Summary
Genetically engineered human pluripotent stem cells (hPSCs) have been proposed as a source for transplantation therapies and are rapidly becoming valuable tools for human disease modeling. However, many applications are limited due to the lack of robust differentiation paradigms that allow for the isolation of defined functional tissues. Here, using an endogenous LGR5-GFP reporter, we derived adult stem cells from hPSCs that gave rise to functional human intestinal tissue comprising all major cell types of the intestine. Histological and functional analyses revealed that such human organoid cultures could be derived with high purity and with a composition and morphology similar to those of cultures obtained from human biopsies. Importantly, hPSC-derived organoids responded to the canonical signaling pathways that control self-renewal and differentiation in the adult human intestinal stem cell compartment. This adult stem cell system provides a platform for studying human intestinal disease in vitro using genetically engineered hPSCs.
Highlights
-
•
We isolate intestinal organoid cultures from hPSCs through teratoma differentiation
-
•
Organoids comprise all cell types of the intestinal epithelium
-
•
RNA-seq reveals a high similarity to organoids isolated form primary tissue
-
•
Organoids can respond to pathways that control self-renewal and differentiation
Hockemeyer, Jaenisch, and colleagues developed a method for deriving human intestinal organoid cultures from hPSCs. Isolating intestinal cells from hPSC-derived teratomas yields intestinal cultures similar to organoids isolated from primary tissue. Molecular and histological analyses demonstrate that hPSC-derived organoids comprise the major cell types of the intestinal epithelium, including adult stem cell-like cells. Transcriptional profiling reveals that hESC-derived organoids are highly similar to organoids isolated from primary tissue.
Introduction
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs) (Takahashi et al., 2007), collectively referred to as human pluripotent stem cells (hPSCs), are currently used in disease modeling to address questions specific to humans and to complement insights gained from other model organisms (Soldner and Jaenisch, 2012; Soldner et al., 2011). Genetic engineering using site-specific nucleases was recently established in hPSCs (Dekelver et al., 2010; Hockemeyer et al., 2009, 2011; Yusa et al., 2011; Zou et al., 2009), allowing a level of genetic control that was previously limited to model systems. We can now target gene knockouts, generate tissue-specific cell lineage reporters, overexpress genes from a defined locus, and introduce or repair single-point mutations in hPSCs. Realizing the full potential of hPSCs will require robust differentiation protocols. Most current protocols isolate individual cell types rather than establish functional tissues. Although the former methods can identify cell-autonomous phenotypes, the study of cell-nonautonomous disease mechanisms necessitates a defined tissue context in which individual cell types are represented with the same stoichiometry and architecture as occur in vivo. The recent establishment of human intestinal tissue as in vitro organoid cultures from hPSCs and primary tissue represents a major advance toward creating such a model system for human tissue (Jung et al., 2011; McCracken et al., 2011; Ootani et al., 2009; Sato et al., 2009, 2011b; Spence et al., 2011). Intestinal organoid cultures comprise tissue-specific differentiated cell types and adult stem-like progenitor cells that self-renew and differentiate, by growth factor induction, into the respective cell types of the intestinal epithelium. Here, we establish a protocol that can enrich for intestinal cells with adult stem character. We first generated an hESC line using gene editing that specifically labeled intestinal adult stem cells using a fluorescent reporter placed into an endogenous gene, and then used this cell line to identify and isolate adult stem cells from a pool of heterogeneous cell types during the differentiation of hPSCs.
We focused on a member of the leucine-rich repeat-containing G protein-coupled receptor (LGR) protein class, LGR5 (McDonald et al., 1998). LGR5 functions within the Wingless-related integration site (WNT) signaling cascade, which maintains the adult intestinal stem cell compartment (de Lau et al., 2011). LGR5 is activated by its ligand, R-spondin (RSPO1) (Carmon et al., 2011; de Lau et al., 2011; Kim et al., 2005; Ruffner et al., 2012), and has been shown by genetic lineage tracing experiments to mark intestinal stem cells (Barker et al., 2007). LGR5-expressing cells at the base of the intestinal crypt exhibit WNT-dependent self-renewal and can differentiate into all cell types of the adult intestine (Snippert et al., 2010). Together, LGR5-expressing cells and Paneth cells form the adult stem cell niche and are sufficient to establish in vitro organoid cultures from mice (Sato et al., 2011b). Such murine in vitro organoids can be maintained over time in 3D Matrigel cultures under defined conditions that support either WNT-dependent self-renewal of adult stem cells or differentiation by the withdrawal of WNT and Notch signaling (Korinek et al., 1998; Pellegrinet et al., 2011; van Es et al., 2005). Similarly, human organoid cultures lacking stromal components can be derived from primary tissue biopsies when supplemented with additional small-molecule signals (Jung et al., 2011; Sato et al., 2009, 2011a), and in vitro hPSC-derived organoids can be maintained under a variety of conditions (Jung et al., 2011; McCracken et al., 2011; Sato et al., 2011a; Spence et al., 2011; Wang et al., 2013) and used in human disease modeling (Dekkers et al., 2013). Importantly, LGR5-positive mouse colon cells can form organoids that can be expanded ex vivo and allogenically transplanted into colitis models (Fordham et al., 2013; Yui et al., 2012), suggesting that human intestinal tissue might be amenable to transplantation therapies.
Here, we report tools that allow for the isolation of adult intestinal stem cells and intestinal organoid cultures from direct differentiation of hPSCs using standard teratoma differentiation assays. Cultures with posterior gut qualities and expression profiles closely resembling those of human intestinal tissue can be derived and progressively enriched. Our strategy is based on direct isolation of LGR5-positive intestinal cells, a cell type that is only acquired over extended periods by other protocols (Cao et al., 2011; McCracken et al., 2011; Ogaki et al., 2013; Spence et al., 2011; Wang et al., 2011). Functional analysis and mRNA expression profiling of these organoid cultures confirmed the presence of a subset of adult stem cells. We expect that genetically engineering the parent hPSCs of an in vitro human intestinal tissue will provide a model for investigating human intestinal pathophysiology.
Results
Genome Editing the Endogenous Human LGR5 Locus in hPSCs to Isolate Adult Intestinal Stem Cells
Based on previous studies that used LGR5 expression to identify (Barker et al., 2007) and isolate (Sato et al., 2011b) adult intestinal stem cells in mice, we tested whether a similar approach could be used to isolate and characterize gastrointestinal tissue derived from hPSCs. We developed two Zinc Finger Nuclease (ZFN) pairs that target the LGR5 gene in either the first (LGR5-GFPN-term) or last coding exon (LGR5-GFPC-term; Figure 1A; Figure S1A available online). We coelectroporated these ZFNs and their corresponding donor plasmids into WIBR3 hESCs (Lengner et al., 2010) to integrate a GFP cassette together with a PGK-Puromycin into the LGR5 locus (Figure 1A). Southern blot analysis using external and internal probes (Figures S1A and S1B) and sequence analysis in subsequent RNA sequencing (RNA-seq) confirmed the correct junction of the LGR5 gene with the GFP reading frame at either position. The targeting efficiencies for the generation of LGR5-GFPN-term and LGR5-GFPC-term cell lines were 18.75% and 6.25%, respectively.
Figure 1.
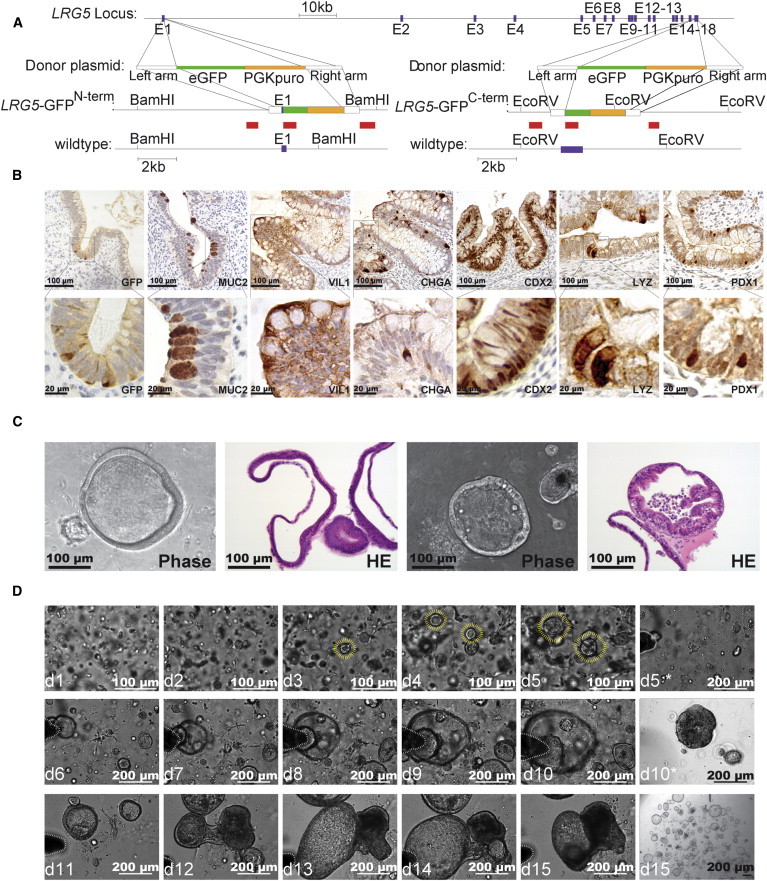
Generation of LGR5-GFP Reporter hESCs Using ZFNs
(A) Schematic overview depicting the gene-editing strategy for the LGR5 locus using a ZFN targeted to either the first or last coding exon of LGR5. Southern blot probes are shown as red boxes, exons are shown as blue boxes. Below are the donor plasmids used to target the LGR5 locus to generate either an N-terminal (LGR5-GFPN-term) or a C-terminal (LGR5-GFPC-term) LGR5-GFP reporter. pA, polyadenylation sequence; PGK, phosphoglycerate kinase promoter; Puro, puromycin resistance gene; eGFP, enhanced green fluorescent protein. Shown below the donor plasmids is the LGR5 locus after targeting with the respective donor plasmids.
(B) IHC staining for indicated proteins in teratoma sections derived from LGR5-GFPN-term hESC reporter cells. MUC2, mucin2; VIL1, villin1; CHGA, chromogranin A; CDX2, caudal type homeobox 2; LYZ, lysozyme; PDX1, pancreatic and duodenal homeobox 1.
(C) Phase contrast and H&E staining of epithelial organoid cultures arising from LGR5-GFP sorting experiments. See quantification in Table 1.
(D) Bright-field images of developing organoids (outlined by yellow lines) derived from nonsorted single-cell suspensions after Matrigel embedding. The demarcation outlined by white lines in bright-field images taken on days 5∗–15 was used as a reference point for tracking organoids clustered near this region. Cells were passaged every 5 days; here, an image before and after (∗) passaging is shown. All bars, 200 μm.
See also Figure S1.
Initially, we characterized this new LGR5 reporter system using a previously established protocol for the direct differentiation of hPSCs to intestinal cells (McCracken et al., 2011; Spence et al., 2011; data not shown). In order to enrich more of the later steps of differentiation with increased LGR5 expression, we capitalized on our previous observation that in teratoma formation assays, some regions of the teratoma differentiate into intestinal-like tissues (Figures 1B, S1C, and S2D). When we performed these teratoma formation assays with the LGR5-GFP reporter cells, we were able to specifically detect GFP expression by immunohistochemistry (IHC) staining in regions that formed a polarized and specialized epithelium (Figure 1B). GFP expression was restricted to those intestinal tissues that uniformly stained positive for CDX2 and VIL1 (Figures 1B, S1C, and S1D), and a subset of cells stained positive for intestinal markers mucin 2 (MUC2), lysozyme (LYZ), chromogranin A (CHGA), and at a lower frequency for PDX1 (Figure 1B, S1C, and S1D). No significant GFP-positive staining in any other tissue type present in the teratoma, such as neuronal rosettes, cartilage, and smooth muscle, was detected (Figure S1C). Further, we could only detect GFP-positive cells in teratomas derived from LGR5-GFPN-term (n = 3) by IHC, and not in those derived from wild-type cells (n = 3) or LGR5-GFPC-term cells (n = 3), although those cells did functionally sort by fluorescence-activated cell sorting (FACS). These observations may suggest a functional difference between the two reporter systems, or they may reflect differential stability, processing, or localization of the LGR5-GFP fusion protein.
Isolation of Intestinal Organoids from LGR5-GFP hPSCs
The detection of LGR5-GFP-positive cells in teratomas prompted us to isolate these cells and test their ability to generate intestinal organoid cultures. We dissociated teratomas derived from LGR5-GFP hPSCs to a single-cell suspension and used FACS to isolate individual cells based on their GFP expression (Tables 1, top, and S2B). LGR5-GFP-positive cells were embedded in Matrigel and cultured in conditions previously described for human intestinal organoid cultures (Jung et al., 2011; Sato et al., 2011a, 2011b; Spence et al., 2011; Wang et al., 2013). Under these conditions, we observed multicellular epithelial structures with a central lumen after 3 days that proliferated into uniform, long-lived organoids with a morphology previously described for human intestinal organoids isolated from primary tissue (Fordham et al., 2013; Jung et al., 2011; Sato et al., 2011a; Figures 1C, 1D, S1E, and S4A–S4D).
Table 1.
| WIBR3-hESC Derived | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Teratoma-derived organoidsa | |||||||||
| Teratomas | No. of organoids formed |
GFP Negative |
GFP Positive |
Total |
|||||
| No. of cells sorted | Cells forming organoids (%) | No. of organoids formed | No. of cells sorted | Cells forming organoids (%) | No. of organoids formed | No. of cells sorted | Cells forming organoids (%) | ||
| Lgr5-GFP N-term | 0 | 1,000 | 0 | 23 | 1,000 | 2.3 | ∼50 | >50,000 | <0.1% |
| Lgr5-GFP N-term | 0 | 1,710 | 0 | 101 | 1,710 | 5.91 | ∼50 | >50,000 | <0.1% |
| Lgr5-GFP N-term | 0 | 1,675 | 0 | 21 | 1,675 | 1.25 | ∼50 | >50,000 | <0.1% |
| Lgr5-GFP C-term | 0 | 4,203 | 0 | 38 | 4,203 | 0.9 | ∼50 | >50,000 | <0.1% |
| Lgr5-GFP C-term | 0 | 6,613 | 0 | 81 | 6,613 | 1.22 | ∼50 | >50,000 | <0.1% |
| Lgr5-GFP N-term | 0 | 6,046 | 0 | 3 | 6,046 | 0.05 | ∼50 | >50,000 | <0.1% |
| Organoid-rederived organoidsb | |||||||||
| Organoids | No. of organoids formed | GFP Negative | GFP Positive | Total | |||||
| No. of cells sorted | Cells forming organoids (%) | No. of organoids formed | No. of cells sorted | Cells forming organoids (%) | No. of organoids formed | No. of cells sorted | Cells forming organoids (%) | ||
| Lgr5-GFP N-term | 0 | 1,500 | 0 | 7 | 1,500 | 0.47 | 7 | 15,000 | 0.05 |
| Lgr5-GFP N-term | 24 | 6,700 | 0.36 | 140 | 6,700 | 2.09 | 65 | 13,000 | 0.5 |
| Lgr5-GFP N-term | 3 | 7,960 | 0.04 | 20 | 7,960 | 0.26 | 15 | 38,450 | 0.04 |
Six independent teratomas FAC-sorted and assayed for organoid formation.
Three organoids derived from three independent unsorted teratomas (dissociated, FAC-sorted, and assayed for organoid formation).
It is important to note that in independent experiments, these organoids were generated with high frequency from GFP-positive cells (Table 1, top), although they could also be robustly generated from nonsorted teratoma cells by enrichment through serial passage (Figure 1D). Undifferentiated hESCs (Figure S4A) as well as the sorted GFP-negative fraction failed to give rise to organoids (Table 1, top), although this does not exclude the possibility that LGR5-negative cells could give rise to organoids under alternative conditions. The most prominent nonorganoid cells found to grow in the 3D matrix, regardless of whether the cells were GFP sorted or not, were single cells with neuronal precursor morphology. These contaminating cells were removed at each passage, enriching for nearly homogeneous long-term (>150 days) organoid cultures that could be maintained regardless of whether they were derived from GFP-positive cells or from nonsorted teratomas (Figures 1C, 1D, and S4A–S4D).
hPSC-Derived Intestinal Organoids Comprise All Major Cell Types of the Intestine
To characterize hPSC-derived intestinal organoids, we performed quantitative RT-PCR (qRT-PCR) analysis of cultures derived from GFP-FACS experiments, as well as from nonsorted teratoma cells, followed by RNA-seq analysis of samples of tissues from different stages of our differentiation paradigm and primary intestinal biopsy-derived organoids (Table S1). These analyses revealed the specific upregulation of intestinal stem cell markers such as LGR5, KLF5, and SOX9, and intestine-specific genes such as VIL1, MUC2, and TFF3 (Figures S5E and S5F). Marker gene expression for FGF8 and other nonintestinal cell types such as Nestin and Sox1 (both neural) and MIXL1 (mesoderm) was significantly decreased in hPSC-derived intestinal organoids. Genes that were not associated with high intestinal stem cell expression showed low and variable expression levels in the GFP-sorted organoids, and pluripotency genes such as Nanog were absent. These data indicate that hESC-derived organoids were no longer pluripotent, had an intestinal expression pattern, and could be isolated without LGR5-GFP sorting (Figures 2, S2A, and S5F). Organoid cultures failed to form tumors when injected subcutaneously into immunocompromised mice (data not shown).
Figure 2.
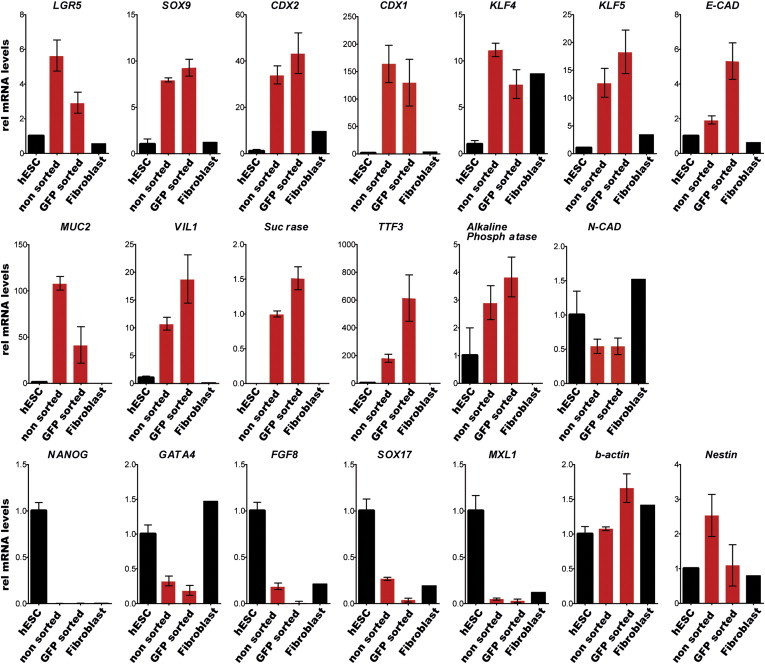
Isolation of Intestinal Organoids Independent of the LGR5-GFP Reporter System
Expression profiling of organoids from GFP-positive and nonsorted LGR5-GFP teratoma cells compared with hESCs and fibroblast-like cells. qRT-PCR for the indicated genes in hESCs, organoids derived from eGFP-positive cells (n = 3), and nonsorted (n = 3) cells of LGR5-GFPN-term, hESCs, and fibroblast-like cells (derived from hESCs as described previously [Hockemeyer et al., 2008], expressing telomerase from the AAVS1 locus). Relative expression levels were normalized to baseline expression of these genes in hESCs. Data are biological replicates of independent experiments; bars represent the SEM. See also Figure S2.
We found a nearly identical expression profile between organoid cultures that had been generated using either the LGR5-GFP marker or unsorted teratoma cells, suggesting that our culture method was selective for intestinal stem/progenitor cells, and that although the LGR5-GFP reporter was functional, it was dispensable for isolating intestinal organoid cultures by this method. These observations indicate that our protocol may be generalized to other genetically engineered hPSCs without the LGR5 reporter.
The major differentiated cell types of the intestinal epithelium include Paneth cells, enterocytes, goblet cells, and enteroendocrine cells. We asked whether these cell types could also be detected in our hPSC-derived intestinal organoids. IHC analysis showed that the hPSC-derived organoids comprise a mostly polarized epithelium that includes differentiated cell types with distinct morphologies and specific marker gene expression (Figure 3). We identified cells with robust expression of CHGA, a marker for enteroendocrine cells; MUC2, a marker for goblet cells; and VIL1, which marks the villi of enterocytes (Figure 3). Furthermore, we observed a subset of organoids containing cells that were enriched for lysozyme staining (Figure S3; Movie S1).
Figure 3.
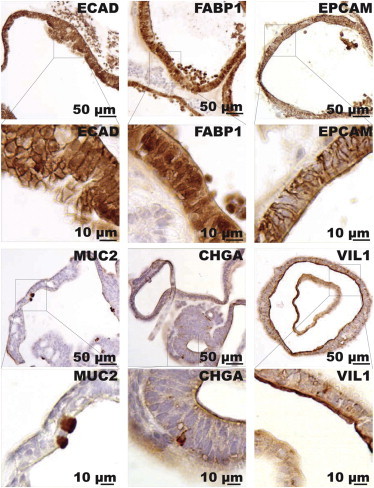
hESC-Derived Organoids Comprise Specific Cell Types Characteristic of the Human Intestinal Epithelium
(A) IHC staining for intestinal marker proteins in sections of hESC-derived organoids generated from wild-type hESC reporter cells. ECAD1, E-cadherin; FABP1, fatty acid-binding protein 1; EPCAM, epithelial cell adhesion molecule; MUC2, mucin2; CHGA, chromogranin A; VIL1, villin1.
See also Figure S3.
The expression of adult intestinal marker proteins for the major cell types of the adult intestinal tissue prompted us to further analyze the composition and subcellular organization of hESC-derived organoids by electron microscopy (EM). Micrographs showed that organoids were organized in a polarized epithelium, with nuclei positioned proximal to the basolateral membrane (Figure 4A). The apical cellular membranes were characteristic of enterocytes with highly organized microvilli (Figures 4B-II and 4D-III) and tight junctions connecting neighboring enterocyte-like cells (Figures 4B-I and 4D-III). Goblet-shaped secretory cells (Figure 4C) that contained large vesicles coming from a well-defined rough endoplasmic reticulum (Figures 4D-I and 4D-II) were embedded in the enterocytes. Vesicles of these cells were localized at the apical side of the cell, suggesting secretion toward the lumen of the organoid (Figure 4).
Figure 4.
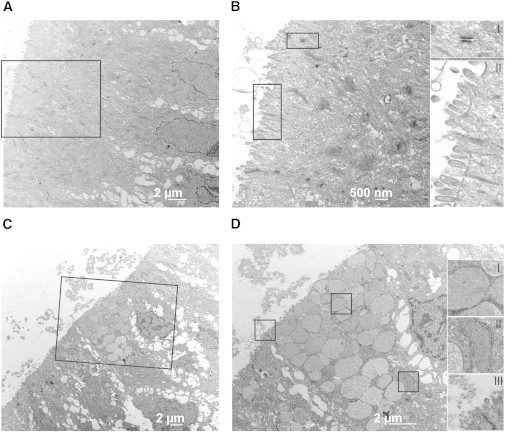
Subcellular Organization of Organoids Resembles the Structure of the Human Intestinal Epithelium
(A) Electron micrograph of hESC-derived organoids. Shown is a representative image with enterocyte-like cells forming a polarized epithelium. The lumen of the organoid is oriented to the left (apical), while the nuclei are aligned along the right (basal).
(B) Higher-magnification images of the micrograph shown in (A). Insets indicate areas of increased magnification. Images show the microvilli lining the luminal cell surface, vesicles in the luminal space (II), and the tight junctions connecting adjacent enterocyte-like cells (I).
(C) Electron micrograph of hESC-derived organoids. Shown is a representative image of a goblet-like secretory cell that is embedded into a sheet of enterocyte-like cells. Orientation of the image as in (A).
(D) Higher-magnification images of the micrograph shown in (C). Insets indicate areas of increased magnification. Images show magnification of vesicles (I), rough endoplasmic reticulum at the base of the vesicles (II), and tight junctions and cellular debris shed into the organoid’s lumen (III).
hPSC-Derived Intestinal Organoids Contain Cells with Characteristics of Adult Intestinal Stem Cells
Adult stem cells have the ability to self-renew and a restricted capacity to differentiate into defined cell types. The human intestinal epithelium is a highly proliferative tissue with a turnover rate of ∼5 days in differentiated cells outside the crypt. We found that single-cell-derived intestinal organoids could be maintained in long-term cultures (>6 months), suggesting the presence of a self-renewing cell population. To further test for the presence of such adult stem cell-like cells, we assayed for the minimum growth factors required to establish and maintain the organoid cultures. As expected for a system that originates from adult intestinal stem cells, we found a phenotypic growth dependence on epidermal growth factor (EGF), Noggin, and RSPO1 (ENR; Figures 5A and 5B), factors that are also essential in the mouse organoid system and human primary cultures (Barker et al., 2007; Sato et al., 2009; Spence et al., 2011). Alternative conditions that were previously reported to support human organoid cultures (Jung et al., 2011; McCracken et al., 2011; Spence et al., 2011; Wang et al., 2013) also allowed us to derive organoids from teratoma samples with a morphology similar to that described in previous studies (Finkbeiner and Spence, 2013; Sato et al., 2009; Stelzner et al., 2012; Figures S4B and S4C).
Figure 5.
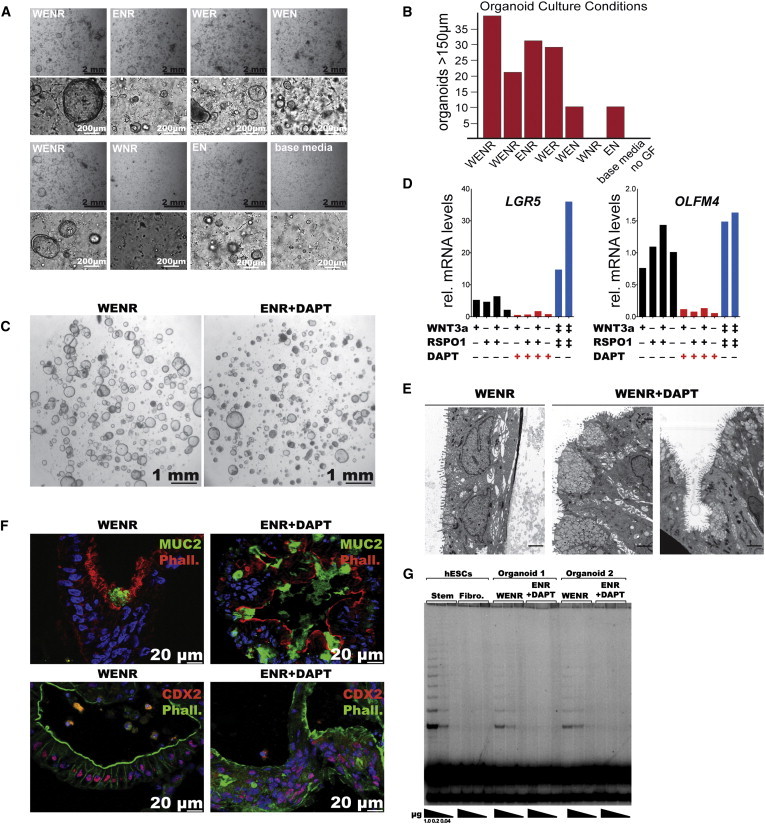
WNT and Notch Signaling Is Required to Establish and Maintain Cells with Adult Stem Cell Properties in hESC-Derived Intestinal Organoids
(A) Bright-field image of organoids at day 5 of their derivation under the indicated culture conditions at two different magnifications. Growth factors (GFs) : WENR [W, Wnt3a; E, EGF; N, Noggin; R, R-spondin-1] were supplemented in the combinations indicated. No GFs were added to “base media.”
(B) Quantification of images shown in (A). The graph shows the number of organoids larger than 150 μm from a single teratoma isolation cultured in parallel under the conditions described in (A), where organoids were grown in 50 μl solidified Matrigel in 500 μl media/well in a 24-well plate.
(C) Bright-field image of organoids at day 15. The image to the left shows a culture at day 15 when grown in WENR, and the right image shows the same cells when switched at day 10 to differentiation media.
(D) qRT-PCR for the intestinal stem cell markers LGR5 and OLFM4 in a single organoid culture treated in parallel with different culture conditions. The ‡ symbol indicates 1,000 ng/ml and 200 ng/ml of RSPO1 and WNT3a, respectively, and + indicates 200 μg/ml and 50 ng/ml of RSPO1 and WNT3a, respectively. Where indicated, DAPT was added at 10 μM.
(E) Electron micrographs of the cells shown in (C). Size bars indicate 2 μm.
(F) IF staining of cryosectioned organoids cultured in either stem cell media (WENR) or differentiation media (10 μM ENR+DAPT added for 4 consecutive days). Top: MUC2 (green), phalloidin (red), and DAPI (blue). Bottom: CDX2 (red), phalloidin (green), and DAPI (blue).
(G) TRAP assay (Kim and Wu, 1997) of hESCs, hESCs differentiated into fibroblast-like cells (Hockemeyer et al., 2008), intestinal organoids grown in stem cell media (WENR, n = 2) or differentiation media (10 μm ENR+ DAPT, n = 2). Shown is a 32P autoradiogram of TRAP activity described for decreasing amounts of protein extracts (1.0–0.4 μg) for the indicated cell types (hESCs [WIBR3]; Fibro., fibroblast-like cells derived from WIBR3).
See also Figure S4.
To further validate the presence of cells with adult stem cell-like characteristics in hPSC-derived intestinal organoids, we took advantage of the LGR5-GFP reporter system. First, we isolated organoid cultures from LGR5-GFP-derived teratomas without FACS sorting for GFP. After maintaining these cells for 5 days in 3D cultures, we investigated whether LGR5-GFP-positive cells were still present and able to form organoids after single-cell dissociation. We found that sorting for GFP fluorescence enriched for cells competent to form organoids when compared with the GFP-negative cell fraction (Table 1). Since LGR5 in mice marks intestinal stem, but not differentiated, cell types, this finding suggested that our culture conditions maintained cells with adult stem cell properties sufficient to form new organoids from single cells (Table 1; Figures 1D and S1).
To further substantiate the presence of functional adult stem cell-like cells in the hPSC-derived organoid system, we cultured hPSC-derived organoids under conditions known to either promote self-renewal (high WNT signaling) or induce differentiation (Notch inhibition) of intestinal stem cells. We found that reduced WNT signaling by withdrawal of either WNT or RSPO1 reduced proliferation and decreased expression of the stem cell markers LGR5 and OLFM4 (Figures 5C and 5D), and induced differentiation into secretory cells as indicated by a shift in cellular morphology detected by EM and immunofluorescence (IF) staining (Figure 5E and 5F). These cells showed an increased number of vesicles at the apical/luminal border (Figure 5E), maintained CDX2, increased MUC2, and a density of filamentous actin at the apical border by IF (Figures 4, 5E, and 5F).
Long-term maintenance of adult stem cells requires telomerase expression and enzymatic activity (Günes and Rudolph, 2013). Telomerase activity in the human intestine is restricted to the stem cell compartment and is silenced upon differentiation of these cells (Schepers et al., 2011). We found that hPSC-derived organoid cultures could be maintained for several months, suggesting the presence of a self-renewing cell type. To functionally test whether hPSC-derived organoids contained telomerase-positive cells, we measured telomerase in vitro. As a positive control, we confirmed that the high level of telomerase activity in hESCs was significantly downregulated upon differentiation into fibroblast-like cells (Figure 5G). Notably, we could detect robust telomerase activity in three out of four independently derived intestinal organoid cultures when grown under adult stem cell conditions by telomeric repeat amplification protocol (TRAP) assay (Kim and Wu, 1997; Figure 5G). However, this activity was abrogated upon terminal differentiation of organoids by WNT-signaling withdrawal and inhibition of γ-secretase by DAPT. These functional data suggest that cells with adult stem cell properties, such as self-renewal and multipotency, represent a subpopulation of organoid cells that can respond appropriately to differentiation cues.
hPSC-Derived Intestinal Tissue Organoids Are Closely Related to Human Primary Intestinal Tissue-Derived Organoids
To determine the degree to which hPSC-derived organoids could recapitulate primary cultures established from adult intestinal tissue, we evaluated the expression profile using next-generation RNA-seq of hPSC-derived intestinal organoids cultured in media that either supported stem cell maintenance (n = 10) or induced terminal differentiation (n = 4). We compared these organoids with the parent hESC lines (n = 4) and teratoma samples (n = 4), and with organoids generated from adult human tissue samples biopsied from the duodenum (n = 2), ileum (n = 2), or rectum (n = 2). Unbiased complete linkage clustering for the top 5,000 most differentially expressed transcripts across these samples revealed that hESC-derived organoids closely resembled organoids isolated from the intestines of human subjects (Figures 6A and S5A). In contrast, teratoma samples were most similar to hESCs (Figures 6A, S5A, and S5C). Transcriptional profiling of organoids derived from an independent hESC line (BG01) confirmed that organoids could be derived irrespective of the genetic background of the hPSC cell line with high similarity to primary tissue-derived organoids (Figures 6A, 6B, and S5A–S5D). hESC-derived intestinal organoids grown in WENR (Wnt3a, EGF, Noggin, R-spondin-1) media expressed levels of intestinal adult stem cell marker genes (LGR5, OLFM4, KLF4, KLF5, SOX9, and TERT) and differentiated/secretory linage genes (e.g., KRT20, VIL1, MUC2, LYZ, CA2, SI, and PLA2GA) analogous to those found in organoids derived from primary tissue, with the greatest similarity to rectum-derived samples (Figures 6B, 6E, and S5D–S5G).
Figure 6.
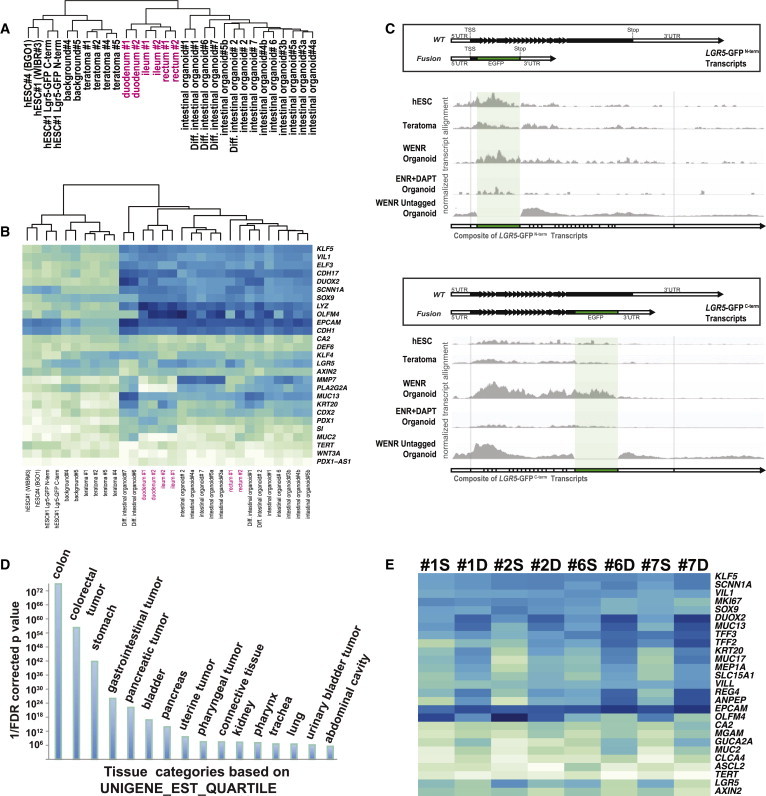
hESC-Derived Organoids Share a Transcriptional Profile with Primary Intestinal Tissue-Derived Organoids and Display the Characteristic Responses to Differentiation Stimuli
(A) Cluster analysis of hESCs (n = 4), teratoma samples (n = 4), bulk hESC-derived organoid cultures grown in WENR (n = 10), bulk cultures of hESC-derived organoids grown in differentiation medium (ENR+DAPT, n = 4), and primary duodenum- (n = 2), rectum- (n = 2) and ileum- (n = 2) derived organoids. Euclidian distances calculated from the abundance levels of the top 5,000 differentially expressed transcripts are also shown in Figure S1A.
(B) Heatmap displaying an unbiased cluster analysis of samples analyzed in (A) for a limited selection of genes relevant to intestinal expression and, in most cases, reported functions. Shown are the Euclidian distances calculated from the relative expression of the genes as determined by next-generation RNA-seq expression analysis.
(C) Density maps of RNA-seq reads mapped to the genetically engineered LGR5-GFPN-term and LGR5-GFPC-term locus. The top boxes show the predicted transcripts for each allele based on validated gene editing described in Figure 1A. Shaded in green is the region mapping to the coding sequence for the eGPF fusion reporter. Shown across each histogram are the alignments to Lgr5-eGFP fusions predicted for each cell line. RNA was collected from the parent hESC cell lines, the intermediate teratoma samples, intestinal organoids, and intestinal organoids differentiated by the withdrawal of WNT3a and the addition of DAPT.
(D) Gene Ontology analysis for tissue-specific expression using the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/home.jsp) and the “UNIGENE_EST_QUARTILE” expression profile database. Analyzed were genes significantly (FDR corrected p value < 0.05) unregulated (>22.5-fold) in hESC-derived intestinal organoids (n = 10) compared with hESCs (n = 4).
(E) Heatmap displaying an unbiased cluster analysis of organoid samples (from groups 1, 2, 6, and 7 as described in Table S1) analyzed in (A) for selected genes that are associated with the differentiation of adult intestinal stem cells. Shown are pairwise hESC-derived intestinal organoid cultures (n = 4) grown in either WENR stem cell media (S) or differentiated in ENR+DAPT (D).
See also Figure S5.
In contrast to previous results obtained by established protocols, our intestinal organoids did not result in a significant budding of crypt-like structures, which are characteristic of mouse small intestine cultures. The cystic morphology of our organoids was previously described for developmental immature stages of enterosphere differentiation, as well as for adult colon-derived organoids (Fordham et al., 2013; Jung et al., 2011; Sato et al., 2009; Stelzner et al., 2012). Whereas the expression levels of several marker proteins for intestinal function, such as PLAG2A, TFF1, and TFF2 (Figures S5D–S5F) offer evidence of cell maturation of hESC-derived organoids, the full complexity of intestinal maturation was not recapitulated by this in vitro culture. The complex maturation signals required from the neural and vascular plexus and the mesenchyme have yet to be defined in an in vitro culture (Wells and Spence, 2014). Nevertheless, our protocol results in organoids that express adult stem cell markers to the same extent as primary tissue-derived organoids, suggesting that they are beyond a developmental stage that was recently described for human embryonic-like intestinal progenitor cells (Fordham et al., 2013; Figures S5B and S5D–S5F).
Next, we attempted to address the positional identity of these organoids with respect to anterior/posterior gut development. We found expression of the caudal-type homeobox gene CDX1 and the hindgut markers CDX2 (Sherwood et al., 2009) PLA2G2A, CA2, and MUC2 in hPSC-derived organoids and in rectum-derived tissues (Figures 6, S5D, and S5E). The midgut gene PDX1 was enriched in duodenal-derived samples only, and organoids expressed levels more comparable to the distal gut (ileum/rectum; Figure 6B). This analysis suggests that our differentiation protocol may bias for relatively mature hindgut/large intestinal-like tissue.
To further substantiate that hESC-derived organoids specifically express intestinal adult stem cell markers, we analyzed the expression profile of the LGR5-GFP reporter gene in hESC-derived organoids by RNA-seq. Here we found that both the LGR5-GFPN-term and LGR5-GFPC-term reporter recapitulated the expression of the endogenous LGR5 gene and contained a sequence that aligned across the expected fusion junction by RNA-seq (Figures 1A and 6C). Although the LGR5 reporter was expressed in hESCs and slightly reduced in teratoma cells, it was higher in the bulk analysis of organoids that were maintained under stem cell conditions. Importantly, LGR5-GFP expression was significantly repressed in organoid cultures under differentiation conditions (Figure 6C). These data not only validate our LGR5-GFP reporter systems, even though they showed minimal fluorescence by microscopy, but also support the hypothesis that a defined organoid system containing adult stem cells can be successfully generated from hPSCs by this method. Furthermore, a Gene Ontology analysis for genes that were differentially expressed between hESCs (n = 4) and hESC-derived intestinal organoids (n = 10) revealed a significant enrichment of genes associated with the gastrointestinal system (Figure 6D). Categories that were enriched included Epithelial Differentiation and Digestion for biological processes, and various Peptidase Activities for biological function. We also analyzed the genes that were differentially expressed between the human primary-derived samples and the hESC-derived organoids (see Supplemental Experimental Procedures for further discussion of the RNA-seq data).
Finally, we analyzed independently derived organoid cultures that were grown in the absence of WNT3a and in the presence of DAPT. Transcriptional changes showed that this specific change in WNT and Notch signaling resulted in the induction of intestinal-specific differentiation markers and a reduction of adult intestinal stem cell markers (Figures 6E and S5D).
In summary, our genome-wide expression analysis confirmed the following hypotheses: (1) organoids are similar to primary intestinal tissue, specifically the large intestine; (2) organoids express adult intestinal stem cell markers such as SOX9, OLFM4, KLF4, KLF5, LGR5, and TERT; (3) the genes expressed indicate that organoid cultures comprise a relatively mature hindgut-like tissue; (4) expression of LGR5 and the LGR5-GFP reporter is upregulated in mature organoids; and (5) organoids respond to the canonical cues that also determine differentiation and self-renewal in the intestine of adult humans.
Discussion
A Human LGR5-GFP Reporter System Allows the Isolation of Intestinal Tissue Organoids
Here, we report on the development of a method that allows robust derivation of human intestinal adult stem cells and organoid tissue from hPSCs. We used ZFN-mediated genome editing of the endogenous LGR5 locus to generate a reporter system based on a previous study in mice that established LGR5 as a bona fide marker of adult intestinal stem cells (Barker et al., 2007). Analogous to what was observed in previous mouse experiments, we demonstrate here that generating either an N- or C-terminal reporter of human LGR5 allows the specific isolation and enrichment of cells with the ability to form organoid cultures from hPSCs despite showing minimal fluorescence by microscopy. Our results provide proof of concept for using site-specific gene-editing technology to isolate novel human cell types from hPSCs.
The intestinal organoids that we generated are similar to tissue isolated directly from humans, as they express gastrointestinal-specific genes and contain highly organized and differentiated cell types such as goblet-, enterocyte-, and enteroendocrine-like cells. In addition to the tissue organization, the cellular and subcellular morphology of hESC-derived organoids is reminiscent of primary organoid cultures. When grown under conditions that promote differentiation, organoids consist of an epithelial sheet of polarized cells with a subcellular organization characteristic of specific cell subtypes, such as secretory cells. Expression profiling suggests that our differentiation protocol may bias for relatively mature hindgut/large intestinal-like tissue. We do not know if this is an intrinsic feature of the teratoma or a function of our culture conditions. However, we find that a subset of organoids contain cells that stain positive for the small-intestine marker gene lysozyme, suggesting that our protocol does not strictly discriminate against more anterior regions of the intestine. As we can detect lysozyme- and PDX1-positive cells in the parent teratoma samples, it seems possible that our method could also be refined to specifically isolate and enrich for small-intestine cellular subtypes.
Modeling Intestinal Function and Disease Using hESC-Derived Intestinal Tissue
For many research applications, including nutrient uptake, drug delivery, metabolic regulation, and human disease modeling (e.g., intestinal cancer), rodent models do not fully recapitulate human intestinal physiology. However, organoids derived from hPSCs may provide a novel platform for in vitro disease modeling and drug screening to complement experiments in conventional animal models.
The organoid system established here will advance disease modeling because the differentiation protocol does not rely on the LGR5 reporter and because the committed intestinal stem cells can be derived from hiPSCs and hESCs. We demonstrate that hESCs that are genetically engineered by TALEN and ZFN-mediated genome editing can subsequently be used to generate intestinal organoids, suggesting that genome-editing approaches such as ZFN, TALEN, and CRISPR-mediated gene knockout and transgenesis will facilitate the study of intestinal biology. Gene editing will allow the generation of isogenic disease-specific cell lines, circumventing the problem of heterogeneity in human samples and eliminating the phenotypic heterogeneity of in vitro phenotypes (Soldner and Jaenisch, 2012; Soldner et al., 2011), thereby complementing current approaches that directly genetically modify intestinal organoid cultures (Miyoshi and Stappenbeck, 2013; Schwank et al., 2013).
hESC-Derived Organoids Represent a Novel In Vitro Adult Stem Cell System
Adult intestinal stem cells are a highly specialized and differentiated cell type that is formed through a well-timed and coordinated developmental process. Our isolation procedure for hPSC-derived intestinal adult stem cells suggests that these developmental cues can be sufficiently recapitulated when hPSCs form a teratoma. Key features of the differentiation process during teratoma formation that may allow for the establishment of adult stem cells include the extended period of time in which the tissue is allowed to form without perturbation, the optimal nutrient/hormonal conditions, and the unconstrained 3D growth in the subcutaneous compartment. In the future, these particular characteristics will have to be recapitulated in a xeno-free differentiation paradigm. Considering that LGR5 has recently also been implicated in adult stem cell function and maintenance of other tissue types, it will be interesting to investigate whether this strategy of genetically engineering hPSCs to carry reporter genes can be applied to the culture of other adult stem cell types that previously could not be isolated.
Experimental Procedures
Teratoma Formation and Analysis
hESCs were collected by collagenase treatment (1.5 mg/ml) and separated from feeder cells by sedimentation. Cells were resuspended in 250 μl of PBS and injected subcutaneously into NOD-SCID mice (Taconic). All animal protocols were approved by the ACUC of UC Berkeley. Tumors (<1.5 cm) formed within 4–8 weeks, at which time teratomas were isolated and disaggregated to single cells or fixed in formalin for analysis of hematoxylin and eosin (H&E) staining. Immunostaining of paraffin sections was performed with standard techniques using a rabbit polyclonal anti-GFP antibody (Abcam 290).
Culture of Intestinal Epithelial Organoids from hESCs
Single cells isolated from teratomas were embedded at 4°C in 50 μl Matrigel containing 1 μM JAG-1 (No. 61298; AnaSpec) and incubated at 37°C for 10 min. The medium (500 μl/well) consisted of 1:1 Dulbecco’s modified Eagle’s medium F-12 and Neurobasic with N2, B27, L-glutamine, nonessential amino acids, penicillin/streptomycin, and growth factors WNT3A (200 ng ml−1), RSPO1 (1 μg ml−1), EGF (50 ng ml−1), Noggin (100 ng ml−1), and ROCK inhibitor (10 μM), and was changed every 2 days. Organoids were passaged using 5 min dispase digestion, subcultured, and gravity separated from single cells using a PBS 0.5% BSA wash. After a 5 min incubation in PBS, 0.5% BSA, and 2 mM EDTA, and hESC media inactivation, the tissue was pelleted and resuspended in 4°C Matrigel as described above. When indicated, WNT was withdrawn and 10 μM DAPT was added to the cultures for 4 days to induce differentiation.
Human Subject Tissues
The ethics committee of the University Medical Centre Utrecht approved this study and written informed consent was obtained from the human subjects. Organoids were generated from biopsies obtained for diagnostic purposes and maintained in culture (for 1–6 months) as described previously (Sato et al., 2011a).
IF Staining
Organoids were fixed in 3% paraformaldehyde with 5% sucrose in PBS (pH 7.4) for 30 min. Samples were embedded for cryosectioning, sectioned at 10 μM, and mounted on poly-L-lysine-coated slides. Samples were permeabilized with TBST (Tris-buffered saline with 0.5% Triton X-100) and incubated at 4°C, 1:5,000 dilution, with shaking overnight. The slides were washed three times with TBST at 4°C and then incubated with the appropriate Alexa Fluor secondary antibodies (1:1,000) and a phalloidin conjugate (1:1,000). Afterward, the slides were washed as before, stained for DAPI, and mounted.
Transcriptional Profiling
Details regarding the protocol used for transcriptional profiling are provided in Supplemental Experimental Procedures. Briefly, an RNA-seq library was created using the standard Illumina library preparation protocol. Ribosomal RNAs were depleted using a oligo(dT) 25 magnetic bead kit (Life Technologies), and the libraries were generated using the PrepX SPIA RNA-Seq and PrepX Library kit (IntegenX) according to the manufacturer’s protocol. Reads were mapped to the Ensembl cDNA release 72 (Flicek et al., 2013) with Bowtie2 version 2.1.0 (Langmead and Salzberg, 2012) using the parameters –rdg 6,5, –rfg 6,5, and –score-min L,-.6,-.4. Transcript abundances were calculated with eXpress version 1.4.0 (Roberts and Pachter, 2013) and the resulting effective counts for each transcript were used to calculate fold changes. Effective counts were also processed with DESeq version 1.12.0 (Anders and Huber, 2010) to identify statistically significant differentially expressed genes and transcripts. Fragments per kilobase of transcript per million mapped reads (FPKM) results are available in Supplemental Experimental Procedures and the raw data for RNA-seq are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56930.
Additional details regarding the methods used in this work can be found in Supplemental Experimental Procedures.
Acknowledgments
We thank R. Alagappan, P. Xu, Dong Dong Wu, and Lei Zhang and the Sangamo Production group for expert technical assistance. We thank Frank Soldner, Thomas Sandmann, and Helen Bateup for helpful comments during the design of the experiment. We thank Nicki Watson from the Keck Imaging Facility at the Whitehead Institute for performing the EM analysis. R.F. is supported by the National Science Foundation Graduate Research Fellowship Program (GRFP) under grant DGE 1106400 and NIH training grant 2T32GM007232-36. K.C. was supported by a fellowship from the Nakajima Foundation. R.J. was supported by NIH grants R37-CA084198, RO1-CA087869, and RO1-HD045022, and by a grant from the HHMI. R.J. is an adviser to Stemgent and a cofounder of Fate Therapeutics. D.H. is a New Scholar in Aging of the Ellison Medical Foundation and is supported by the Glenn Foundation and the Shurl and Kay Curci Foundation. G.J.C., A.C., E.J.R., P.D.G., and F.D.U. are full-time employees of Sangamo BioSciences.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Rudolf Jaenisch, Email: jaenisch@wi.mit.edu.
Dirk Hockemeyer, Email: hockemeyer@berkeley.edu.
Accession Numbers
The GEO accession number for the RNA-seq data reported in this paper is GSE56930.
Supplemental Information
Fragments per kilobase of transcript per million mapped reads (FPKM) values for all RNA-seq samples generated as described in Experimental Procedures. The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE56930 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56930).
Movie of a sequential z stack of an hESC-derived human intestinal organoid stained for lysozyme (green) and DAPI (red).
References
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Cao L., Gibson J.D., Miyamoto S., Sail V., Verma R., Rosenberg D.W., Nelson C.E., Giardina C. Intestinal lineage commitment of embryonic stem cells. Differentiation. 2011;81:1–10. doi: 10.1016/j.diff.2010.09.182. [DOI] [PubMed] [Google Scholar]
- Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.-K., Li V.S.W., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- DeKelver R.C., Choi V.M., Moehle E.A., Paschon D.E., Hockemeyer D., Meijsing S.H., Sancak Y., Cui X., Steine E.J., Miller J.C. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J.F., Wiegerinck C.L., de Jonge H.R., Bronsveld I., Janssens H.M., de Winter-de Groot K.M., Brandsma A.M., de Jong N.W.M., Bijvelds M.J.C., Scholte B.J. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S.R., Spence J.R. A gutsy task: generating intestinal tissue from human pluripotent stem cells. Dig. Dis. Sci. 2013;58:1176–1184. doi: 10.1007/s10620-013-2620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham R.P., Yui S., Hannan N.R., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günes C., Rudolph K.L. The role of telomeres in stem cells and cancer. Cell. 2013;152:390–393. doi: 10.1016/j.cell.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Cook E.G., Gao Q., Mitalipova M., Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P., Sato T., Merlos-Suárez A., Barriga F.M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M.A., Sancho E. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Kim N.W., Wu F. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP) Nucleic Acids Res. 1997;25:2595–2597. doi: 10.1093/nar/25.13.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-A., Kakitani M., Zhao J., Oshima T., Tang T., Binnerts M., Liu Y., Boyle B., Park E., Emtage P. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P.J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C.J., Gimelbrant A.A., Erwin J.A., Cheng A.W., Guenther M.G., Welstead G.G., Alagappan R., Frampton G.M., Xu P., Muffat J. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- McCracken K.W., Howell J.C., Wells J.M., Spence J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald T., Wang R., Bailey W., Xie G., Chen F., Caskey C.T., Liu Q. Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem. Biophys. Res. Commun. 1998;247:266–270. doi: 10.1006/bbrc.1998.8774. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Stappenbeck T.S. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogaki S., Shiraki N., Kume K., Kume S. Wnt and Notch signals guide embryonic stem cell differentiation into the intestinal lineages. Stem Cells. 2013;31:1086–1096. doi: 10.1002/stem.1344. [DOI] [PubMed] [Google Scholar]
- Ootani A., Li X., Sangiorgi E., Ho Q.T., Ueno H., Toda S., Sugihara H., Fujimoto K., Weissman I.L., Capecchi M.R., Kuo C.J. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat. Med. 2009;15:701–706. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrinet L., Rodilla V., Liu Z., Chen S., Koch U., Espinosa L., Kaestner K.H., Kopan R., Lewis J., Radtke F. Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140:1230–1240. doi: 10.1053/j.gastro.2011.01.005. e1–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner H., Sprunger J., Charlat O., Leighton-Davies J., Grosshans B., Salathe A., Zietzling S., Beck V., Therier M., Isken A. R-Spondin potentiates Wnt/β-catenin signaling through orphan receptors LGR4 and LGR5. PLoS ONE. 2012;7:e40976. doi: 10.1371/journal.pone.0040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A.G., Vries R., van den Born M., van de Wetering M., Clevers H. Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 2011;30:1104–1109. doi: 10.1038/emboj.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Sherwood R.I., Chen T.Y., Melton D.A. Transcriptional dynamics of endodermal organ formation. Dev. Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H.J., van der Flier L.G., Sato T., van Es J.H., van den Born M., Kroon-Veenboer C., Barker N., Klein A.M., van Rheenen J., Simons B.D., Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Soldner F., Jaenisch R. Medicine. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Soldner F., Laganière J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzner M., Helmrath M., Dunn J.C., Henning S.J., Houchen C.W., Kuo C., Lynch J., Li L., Magness S.T., Martin M.G., NIH Intestinal Stem Cell Consortium A nomenclature for intestinal in vitro cultures. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Wang P., Rodriguez R.T., Wang J., Ghodasara A., Kim S.K. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Scoville D., He X.C., Mahe M.M., Box A., Perry J.M., Smith N.R., Lei N.Y., Davies P.S., Fuller M.K. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383–395. doi: 10.1053/j.gastro.2013.04.050. e1–e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.M., Spence J.R. How to make an intestine. Development. 2014;141:752–760. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Yusa K., Rashid S.T., Strick-Marchand H., Varela I., Liu P.-Q., Paschon D.E., Miranda E., Ordóñez A., Hannan N.R.F., Rouhani F.J. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–394. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Maeder M.L., Mali P., Pruett-Miller S.M., Thibodeau-Beganny S., Chou B.K., Chen G., Ye Z., Park I.H., Daley G.Q. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fragments per kilobase of transcript per million mapped reads (FPKM) values for all RNA-seq samples generated as described in Experimental Procedures. The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSE56930 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56930).
Movie of a sequential z stack of an hESC-derived human intestinal organoid stained for lysozyme (green) and DAPI (red).


