitochondrial haplogroups in North and South Romania and their neighboring populations (PCA, Figure 2) cannot be explained by a North–south differentiation, but most likely due to a differential genetic influence in Europe from Near Eastern populations. The differential influence of population movements from the Near East in Romania can be detected by the presence of higher frequency for the haplogroup J and haplogroup K2a in the sample of southern Romania (Figure 5 and Figure 6). These haplogroups, given their coalescence age, were considered as markers of the Neolithic expansion in Europe from the Near East [10,15,16].Despite these differences between North and South Romania, when a MDS analysis encompassing all mitochondrial variability of the populations (Figure 3) is considered, it appears that both Romanian populations are included within the range of the European mitochondrial variability, rather than being closer to the Near Eastern populations. However, the North Romanian population is slightly separated from the rest of the populations included in the (MDS) (Figure 3). Interestingly, the results of the Monmonier’s algorithm (Figure 4) showed four genetic boundaries in Eastern Europe, with the Carpathians range being the weakest of them.
The mtDNA differences between North Romania and South Romania may reflect the fact that they suffered a different genetic impact of past demographical events from prehistory to the present. Thus, we suggest that the Carpathian mountain range that crosses the country would act as a weak geographical barrier partially limiting the contact between the two Romanian regions. However, this limitation is weak at most, without a strong Northern and Southern Carpathian Mountains mitochondrial haplogroup differentiation. Instead, the haplogroup composition of South Romania reflects the trace of the demographic movements that also affected other Southern European populations. In contrast, the mitochondrial haplogroup diversity found in North Romania may reflect other prehistoric (Celtic influence) or historic (Eastern Asian migrations) events that differentiate the region of Transylvania compared to the rest of Romania.
Conclusions
In the present study we evaluated whether Carpathian Mountain represent a genetic barrier in East Europe. Regarding the mtDNA haplogroup and haplotype distributions the populations living outside the Carpathian range (South of Romania) displayed some degree of genetic differentiation compared to those living within the Carpahian range (North of Romania). However, this differentiation can be mostly attributed to the demographic movements from East to West (prehistoric or historic) that differently affected in North and South Romania.
Methods
Populations
In the present work, we analyze the mtDNA variability of 128 individuals from two different Romanian regions separated by the Carpathian mountains, including 62 samples from Cluj-Napoca (North of Romania) and 66 from Dolj and Mehedinti (South of Romania) (Figure 1). Selection criteria included the Romanian origin of the donors, and the geographical origin of their parents and grandparents within the Romanian region. Participants were carefully selected by the authors in the field to avoid inclusion of relatives in the sample. The study was approved by the Craiova University Ethical Committee, we have indeed obtained written informed consent from all the volunteers, and none of them were children.
Mitochondrial DNA analysis
The maternal ancestry of the 128 Romanian individuals was explored by the D-loop region sequence variability, including the analysis of the HVS-I (nts 16,000-16,399, as per Andrews [18]. In the individuals with an HVS-I haplotype corresponding to more than one possible haplogroup, we analyzed the sequence of HVS-II (nts 1–425, as per Andrews [18]). Likewise, in order to verify the mtDNA haplogroups obtained, nucleotide positions of the coding region were determined by means of PCR-RFLPs, as described in Izagirre and de la Rua [19].
PCRs were performed in 25 μl of reaction mixture containing 10 mM Tris–HCl pH 8.3, 2 mM of MgCl2, 0.1 μM of each dNTP, 0.4 μM of each primer, 5 units of Taq (Bioline) and 10 μl of diluted DNA (1 μl of DNA extract in 10 μl of distilled water). Cycling parameters were 95°C for 5 min, followed by 35 cycles of 95°C for 10 sec, 58°C for 30 sec and 72°C for 30 sec, and a final cicle of 72°C for 10 min. The sequence of the primers to amplify the HVS-I and HVS-II, were those in Hervella et al.[20]. In the case of positive amplification and absence of contamination, the amplification products were purified by ExoSAP-IT (USB Corporation), with subsequent sequencing in an ABI310 automatic Sequencer using Big Dye 1.1 chemistry (Life Technologies). The results obtained were edited with BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and the sequences were manually aligned.
Finally, in order to classify the mitochondrial variability of the individuals analyzed in this study, we proceeded to amplify 11 markers to define the 10 Western Eurasian haplogroups [21]. The protocol and primers are described in [19,22]. The digestion patterns were analyzed using a fragment Bioanalyzer (Agilent Technologies).
Statistical analysis
Intrapopulation genetic diversity parameters such as the number of different sequences (K), sequence diversity (Ĥ) [23], number of polymorphic sites (S) and nucleotide diversity (π) [23,24] were calculated for the HVS-I using the DnaSP software v5.10.01 [25] and the Arlequin software v3.11 [26]. The genetic distances (FST distances) were calculated on the basis of haplogroup and haplotypes frequencies using Arlequin software v3.11 [26]. In addition, we have analyzed the mtDNA variability of both Romanian samples in the context of other European and Near Eastern populations [10-14,27-50] by means of Principal Component analysis (PCA) (SPSS 17). The distance matrix between all the populations was calculated by means of Arlequin v3.11 [26]. This distance matrix has been depicted in two dimensions by means of a Multidimensional Scaling (MDS) analysis (SPSS 17 Software). Furthermore, a Median Joining Network (MJN) was generated to infer genealogical relationships between the mtDNA lineages (HVS-I) from North and South Romanian, North East Europe and Near Eastern by means of Network software v4.5.0.0 (http://www.fluxus-engineering.com/). Given the high mutation rate of HVS-I, the substitution rates obtained by Meyer et al.[51,52] have been applied for assignation of mutational weight between 0–10, corresponding the value of 10 to those positions with substitution rates of 0–1, and the value of 0 to those of rates of 4–5.
The genetic barriers associated with each geographical location including in Figure 1 and population were investigated using Monmonier’s maximum-difference algorithm [53] in BARRIER version 2.2 [54].
Competing interests
The authors would like to declare no competing financial or personal interests in the preparation of this manuscript.
Authors’ contributions
Conceived and designed the experiments: CR, MGN, MI, SA, NI. Performed the experiments: MH, SA, NI, CR. Analyzed the data: MH, NI, SA, CR, MGN. Contributed reagents/materials/analysis tools: CR. Wrote the paper: MH, CR, MGN. Revised the manuscript critically for important intellectual content: SA, NI, MI. Approved final version to be published: MH, SA, NI, CR, MGN, MI. All authors read and approved the final manuscript.
Supplementary Material
The different mitochondrial sequences (nps 16000–16399) obtained in this study as FASTA file.
FST analysis: FST values (upper the diagonal) and p-values with standard deviation (p ± sd) (under the diagonal) based on mitochondrial haplogroup frequencies (P < 0.0002, in grey).
Contributor Information
Montserrat Hervella, Email: montse.hervella@ehu.es.
Neskuts Izagirre, Email: neskuts.izagirre@ehu.es.
Santos Alonso, Email: santos.alonso@ehu.es.
Mihai Ioana, Email: mihaiioana_romania@yahoo.com.
Mihai G Netea, Email: M.Netea@aig.uman.nl.
Concepción de-la-Rua, Email: conchi.delarua@ehu.es.
Acknowledgements
We are indebted to all the people who have contributed samples to this study. On the other hand, we are very grateful to David Comas for fruitful discussion and to Gartze Mentxaka for technical support. This work was supported by a Vici grant of the Netherlands Organization for Scientific Research to MGN, and by the Spanish Ministry of Science and Innovation, GCL2011-29057/BOS and grant IT542-10 from the Basque Government to Research Groups of the Basque University System to CR, and (UFI 11/09) from the University of the Basque Country, UPV/EHU. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Freedman FM, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Petryshen TL, Kolone LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D. Assessing the impact of population stratification on genetic association studies. Nat Genetics. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genetics. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, Stephens M, Bustamante CD. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, Fabianova E, Foretova L, Georges M, Janout V, Kabesch M, Krokan HE, Elvestad MB, Lissowska J, Mates D, Rudnai P, Skorpen F, Schreiber S, Soria JM, Syvänen AC, Meneton P, Herçberg S, Galan P, Szeszenia-Dabrowska N, Zaridze D, Génin E, Cardon LR, Lathrop M. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genetics. 2008;16:1413–1429. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- Stefan M, Stefanescu G, Gavrila L, Terrenato L, Jobling MA, Malaspina P, Novelletto A. Y chromosome analysis reveals a sharp genetic boundary in the Carpathian region. Eur J Hum Genetics. 2001;9:27–33. doi: 10.1038/sj.ejhg.5200580. [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, González-Neira A, Flaiz C, Mateu E, Scheil HG, Huckenbeck W, Efremovska L, Mikerezi I, Xirotiris N, Grasa C, Schmidt H, Comas D. Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Ann Hum Genetics. 2006;70:459–487. doi: 10.1111/j.1469-1809.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- Varzari A, Stephan W, Stepano V, Raicu F, Cojocaru R, Roschin Y, Glavce C, Dergachev V, Spiridonova M, Schmidt HD. Population history of the Dniester-Carpathians: evidence from Alu markers. J Hum Genet. 2007;52:308–316. doi: 10.1007/s10038-007-0113-x. [DOI] [PubMed] [Google Scholar]
- Zirra V. The Eastern Celts of Romania pp. 1–4. J Indo Eur Stud. 1976;4:1. [Google Scholar]
- The History of Romanians. Romanian Academy. Bucharest: Editura Enciclopedica; 2010. [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, Gölge M, Dimitrov D, Hill E, Bradley D, Romano V, Calì F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, Stefanescu G, Hatina J, Belledi M, Di Rienzo A, Novelletto A. et al. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. doi: 10.1086/321197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karachanak S, Carossa V, Nesheva D, Olivieri A, Pala M, Hooshiar Kashani B, Grugni V, Battaglia V, Achilli A, Yordanov Y, Galabov AS, Semino O, Toncheva D, Torroni A. Bulgarians vs the other European populations: a mitochondrial DNA perspective. Int J Legal Med. 2012;126:497–503. doi: 10.1007/s00414-011-0589-y. [DOI] [PubMed] [Google Scholar]
- Malyarchuck BA, Grzybowski T, Derenko MV, Czarny J, Woźniak M, Miścicka-Sliwka D. Mitochondrial DNA variability in Poles and Russians. Ann Hum Genet. 2002;66:261–283. doi: 10.1046/j.1469-1809.2002.00116.x. [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Perkova MA, Derenko MV, Vanecek T, Lazur J, Gomolcak P. Mitochondrial DNA variability in Slovaks, with application to the Roma origin. Ann Hum Genet. 2008;72:228–240. doi: 10.1111/j.1469-1809.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- Malyarchuk BA, Vanecek T, Perkova MA, Derenko MV, Sip M. Mitochondrial DNA variability in the Czech population, with application to the ethnic history of Slavs. Hum Biol. 2006;78:681–696. doi: 10.1353/hub.2007.0014. [DOI] [PubMed] [Google Scholar]
- Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, Torroni A, Richards MB. The archaeogenetics of Europe. Curr Biol. 2010;20:174–183. doi: 10.1016/j.cub.2009.11.054. [DOI] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Röhl A, Salas A, Oppenheimer S, Macaulay V, Richards MB. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:386–394. doi: 10.1002/humu.20921. http://www.phylotree.org. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Izagirre N, de la Rúa C. An mtDNA analysis in ancient Basque populations: implications for haplogroup V as a marker for a major Paleolithic expansion from southwestern Europe. Am J Hum Genet. 1999;65:199–207. doi: 10.1086/302442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervella M, Izagirre N, Alonso S, Fregel R, Alonso A, Cabrera VM, de-la-Rúa C. Ancient DNA from hunter-gatherer and farmer groups from Northern Spain supports a random dispersion model for the Neolithic expansion into Europe. PLoS One. 2012;7:e34417. doi: 10.1371/journal.pone.0034417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A. The emerging tree of West Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzualde A, Izagirre N, Alonso S, Alonso A, de la Rúa C. Temporal mitochondrial DNA variation in the Basque Country: influence of post-neolithic events. Ann Hum Genet. 2005;69:665–679. doi: 10.1046/j.1529-8817.2005.00170.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;2:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso-Sánchez MA, Cardoso S, Martínez-Bouzas C, Peña JA, Herrera RJ, Castro A, Fernández-Fernández I, de Pancorbo MM. Mitochondrial DNA haplogroup diversity in Basques: a reassessment based on HVI and HVII polymorphisms. Am J Hum Biol. 2008;20:154–156. doi: 10.1002/ajhb.20706. [DOI] [PubMed] [Google Scholar]
- Alvarez-Iglesias JC, Johnson DL, Lorente JA, Martinez-Espin E, Martinez-Gonzalez LJ, Allard M, Wilson MR, Budowle B. Characterization of human control region sequences for Spanish individuals in a forensic mtDNA data set. Leg Med. 2007;9:293–304. doi: 10.1016/j.legalmed.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Comas D, Calafell F, Mateu E, Perez-Lezaun A, Bertranpetit J. Geographic variation in human mitochondrial DNA control region sequence: the population history of Turkey and its relationship to the European populations. Mol Biol Evol. 1996;13:1067–1077. doi: 10.1093/oxfordjournals.molbev.a025669. [DOI] [PubMed] [Google Scholar]
- Behar, van Oven M, Rosset S, Metspalu M, Loogväli EL, Silva NM, Kivisild T, Torroni A, Villems R. A Copernican reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet. 2012;9:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belledi M, Poloni ES, Casalotti R, Conterio F, Mikerezi I, Tagliavini J, Excoffier L. Maternal and paternal lineages in Albania and the genetic structure of Indo-European populations. Eur J Hum Genet. 2000;8:480–486. doi: 10.1038/sj.ejhg.5200443. [DOI] [PubMed] [Google Scholar]
- Bertranpetit J, Sala J, Calafell F, Underhill PA, Moral P, Comas D. Human mitochondrial DNA variation and the origin of Basques. Ann Hum Genet. 1995;59:63–81. doi: 10.1111/j.1469-1809.1995.tb01606.x. [DOI] [PubMed] [Google Scholar]
- Corte-Real HB, Macaulay VA, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit J, Sykes BC. Genetic diversity in the Iberian Peninsula determined from mitochondrial sequence analysis. Ann Hum Genet. 1996;60:331–350. doi: 10.1111/j.1469-1809.1996.tb01196.x. [DOI] [PubMed] [Google Scholar]
- di Rienzo A, Wilson AC. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 1991;88:1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francalacci P, Bertranpetit J, Calafell F, Underhill PA. Sequence diversity of the control region of mitocnondrial DNA in Tuscany and its implications for the peopling of Europe. Am J Phys Anthropol. 1996;100:443–460. doi: 10.1002/(SICI)1096-8644(199608)100:4<443::AID-AJPA1>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- García O, Fregel R, Larruga JM, Álvarez V, Yurrebaso I, Cabrera VM, González AM. Using mitochondrial DNA to test the hypothesis of a European post-glacial human recolonization from the Franco-Cantabrian refuge. Heredity. 2011;106:37–45. doi: 10.1038/hdy.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González AM, Brehm A, Pérez JA, Maca-Meyer N, Flores C, Cabrera VM. Mitochondrial DNA affinities at the Atlantic Fringe of Europe. Am J Phys Anthropol. 2003;120:391–404. doi: 10.1002/ajpa.10168. [DOI] [PubMed] [Google Scholar]
- Grzybowski T, Malyarchuk BA, Derenko MV, Perkova MA, Bednarek J, Woźniak M. Complex interactions of the Eastern and Western Slavic populations with other European groups as revealed by mitochondrial DNA analysis. Forensic Sci Int Genet. 2007;1:141–147. doi: 10.1016/j.fsigen.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Helgason A, Sigurethardottir S, Gulcher JR, Ward R, Stefánsson K. mtDNA and the origin of the Icelanders: deciphering signals of recent population history. Am J Hum Genet. 2000;66:999–1016. doi: 10.1086/302816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larruga JM, Diez F, Pinto FM, Flores C, Gonzalez AM. Mitochondrial DNA characterisation of European isolates: the Maragatos from Spain. Eur J Hum Genet. 2001;9:708–716. doi: 10.1038/sj.ejhg.5200693. [DOI] [PubMed] [Google Scholar]
- Martinez-Cruz B, Harmant C, Platt DE, Haak W, Manry J, Ramos-Luis E, Soria-Hernanz DF, Bauduer F, Salaberria J, Oyharçabal B, Quintana-Murci L, Comas D. Genographic Consortium. Evidence of Pre-roman tribal genetic structure in Basques from uniparentally inherited markers. Mole Biol Evol. 2012;29:2211–2222. doi: 10.1093/molbev/mss091. [DOI] [PubMed] [Google Scholar]
- Piechota J, Tońska K, Nowak M, Kabzińska D, Lorenc A, Bartnik E. Comparison between the Polish population and European populations on the basis of mitochondrial morphs and haplogroups. Acta Biochim Pol. 2004;51:883–895. [PubMed] [Google Scholar]
- Piercy R, Sullivan KM, Benson N, Gill P. The application of mitochondrial DNA typing to the study of white Caucasian genetic identification. Int J Legal Med. 1993;106:85–90. doi: 10.1007/BF01225046. [DOI] [PubMed] [Google Scholar]
- Poetsch M, Wittig H, Krause D, Lignitz E. Mitochondrial diversity of a northeast German population sample. Forensic Sci Int. 2003;137:125–132. doi: 10.1016/j.forsciint.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Pult I, Sajantila A, Simanainen J, Georgiev O, Schaffner W, Pääbo S. Mitochondrial DNA sequences from Switzerland reveal striking homogeneity of European populations. Biol Chem Hoppe Seyler. 1994;375:837–840. [PubMed] [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Gedde-Dahl T, Issel-Tarver L, DiRienzo A, Pääbo S. Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res. 1995;5:42–52. doi: 10.1101/gr.5.1.42. [DOI] [PubMed] [Google Scholar]
- Sajantila A, Salem AH, Savolainen P, Savolainen P, Bauer K, Gierig C, Pääbo S. Paternal and maternal DNA lineages reveal a bottleneck in the founding of the Finnish population. Proc Natl Acad Sci U S A. 1996;93:12035–12039. doi: 10.1073/pnas.93.21.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A. mtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet. 1998;6:365–375. doi: 10.1038/sj.ejhg.5200202. [DOI] [PubMed] [Google Scholar]
- Tömöry G, Csányi B, Bogácsi-Szabó E, Kalmár T, Czibula A, Csosz A, Priskin K, Mende B, Langó P, Downes CS, Raskó I. Comparison of maternal lineage and biogeographic analyses of ancient and modern Hungarian populations. Am J Phys Anthropol. 2007;134:354–368. doi: 10.1002/ajpa.20677. [DOI] [PubMed] [Google Scholar]
- Torroni A, Bandelt HJ, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savontaus ML, Bonné-Tamir B, Scozzari R. mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Weiss G, von Haeseler A. Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics. 1999;152:1103–1110. doi: 10.1093/genetics/152.3.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C, Montiel R, Sierra B, Bettencourt C, Fernandez E, Alvarez L, Lima M, Abade A, Aluja MP. Understanding differences between phylogenetic and pedigree-derived mtDNA mutation rate: a model using families from the Azores Islands (Portugal) Mol Biol Evol. 2005;22:1490–1505. doi: 10.1093/molbev/msi141. [DOI] [PubMed] [Google Scholar]
- Monmonier M. Maximum-difference barriers: an alternative numerical regionalization method. Geogr Anal. 1973;3:245–261. [Google Scholar]
- Manni F, Guerard E, Herer E. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by “Monmonier’s algorithm”. Hum Biol. 2004;76:173–190. doi: 10.1353/hub.2004.0034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The different mitochondrial sequences (nps 16000–16399) obtained in this study as FASTA file.
FST analysis: FST values (upper the diagonal) and p-values with standard deviation (p ± sd) (under the diagonal) based on mitochondrial haplogroup frequencies (P < 0.0002, in grey).


