Summary
Lineage tracing using Cre/lox transgenic mice provides a powerful tool for studying normal mammary epithelial cell (MEC) development and the cellular origins of mammary tumors under physiological settings. However, generation of new transgenic mice for lineage-tracing purposes is often time consuming. Here, we report a lineage-tracing tool for MECs based on intraductal injection of lineage-specific Cre-expressing adenovirus (Ad-Cre). Using well-characterized promoters for Keratin 8 and Keratin 14, we generated lineage-specific Ad-Cre lines for luminal and basal MECs, respectively. By pulse-chase lineage tracing using these Ad-Cre lines, we showed that luminal and basal lineages are largely self-sustained and that IRS1 and IRS2 are essential for maintaining the basal lineage; we also showed that heterogeneous mammary tumors can be induced from luminal MECs in mice carrying the Etv6-NTRK3 fusion gene. Overall, we validated the Ad-Cre system as a promising and efficient tool for fate mapping of normal and malignant cells in adult tissues.
Highlights
-
•
Adenovirus-Cre can be used for pulse-chase lineage tracing of adult stem cells
-
•
Mammary luminal and basal lineages in adults are largely self-sustained
-
•
IRS1 and IRS2 are essential for maintaining the adult mammary basal lineage
-
•
Multiple adult mammary luminal cell types may serve as breast cancer cellular origins
In this study, Li and colleagues used cell-type-specific Adeno-Cre viruses for pulse-chase lineage tracing of adult stem cells. In mammary glands, it was found that adult mammary luminal and basal epithelial lineages were largely self-sustained, IRS1 and IRS2 were essential for maintaining the basal linage, and multiple luminal cell subtypes might serve as cellular origins of breast cancer.
Introduction
Lineage tracing is a powerful tool for studying tissue development, homeostasis, and disease, and has provided unprecedented insights into stem cell biology (Kretzschmar and Watt, 2012). Previous lineage-tracing studies mostly relied on inducible Cre-estrogen receptor fusion protein (CreER)-expressing transgenic mice upon induction by tamoxifen. This inducible system was recently used for fate-mapping studies of mammary epithelial cells (MECs) under the physiological setting (Lafkas et al., 2013; Rios et al., 2014; Šale et al., 2013; van Amerongen et al., 2012; Van Keymeulen et al., 2011). However, wider application of this approach is limited by several factors. First, the choice of specific inducible CreER-expressing lines is often limited, and generating new mouse lines for this purpose can be time consuming. Second, most CreER mice do not target MECs exclusively, and for breast cancer modeling studies, their activities outside of the mammary gland (MG) may lead to systematic deficiency or unwanted tumor induction in other tissues, which could limit their use for studying MECs. Third, administration of tamoxifen may interfere with development of hormone-dependent tumors (e.g., mammary tumors), as well as normal MG development (Rios et al., 2014). Lastly, recent studies showed that the tamoxifen doses commonly used to induce Cre/lox recombination in mice might continue to label significant numbers of cells for weeks after tamoxifen treatment (Reinert et al., 2012) and that tamoxifen could change the behavior of stem cells (Zhu et al., 2013), both of which could affect interpretation of results from lineage-tracing experiments.
Adenovirus is a DNA virus, and it does not integrate into the host genome. It can infect both dividing and nondividing cells, leading to transient high-level protein expression (Anderson et al., 2000). Intraductal injection of adenovirus was previously shown to be an efficient way to transduce genes in MECs (Russell et al., 2003). Cre-expressing adenovirus (Ad-Cre) under the control of the constitutive CMV promoter (Ad-CMV-Cre) has been successfully used to induce cancer development in multiple organs (e.g., lung, ovary, and bladder; [DuPage et al., 2009; Flesken-Nikitin et al., 2003; Puzio-Kuter et al., 2009]). Recently, Sutherland et al. (2011) used cell-type-specific promoters and Ad-Cre to initiate small cell lung cancer development from different subsets of lung cells. We hypothesized that, similarly to the inducible CreER system, transient expression of Cre from adenoviral vectors could offer a temporal and spatial genetic-marking system for pulse-chase lineage-tracing studies in adult cells. In this study, we tested this approach in the MG by generating MEC lineage-specific Ad-Cre lines, and demonstrated that they can be used for MEC fate-mapping, gene loss-of-function, and cancer-induction studies in the native environment. This approach should also be suitable for lineage-tracing studies in other systems in which introduction of Ad-Cre is feasible.
Results and Discussion
Genetic Marking of MECs by Intraductal Injection of Ad-Cre
It was demonstrated previously that mammary epithelium could be effectively transduced in vivo by adenovirus via intraductal injection (Russell et al., 2003). We performed this procedure successfully in MGs of both adult virgin (Figure 1A) and 3-week-old (Figure 1B) female mice. Since puberty in mice occurs at ∼4–6 weeks of age, this approach should allow us to introduce genetic changes into MECs during both pubertal development and adulthood. In pilot experiments, we injected Ad-CMV-Cre into #4 MGs of a conditional Cre-reporter mouse line, Rosa26-lsl-YFP (R26YFP). Cre expression from adenovirus triggered the excision of a floxed Stopper cassette in the R26YFP knockin allele, leading to permanent genetic marking of the infected cells and their progeny by yellow fluorescent protein (YFP; Figures 1C and 1D). The labeling efficiency of MECs, as measured by the percentage of YFP+ cells 3 days after injection, ranged from 0.65% ± 0.05% to 19.23% ± 4.85%, corresponding to titers of Ad-CMV-Cre from 107 to 109 pfu/ml (Figure 1E). All major MEC subpopulations, including mature luminal cells (MLs, CD31−CD45−TER119−(Lin−)CD24hiCD29+CD61−), luminal progenitors (LPs, Lin−CD24hiCD29+CD61+), and basal cells (Lin−CD24medCD29hi), could be effectively labeled (Figure 1F). Only very minimal YFP-marked cells were detected in the stromal gate, which suggests that little viral leakage occurred, thus enabling us to study cell-autonomous effects in MECs (Figure 1F). Since the needle used for injection might have come in contact with skin surrounding the nipple, we performed immunofluorescence (IF) staining of tissues in this area. We only detected YFP+ cells in the mammary ducts directly adjacent to the skin, as well as a few YFP+ stromal cells; no skin cells were found to be YFP+ (Figure S1A available online). Using flow cytometry and PCR for genomic DNA, we did not detect Cre-mediated excision in the R26YFP allele in tissues outside of the MG (Figures S1B–S1D), further confirming the MEC specificity of this approach. Lastly, since it was reported that intratracheal administration of Ad-Cre to the mouse lung could lead to clearance of infected lung cells, possibly due to an immune response (Meuwissen et al., 2001), we checked mammary tissues at various time points after intraductal injection of Ad-Cre. We did not observe any significant inflammatory response (Figure S1E), consistent with a previous study (Russell et al., 2003).
Figure 1.
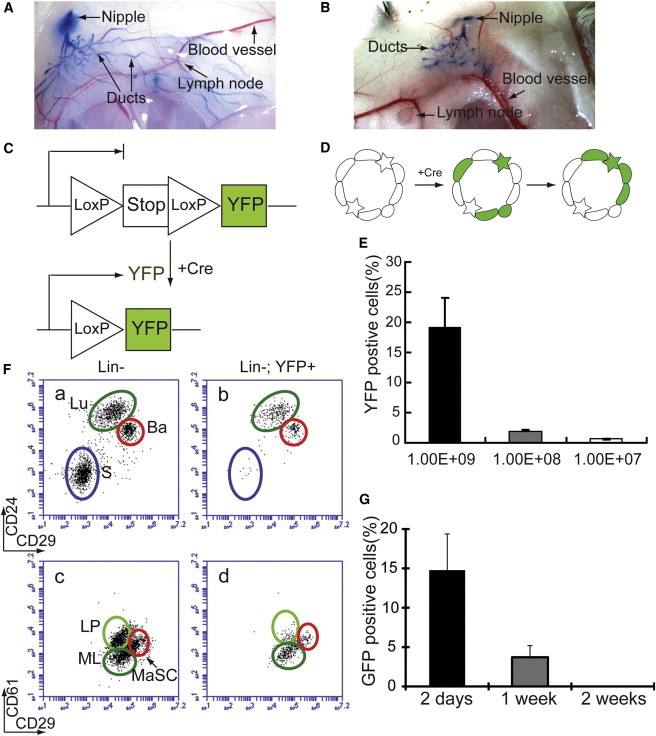
Genetic Marking of MECs by Intraductal Injection of Ad-Cre
(A and B) Intraductal injection of trypan blue dye into adult (A) and 3-week-old (B) female mice.
(C) Schematic diagram of R26YFP reporter activation upon Cre-mediated excision of a floxed Stopper (Stop) cassette.
(D) Schematic diagram of lineage-tracing strategy using intraductal injection of Ad-Cre. Green star designates a YFP-marked stem cell; other green cells refer to more differentiated MECs.
(E) Different efficiencies of Cre-mediated recombination (measured as % YFP+ cells) achieved by using different titers of Ad-CMV-Cre.
(F) Flow-cytometric analysis of R26YFP females 3 days after Ad-CMV-Cre intraductal injection. Luminal cells (Lu), including luminal progenitors (LP) and mature luminal cells (ML), and basal/myoepithelial cells (Ba), including mammary stem cells (MaSCs), can be labeled by YFP upon Ad-Cre induction. Very few stromal cells (S) were labeled by this approach.
(G) Intraductal injection of Ad-CMV-GFP into FVB mice showing transient GFP expression in MGs. In (E) and (G), n = 3 (injected MGs). Data were reported as mean ± SEM.
See also Figure S1.
To assess the transient expression nature of adenovirus in vivo, we measured the duration of adenovirus-mediated gene expression in MECs by intraductally injecting CMV-GFP adenovirus (Ad-CMV-GFP) into wild-type FVB females. We found that at day 2 postinjection, 14.77% ± 4.60% GFP+ cells were detected in the injected MGs; this number quickly dropped to 3.70% ± 1.48% at 1 week after injection, and to zero at 2 weeks postinjection (Figure 1G). We reason that Cre expression from the same adenoviral system should follow a similar kinetics and thus serves a similar purpose as the inducible CreER mice.
MEC Lineage Tracing Using Cell-Type-Specific Ad-Cre Lines
To target different MEC subsets, we generated lineage-specific Ad-Cre lines by using the same adenoviral backbone as Ad-CMV-Cre coupled with a cell-type-specific promoter to drive nlsCre (nls: nuclear localization sequence) expression. For the luminal lineage, we used the mouse Keratin 8 (K8) promoter (Van Keymeulen et al., 2011) and generated Ad-K8-nlsCre virus. Three days after Ad-K8-nlsCre injection into R26YFP females, we found that Ad-K8-nlsCre specifically led to YFP labeling of luminal MECs (Figure 2Ab), including both MLs and LPs (Figure 2Ad). By IF staining of sorted YFP+ cells or MG sections from the injected R26YFP females, we confirmed that the YFP+ cells were exclusively K8+K14− luminal cells (Figures 2C and S2D). To test the long-term labeling efficiency, we chased R26YFP females injected with Ad-K8-nlsCre for 1.5 months. At this stage, we found that YFP+ cells could still be detected robustly and all YFP+ cells remained restricted to the luminal gate (Figure 2Af). A similar observation was made even when the injected mice were chased further to 6 months (Figure S2A). These data suggest that the K8+ luminal lineage contains self-sustained, long-lived MECs. Next, Ad-K8-nlsCre-injected R26YFP females were bred to trigger alveolar differentiation. At midgestation, we found that Ad-K8-nlsCre-marked YFP+ luminal cells contributed to luminal cells in both lobules and main collecting ducts (Figures 2D and S2B). A previous in vivo lineage-tracing study using K8-CreERT2 mice showed that in adult MGs, the luminal and basal lineages were maintained by their own unipotent stem cells (Van Keymeulen et al., 2011). Although a more recent lineage-tracing study provided evidence for a contribution of bipotent basal stem cells to the luminal lineage in vivo, it also revealed long-lived LPs that could contribute to maintenance of the luminal lineage (Rios et al., 2014). The results from our Ad-K8-nlsCre approach agreed with these findings, showing that the luminal lineage, including both ductal and alveolar luminal cells, could be self-sustained by luminal lineage-restricted stem cells or long-lived progenitors.
Figure 2.
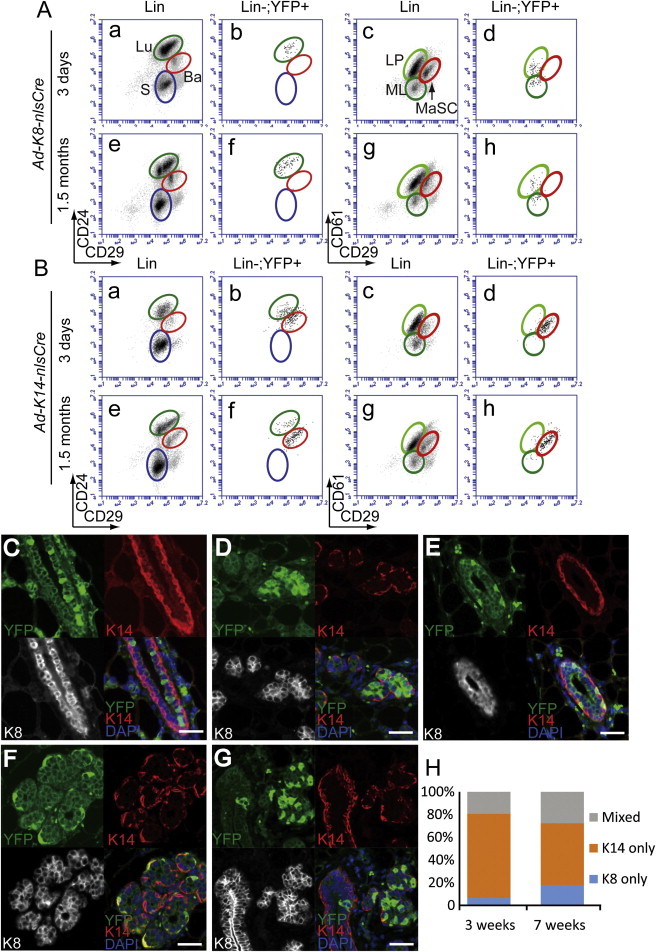
MEC Lineage Tracing Using Cell-Type-Specific Ad-Cre Lines
(A) Ad-K8-nlsCre specifically labeled luminal epithelial cells (Lu), including mature luminal cells (ML) and luminal progenitors (LP), 3 days (a–d) and 1.5 months (e–h) after injection.
(B) Ad-K14-nlsCre-labeled cells were mostly basal/myoepithelial cells (Ba, including MaSC) 3 days (a–d) and 1.5 months (e–h) after injection. A small portion of luminal cells were also labeled by Ad-K14-nlsCre.
(C–G) IF staining of MGs after various chase durations for YFP+ MECs upon pulse labeling by Ad-Cre injection at the nulliparous stage.
(C) Nulliparous MG 3 weeks after Ad-K8-nlsCre injection.
(D) Midgestation MG after Ad-K8-nlsCre injection. Ad-K8-nlsCre-induced YFP+ cells contributed to alveolar formation.
(E) Nulliparous MG 3 weeks after Ad-K14-nlsCre injection.
(F and G) Midgestation MGs after Ad-K14-nlsCre injection. Ad-K14-nlsCre-induced YFP+ cells contributed to alveolar myoepithelial cells (F) and both ductal and alveolar luminal cells (G). Scale bars, 50 μm.
(H) YFP-marked multicell clones (at least three cells per clone) induced by Ad-K14-nlsCre subdivided based on K8 and K14 expression patterns were quantified as K8-only clones, K14-only clones, or mixed clones.
See also Figure S2.
To mark the basal lineage, we used the human Keratin 14 (K14) promoter to direct nlsCre expression via adenovirus (Ad-K14-nlsCre) (Sugihara et al., 2001). By intraductally injecting Ad-K14-nlsCre into R26YFP females, we found that most YFP+ cells were restricted to the basal gate; however, a small population of YFP+ cells could also be detected in the luminal gate (Figure 2Bb). Upon a long-term chase (1.5 months), we found that Ad-K14-nlsCre-marked YFP+ basal and luminal cells were both maintained (Figure 2Bf). To determine the identities of the YFP+ cells, we sorted YFP+ cells in the luminal or basal gate from R26YFP females 3 days after Ad-K14-nlsCre injection, and stained them for K8 and K14. We found that YFP+ cells sorted from the basal gate were exclusively K8−K14+ basal cells (Figure S2E). In contrast, the smaller subpopulation of YFP+ cells sorted from the luminal gate were entirely K8+K14−, suggesting that a small portion of luminal cells were indeed labeled by Ad-K14-nlsCre (Figure S2F). Next, R26YFP females were marked by Ad-K14-nlsCre at the nulliparous stage and either remained virgins or went through pregnancy. By IF staining of their MG sections, we found that Ad-K14-nlsCre-marked basal cells contributed to both alveolar and ductal basal/myoepithelial cells (Figures 2E, 2F, and S2C). A small portion of YFP+ MECs induced by Ad-K14-nlsCre also contributed to lobular and ductal luminal cells (Figure 2G). To determine the cellular origins of these labeled luminal cells, we performed a detailed clonal analysis (Figures 2H and S2G–S2J). We counted YFP+ clones in the injected MGs at 3 days, 3 weeks, and 7 weeks after Ad-K14-nlsCre injection, and classified them into three categories: K14-only clones (K14+K8−), K8-only clones (K8+K14−), and mixed clones containing both cell types. We found that generally, K14+ clones and K8+ clones were labeled independently of each other (Figures 2H and S2G). Upon a 7-week chase, ∼70% K14-only basal clones, 20% K8-only luminal clones, and 10% mixed clones were observed (Figure S2G). These data, combined with those obtained from Ad-K8-nlsCre-based lineage tracing, suggest that the K14+ basal lineage is also self-sustained and may not contribute significantly to the luminal lineage in the short term. However, our data (e.g., showing the presence of mixed clones and a slight increase in the percentages of K8-only and mixed multicell clones upon a 7-week chase; Figure 2H) also do not exclude the possibility that rare K14+ basal MECs can make a long-term contribution to the luminal lineage under the physiological setting, as demonstrated recently (Rios et al., 2014). Since the mixed clones we observed may also have resulted from the fusion of two independently labeled basal and luminal clones that were in a close proximity, three-dimensional analyses at much lower clonal densities will be required before we can precisely assess the cellular origin of these putative mixed clones.
IRS1 and IRS2 Are Essential for Maintaining the Basal Lineage
To determine whether lineage-specific Ad-Cre can be used to study the cell-autonomous function of genes or signaling pathways, we used Ad-K8-nlsCre and Ad-K14-nlsCre to study the role of insulin growth factor/insulin receptor substrate (IGF/IRS) signaling in adult MECs. During development, the IGF pathway is a primary mediator of the Growth Hormone pathway, which stimulates terminal end bud formation and ductal elongation (Kelly et al., 2002; Kleinberg and Ruan, 2008). Intracellular transduction of IGF and Insulin signaling are both mediated by two adaptor proteins, IRS1 and IRS2, through tyrosine phosphorylation (Dearth et al., 2007; Heckman et al., 2007). Abnormal activation of IGF signaling is closely associated with increased breast cancer risk (de Ostrovich et al., 2008).
Both IRS1 and IRS2 were previously shown to play important roles during embryonic MG development (Chan and Lee, 2008; Heckman et al., 2007). Postnatally, both Irs1 and Irs2 null MGs are normal, possibly because they compensate for each other (Chan and Lee, 2008; Heckman et al., 2007). We previously generated Irs1/Irs2 double conditional knockout mice with the R26YFP reporter (Irs1L/L;Irs2L/L;R26YFP) and used K14-Cre and MMTV-Cre mouse lines to disrupt them in MECs (Figure S3A). Although the MGs of these mice looked largely normal by both histological (Figure S3B) and flow-cytometric (Figure S3C) analyses, by characterizing the Lin−YFP+ cells (i.e., Irs1/2 null cells), we found that both models exhibited significantly reduced percentages of Lin−YFP+ Irs1/2 null cells (Figures S3C and S3D), suggesting a growth defect of Irs1/2 double-knockout MECs. In particular, in the K14-Cre model, which induced Irs1/2-loss in a lot more basal cells than the MMTV-Cre model, disruption of IGF/IRS signaling dramatically reduced the percentage of Lin−YFP+ basal cells (Figures S3C and S3E), suggesting that the basal lineage may be more dependent on this pathway for its survival and/or proliferation than luminal cells. However, since both K14 and MMTV promoters are also active during embryonic development, one cannot differentiate the effects of Irs1/2 loss on embryonic MEC development from those of adult MEC maintenance by using conventional K14-Cre or MMTV-Cre mice.
To determine whether IGF/IRS signaling is required for maintenance of each MEC lineage in adults, we injected Ad-K8-nlsCre or Ad-K14-nlsCre into adult Irs1L/L;Irs2L/L;R26YFP nulliparous females. Three days after injection, we found that the Irs1L/L;Irs2L/L;R26YFP mice showed similar YFP-labeling efficiencies compared with R26YFP-only control females (Figures 3A and 3B). However, 3 weeks after injection, whereas a similar level of YFP+ cells (compared with the initial labeling) could be detected in R26YFP control females, in Irs1L/L;Irs2L/L;R26YFP females, the percentages of YFP+ cells induced by either Ad-K8-nlsCre or Ad-K14-nlsCre injection decreased significantly (Figures 3A–3C). These data suggest that IGF/IRS signaling is also essential for maintaining the homeostasis of both the luminal and basal lineages in adult MGs. We also noted that Ad-K14-nlsCre-labeled YFP+ basal cells almost completely disappeared after 3 weeks (Figure 3C, compare red circle in p with those in l and n), whereas a few Ad-K14-nlsCre-labeled or Ad-K8-nlsCre-labeled YFP+ luminal cells were still present after a 3-week chase (Figure 3C, compare green circles in h or p with those in d and f, or in l and n, respectively). These data agree with our findings from the K14-Cre and MMTV-Cre conditional knockout mice, and suggest that basal cells are more sensitive to loss of IGF/IRS signaling and that this pathway is essential for maintaining the basal lineage in vivo. Taken together, these results show that the lineage-specific Ad-Cre system enabled us to quickly study the functions and interactions of different genes and signaling pathways in a cell-autonomous manner.
Figure 3.
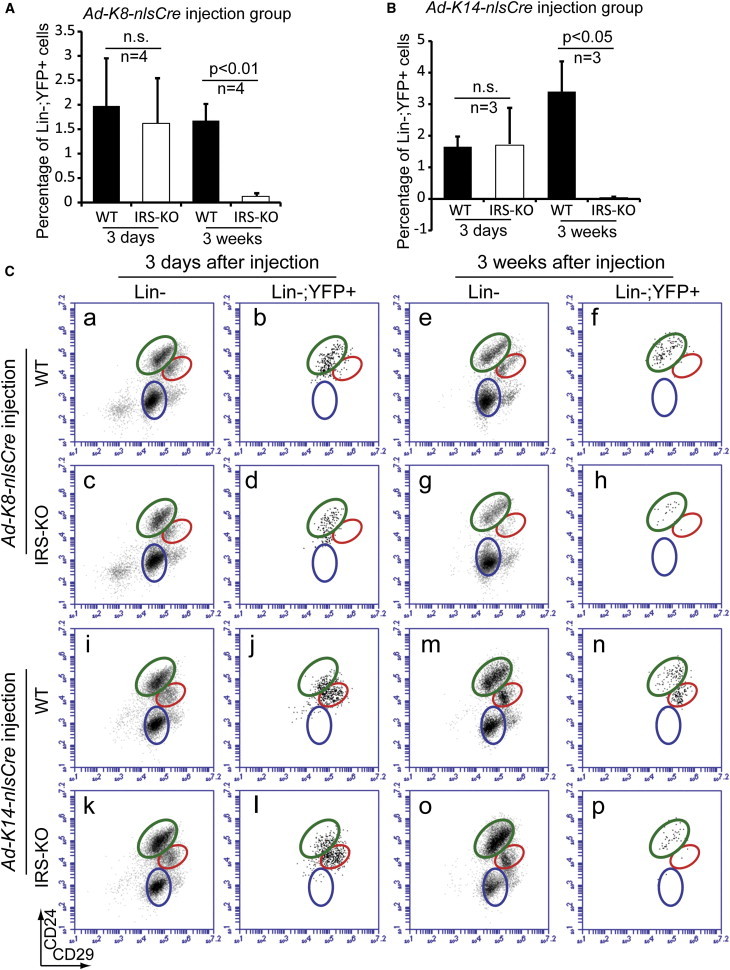
IRS1 and IRS2 Are Essential for Maintaining the Basal Lineage
(A and B) Bar graphs showing the percentages of YFP+ cells. Irs1L/L;Irs2L/L;R26YFP (IRS-KO) and R26YFP-only (WT) control females were injected with Ad-K8-nlsCre (A) or Ad-K14-nlsCre (B), and YFP+ cells were measured by flow cytometry 3 days or 3 weeks after injection. Data were reported as mean ± SEM. n.s., not significant.
(C) Flow-cytometric analysis of YFP+ cells in Irs1L/L;Irs2L/L;R26YFP and R26YFP-only control females injected with Ad-K8-nlsCre (a–h) or Ad-K14-nlsCre (i–p). Both luminal cells (compare f and h) and basal/myoepithelial cells (compare n and p) decreased significantly 3 weeks after Ad-Cre injection. In particular, in Ad-K14-nlsCre injection group (compare n and p), the decrease in YFP+ basal/myoepithelial cells was much more profound than that in YFP+ luminal cells.
See also Figure S3.
Studying the Cellular Origin of Mammary Tumors by Using Lineage-Specific Ad-Cre
Since Sutherland et al. (2011) were able to initiate small cell lung cancer using Ad-Cre, we hypothesized that our cell-lineage-specific Ad-Cre viruses would allow us to study the cellular origin and transformation process of mammary tumorigenesis. To test this, we injected Ad-K8-nlsCre into the mammary ducts of mice carrying the Etv6-NTRK3 (EN) conditional knockin allele we generated previously (Li et al., 2007). EN fusion oncogene, a product from t(12;15)(p13;q25) chromosome translocation, produces a chimeric tyrosine kinase that is characteristic of human secretory breast carcinoma (Tognon et al., 2002). Previously, commonly used MEC-specific Cre-expressing mouse lines, including MMTV-Cre, K14-Cre, and Wap-Cre, were applied to activate EN expression. However, due to activity outside of the MGs, MMTV-Cre or K14-Cre-driven EN activation led to animal lethality before any mammary tumor developed. Only Wap-Cre successfully induced activation of EN in alveolar progenitors in MGs, leading to the development of mammary tumors with 100% penetrance (Li et al., 2007).
We utilized Ad-K8-nlsCre for tumor induction in EN/+ females through intraductal injection. In all injected EN/+ mice, we did not detect any complication or malignancy in other tissues outside of the MG, including skin surrounding the injected nipple area. All injected EN/+ females developed multifocal mammary tumors in the injected glands with a tumor-free latency similar to that observed in Wap-Cre;EN females (Figure 4A). Cre-mediated activation of the EN knockin allele (i.e., removal of a floxed Stopper cassette) was confirmed by PCR using tumor genomic DNA (Figures 4B and S4A), suggesting that the development of mammary tumors was indeed driven by EN. IF staining of the induced tumors for K8 and K14 showed that two tumor types that developed in the previous Wap-Cre;EN model (Li et al., 2007) were recapitulated in the injected females, including type 1 tumors composed of K8+ luminal tumor cells surrounded by K14+K5+ basal tumor cells (“bilineage,” 7 out of 14 tumors), and a type 2 less differentiated tumor comprised of K8+K14+K5− cells (“double positive,” 1 out of 14) (Figures 4C and S4B). Interestingly, in the injected females, a new type of tumor (type 3 “luminal,” 6 out of 14; Figures 4C and S4B) consisting of predominantly K8+K14−K5− luminal tumor cells was also detected. All of these tumors were negative or weakly positive (e.g., type 1) for estrogen receptor (Figure S4B).
Figure 4.
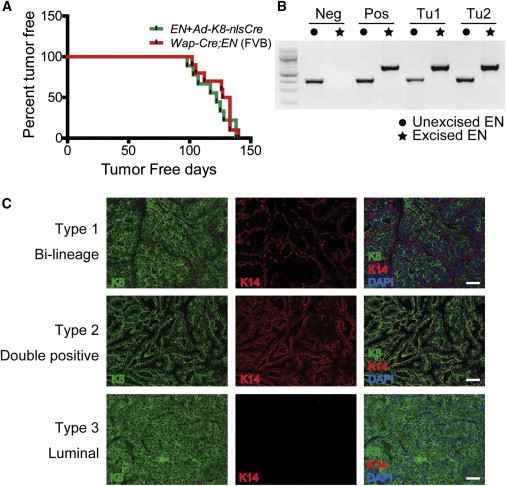
Tumor Induction Using the Ad-Cre Approach
(A) Kaplan-Meier curves showing mammary tumor development in EN knockin virgin female mice injected with Ad-K8-nlsCre (n = 9) and in Wap-Cre;EN virgin females (n = 10). All mice were under the pure FVB background.
(B) Cre-mediated activation of the EN conditional knockin allele in tumor cells was confirmed by PCR. The locations of the primers used to detect unexcised and excised EN alleles are indicated in Figure S4A.
(C) Three types of tumor derived from EN/+ mice injected with Ad-K8-nlsCre as revealed by K8 and K14 IF staining. Scale bars, 50 μm.
See also Figure S4.
In virgin females, Wap-Cre (either from the transgenic mouse or via adenovirus) labeled a population of preexisting unipotent luminal stem or long-lived progenitor cells committed to the alveolar fate, and intraductal injection of Wap-Cre adenovirus into EN/+ females led to the development of type 1 and type 2 mammary tumors identical to those in Wap-Cre;EN females (M.P.A.v.B., L.T., and Z.L., unpublished data). As K8 is a pan-luminal marker, Ad-K8-nlsCre is expected to mark not only Wap-Cre-labeled alveolar luminal stem/progenitor cells but also other luminal cell types. Thus, our Ad-Cre-based tumor study suggests that EN oncoprotein is capable of transforming multiple luminal MEC subsets. Furthermore, due to the short latency of EN-induced mammary tumorigenesis (Figure 4A), our data are consistent with a model in which multiple subpopulations of luminal cells can serve as cells of origin, and distinct cellular origins contribute to the heterogeneity of breast cancer.
Experimental Procedures
Adenovirus Shuttle Vector Construction and Adenovirus Production
Ad-CMV-Cre, Ad-CMV-GFP, promoterless adenoviral shuttle vector (pacAd5 K-NpA), and the packaging vector [pAd5(9.2-100)sub360] were obtained from the Gene Transfer Core of the University of Iowa (Anderson et al., 2000). nlsCre was amplified from pHR-CMV-nlsCRE plasmid (Addgene). Mouse K8 promoter was amplified from mouse genomic DNA. Human K14 promoter was amplified from K14-luciferase plasmid (Sugihara et al., 2001). Crude Ad-K8-nlsCre and Ad-K14-nlsCre were purified using the Adenopure purification kit (Puresyn).
Mice
R26YFP and MMTV-Cre mice were purchased from The Jackson Laboratory. K14-Cre and Wap-Cre mice were obtained from the MMHCC Repository. Etv6-NTRK3 (EN) conditional knockin mice, as well as Irs1 and Irs2 conditional knockout mice, were described previously (Guo et al., 2009; Li et al., 2007). Adenoviruses were introduced into their mammary ducts via intraductal injection. All studies involving mice were approved by our institutional animal care and use committee, and performed in accordance with the relevant protocol.
MEC Preparation, Flow Cytometry, and Sorting
Single-cell suspensions of MECs were prepared as previously described (Shackleton et al., 2006). After antibody staining, flow-cytometric analysis was performed on Accuri 6 and DXP11 flow cytometers. Cell sorting was performed on a BD Aria.
Immunostaining and Clonal Analysis
Mammary tissues were fixed in 10% buffered formalin (Fisher), embedded in paraffin, and sectioned. IF and immunohistochemistry staining were performed on 6 μm paraffin sections for the indicated antibodies by standard procedures. Clonal analysis was performed on MG sagittal sections stained with YFP, K8, and K14 antibodies.
Statistical Analysis
A Student’s t test was used for statistical analysis. Data were reported as mean ± SEM.
Acknowledgments
We are grateful to Grigoriy Losyev and Yiling Qiu for expert technical assistance with FACS sorting, Dr. Beverly Davidson and the University of Iowa Gene Transfer Vector Core for pacAd5 K-NpA, Dr. Bogi Andersen for K14-luciferase plasmid, and Drs. Morris White and Xiaocheng Dong for Irs1/2 conditional knockout mice. This research was supported by a K99/R00 grant from the NCI (CA126980), a Seed Grant and Cancer Program Pilot Grant from Harvard Stem Cell Institute, a Milton Fund Award from Harvard University, a Hearst Foundation Young Investigator Award from Brigham and Women’s Hospital (BWH), startup funds from BWH, and NIH grant R01 HL107663 to Z.L.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Anderson R.D., Haskell R.E., Xia H., Roessler B.J., Davidson B.L. A simple method for the rapid generation of recombinant adenovirus vectors. Gene Ther. 2000;7:1034–1038. doi: 10.1038/sj.gt.3301197. [DOI] [PubMed] [Google Scholar]
- Chan B.T., Lee A.V. Insulin receptor substrates (IRSs) and breast tumorigenesis. J. Mammary Gland Biol. Neoplasia. 2008;13:415–422. doi: 10.1007/s10911-008-9101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ostrovich K.K., Lambertz I., Colby J.K., Tian J., Rundhaug J.E., Johnston D., Conti C.J., DiGiovanni J., Fuchs-Young R. Paracrine overexpression of insulin-like growth factor-1 enhances mammary tumorigenesis in vivo. Am. J. Pathol. 2008;173:824–834. doi: 10.2353/ajpath.2008.071005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearth R.K., Cui X., Kim H.J., Hadsell D.L., Lee A.V. Oncogenic transformation by the signaling adaptor proteins insulin receptor substrate (IRS)-1 and IRS-2. Cell Cycle. 2007;6:705–713. doi: 10.4161/cc.6.6.4035. [DOI] [PubMed] [Google Scholar]
- DuPage M., Dooley A.L., Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flesken-Nikitin A., Choi K.C., Eng J.P., Shmidt E.N., Nikitin A.Y. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res. 2003;63:3459–3463. [PubMed] [Google Scholar]
- Guo S., Copps K.D., Dong X., Park S., Cheng Z., Pocai A., Rossetti L., Sajan M., Farese R.V., White M.F. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Mol. Cell. Biol. 2009;29:5070–5083. doi: 10.1128/MCB.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman B.M., Chakravarty G., Vargo-Gogola T., Gonzales-Rimbau M., Hadsell D.L., Lee A.V., Settleman J., Rosen J.M. Crosstalk between the p190-B RhoGAP and IGF signaling pathways is required for embryonic mammary bud development. Dev. Biol. 2007;309:137–149. doi: 10.1016/j.ydbio.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly P.A., Bachelot A., Kedzia C., Hennighausen L., Ormandy C.J., Kopchick J.J., Binart N. The role of prolactin and growth hormone in mammary gland development. Mol. Cell. Endocrinol. 2002;197:127–131. doi: 10.1016/s0303-7207(02)00286-1. [DOI] [PubMed] [Google Scholar]
- Kleinberg D.L., Ruan W. IGF-I, GH, and sex steroid effects in normal mammary gland development. J. Mammary Gland Biol. Neoplasia. 2008;13:353–360. doi: 10.1007/s10911-008-9103-7. [DOI] [PubMed] [Google Scholar]
- Kretzschmar K., Watt F.M. Lineage tracing. Cell. 2012;148:33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Lafkas D., Rodilla V., Huyghe M., Mourao L., Kiaris H., Fre S. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J. Cell Biol. 2013;203:47–56. doi: 10.1083/jcb.201307046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tognon C.E., Godinho F.J., Yasaitis L., Hock H., Herschkowitz J.I., Lannon C.L., Cho E., Kim S.J., Bronson R.T. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12:542–558. doi: 10.1016/j.ccr.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R., Linn S.C., van der Valk M., Mooi W.J., Berns A. Mouse model for lung tumorigenesis through Cre/lox controlled sporadic activation of the K-Ras oncogene. Oncogene. 2001;20:6551–6558. doi: 10.1038/sj.onc.1204837. [DOI] [PubMed] [Google Scholar]
- Puzio-Kuter A.M., Castillo-Martin M., Kinkade C.W., Wang X., Shen T.H., Matos T., Shen M.M., Cordon-Cardo C., Abate-Shen C. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert R.B., Kantz J., Misfeldt A.A., Poffenberger G., Gannon M., Brissova M., Powers A.C. Tamoxifen-induced Cre-loxP recombination is prolonged in pancreatic islets of adult mice. PLoS ONE. 2012;7:e33529. doi: 10.1371/journal.pone.0033529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios A.C., Fu N.Y., Lindeman G.J., Visvader J.E. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–327. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- Russell T.D., Fischer A., Beeman N.E., Freed E.F., Neville M.C., Schaack J. Transduction of the mammary epithelium with adenovirus vectors in vivo. J. Virol. 2003;77:5801–5809. doi: 10.1128/JVI.77.10.5801-5809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šale S., Lafkas D., Artavanis-Tsakonas S. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat. Cell Biol. 2013;15:451–460. doi: 10.1038/ncb2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Sugihara T.M., Kudryavtseva E.I., Kumar V., Horridge J.J., Andersen B. The POU domain factor Skin-1a represses the keratin 14 promoter independent of DNA binding. A possible role for interactions between Skn-1a and CREB-binding protein/p300. J. Biol. Chem. 2001;276:33036–33044. doi: 10.1074/jbc.M103000200. [DOI] [PubMed] [Google Scholar]
- Sutherland K.D., Proost N., Brouns I., Adriaensen D., Song J.Y., Berns A. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19:754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- Tognon C., Knezevich S.R., Huntsman D., Roskelley C.D., Melnyk N., Mathers J.A., Becker L., Carneiro F., MacPherson N., Horsman D. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- van Amerongen R., Bowman A.N., Nusse R. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Huang Y.F., Kek C., Bulavin D.V. Apoptosis differently affects lineage tracing of Lgr5 and Bmi1 intestinal stem cell populations. Cell Stem Cell. 2013;12:298–303. doi: 10.1016/j.stem.2013.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


