Abstract
Incorporating non-standard amino acids (NSAAs) into proteins enables new chemical properties, new structures, and new functions. In recent years, improvements in cell-free protein synthesis (CFPS) systems have opened the way to accurate and efficient incorporation of NSAAs into proteins. The driving force behind this development has been three-fold. First, a technical renaissance has enabled high-yielding (>1 g/L) and long-lasting (>10 h in batch operation) CFPS in systems derived from Escherichia coli. Second, the efficiency of orthogonal translation systems (OTSs) has improved. Third, the open nature of the CFPS platform has brought about an unprecedented level of control and freedom of design. Here, we review recent developments in CFPS platforms designed to precisely incorporate NSAAs. In the coming years, we anticipate that CFPS systems will impact efforts to elucidate structure/function relationships of proteins and to make biomaterials and sequence-defined biopolymers for medical and industrial applications.
Keywords: non-standard amino acids, cell-free protein synthesis, synthetic biology, sequence-defined polymers, genome engineering
Introduction
The incorporation of non-standard amino acids (NSAAs) into proteins and (poly)peptide-based materials is a key emerging application area in synthetic biology (Liu and Schultz, 2010; Hoesl and Budisa, 2012). In recent years, efforts to incorporate NSAAs using cell-free protein synthesis (CFPS) systems based on Escherichia coli have grown significantly. In this mini-review, we discuss these efforts, beginning with a description of the molecular basis for NSAA incorporation in E. coli using orthogonal translation systems (OTSs). We then describe CFPS and recent improvements in NSAA incorporation in crude cell extract as well as reconstituted systems of purified components. Finally, we discuss emerging frontiers and opportunities for CFPS.
NSAA incorporation
To date, over 100 OTSs have been established for site-specific incorporation of NSAAs into proteins (O'Donoghue et al., 2013). Site-specific NSAA incorporation has been used to expand our understanding of biological systems by enabling studies of protein structure and dynamics with unique IR and X-ray diffraction signatures, fluorescent probes, and photocages (Liu and Schultz, 2010). In other examples, cross-linkable NSAAs have been incorporated to characterize protein-protein and protein-nucleic acid interactions (Liu and Schultz, 2010). In addition to expanding the chemistry of biomolecular systems, NSAA technology has also enabled researchers to mimic post-translational modifications of eukaryotic proteins in bacterial protein expression systems. In an exemplary model, site-specific acetylation of recombinant histones by genetically encoding acetyl-lysine (AcK) elucidated new mechanistic understanding (Neumann et al., 2009).
Beyond fundamental science, NSAA incorporation has also opened the way to novel biopolymer materials, enzymes, and therapeutics which are difficult—if not impossible—to create by other means. Antibody drug conjugates (Zimmerman et al., 2014), modified human therapeutics (Cho et al., 2011), tethered enzymes (Smith et al., 2013), protein polymers (Albayrak and Swartz, in press), phosphoproteins (Park et al., 2011), and selenoproteins (Bröcker et al., 2014) showcase the power of NSAA incorporation. In one example, pegylated human growth hormone showed improved potency and reduced injection frequency (Cho et al., 2011). In another case, an Anti-Her2 antibody bearing p-acetyl-L-phenylalanine enabled precise control of conjugation site and stoichiometry for selective and efficient conjugation to an anti-cancer drug resulting in enhanced tumor regression (Axup et al., 2012). These and other recent breakthroughs highlight exciting opportunities for expanding the chemistry of life.
To incorporate NSAAs site-specifically into proteins, OTSs require (re-)assignment of codons to NSAAs, NSAA-transfer RNA (tRNA) substrates, and ribosome selection of these non-natural substrates into the catalytic center. So far, ribosome accommodation of NSAAs has not been the limiting factor. Rather, strategies to provide for efficient and accurate incorporation of NSAA-tRNA substrates have been the biggest challenge. In practice, this is usually achieved by using orthogonal tRNA (o-tRNA)/aminoacyl-tRNA synthetase (o-aaRS) pairs from phylogenetically distant organisms (Kim et al., 2013). For example, an engineered tRNATyrCUA/TyrRS pair derived from Methanocaldococcus jannaschii is used frequently for NSAA incorporation (Wang et al., 2001). More recent expansions of the technology have used variants of the pyrrolysine translation system, tRNAPylCUA/PylRS from Methanosarcinaceae species (Polycarpo et al., 2006; Wang et al., 2012c). There are many seminal works of orthogonal pairs that have been developed for NSAA incorporation to help drive the field forward (Hughes and Ellington, 2010; Wan et al., 2010; Young et al., 2011; Bianco et al., 2012; Wang et al., 2012a,b; Ko et al., 2013; Lee et al., 2013; Niu et al., 2013; Bröcker et al., 2014; Ma et al., 2014). For codon selection, researchers tend to incorporate NSAAs in response to a non-sense stop codon or quadruplet codon (Wang et al., 2007; Neumann et al., 2010; Niu et al., 2013). The amber codon (TAG) has been the most widely used, because of its low frequency as a stop signal compared to other stop codons (TAA, TGA) (Hoesl and Budisa, 2012).
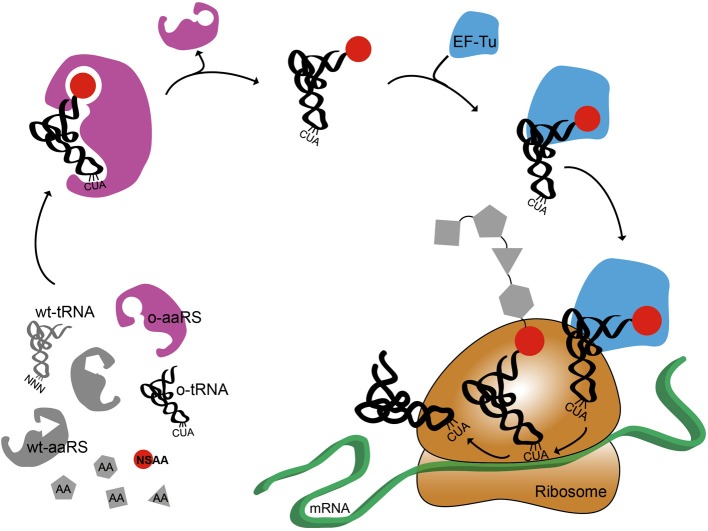
Figure 1 shows a cartoon representation of an OTS for amber suppression. It also highlights the systems biology challenges associated with NSAA incorporation (O'Donoghue et al., 2013). The orthogonal synthetases have poor catalytic efficiency (Tanrikulu et al., 2009; Nehring et al., 2012; Umehara et al., 2012). Elongation Factor Tu (EF-Tu) has a limited capability to incorporate bulky or charged NSAAs (Park et al., 2011; O'Donoghue et al., 2013). The presence of release factor 1 (RF1) can cause early termination of proteins when using amber suppression technology (Johnson et al., 2011; Hong et al., 2014). Recent advances have addressed some of these challenges by improving NSAA incorporation efficiency by engineering o-tRNA (Young et al., 2010; Chatterjee et al., 2012), o-aaRS (Liu et al., 1997; Chatterjee et al., 2012), or EF-Tu (Doi et al., 2007; Park et al., 2011) as well as controlling transcription and translation rate (Young et al., 2010; Chatterjee et al., 2013), and removing RF1 competition (Mukai et al., 2010; Johnson et al., 2011; Loscha et al., 2012; Lajoie et al., 2013). While further efforts to re-engineer translation are still needed, these improvements are accelerating rapid growth in synthetic biology efforts to “upgrade protein synthesis” (O'Donoghue et al., 2013). The bulk of this work is being carried out in vivo; however, complementary in vitro systems are also emerging, which we focus on below.
Figure 1.
Schematic representation of non-standard amino acid incorporation using an orthogonal translation system. Orthogonal aminoacyl tRNA synthetase, o-aaRS; orthogonal tRNA, o-tRNA; wild-type aminoacyl tRNA synthetase, wt-aaRS; wild-type tRNA, wt-tRNA; elongation factor Tu, EF-Tu,; non-standard amino acid, NSAA. Anti-codon sequence on wt-tRNA is NNN, where N is A, C, G, or U. Anti-amber codon sequence on o-tRNA is CUA.
Cell-free protein synthesis
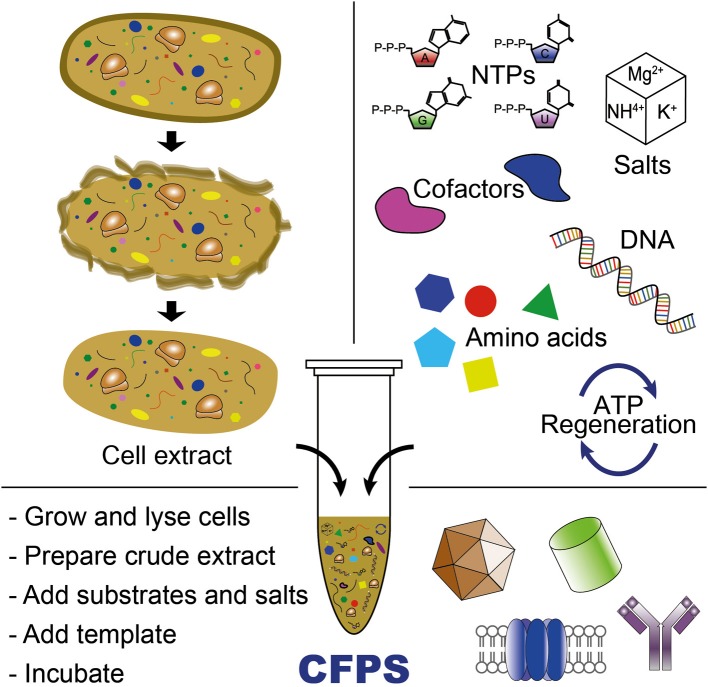
CFPS is the synthesis of proteins in vitro without using intact, living cells (Jewett et al., 2008; Caschera and Noireaux, 2014). Over the last 50 years, CFPS systems have significantly advanced our ability to understand, exploit, and expand the capabilities of biological systems (Carlson et al., 2012; Swartz, 2012; Murray and Baliga, 2013). As a complement to in vivo systems, CFPS systems offer some interesting benefits. First, the open environment of the reaction allows the user to directly influence the biochemical systems of interest and as a result, new components can be added or synthesized and can be maintained at precise concentrations (Figure 2). For example, NSAAs that do not enter the cell can be utilized in CFPS. Second, cell-free systems are not constrained by cell-viability requirements, allowing protein synthesis to proceed with otherwise toxic reagents or protein products. Third, CFPS systems can use linear DNA fragments (e.g., PCR products) for a target gene expression, which avoids time-consuming gene cloning steps commonly required for in vivo protein synthesis. Finally, from a biomanufacturing perspective, cell-free systems separate catalyst synthesis (cell growth) from catalyst utilization (protein production) (Swartz, 2012). This concept represents a significant departure from cell-based processes that rely on microscopic cellular “reactors.”
Figure 2.
Cell-free protein synthesis system for producing proteins or (poly)peptide-based materials. CFPS requires cell extract, an energy regeneration system, and chemical substrates and salts (e.g., NTPs, amino acids, salts, and cofactors). Cell-free transcription and translation is initiated by adding DNA template (plasmid or PCR-amplified linear DNA templates) into the CFPS reaction.
Although CFPS technologies offer many exciting advantages, challenges remain that provide opportunity for improvement. For example, CFPS platforms still have few examples industrially. In addition, cell lysis procedures can be difficult to standardize, leading to different extract performance and limited reaction scales for academic research labs. Thus, while protein yields (mg/L) are often higher in CFPS, the total amount of protein purified from cells in research labs is typically more because the reaction scales are greater. Despite these challenges, the advantages of CFPS are stimulating new application areas. Dominant amongst these are high-throughput protein production (Calhoun and Swartz, 2005; Swartz, 2012; Catherine et al., 2013; Chappell et al., 2013; Murray and Baliga, 2013), clinical manufacture of protein therapeutics (Murray and Baliga, 2013), genetic circuit optimization (Shin and Noireaux, 2012), the construction of synthetic ribosomes (Jewett et al., 2013), and incorporation of NSAAs (Goerke and Swartz, 2009; Bundy and Swartz, 2010; Ugwumba et al., 2010; Mukai et al., 2011; Ugwumba et al., 2011; Loscha et al., 2012; Albayrak and Swartz, 2013a; Hong et al., 2014; Shrestha et al., 2014).
Crude extract-based CFPS for NSAA incorporation
Efforts to use crude extract-based CFPS for the production of proteins containing single and multiple NSAAs are rapidly increasing. Key advances have centered on optimizing the performance of OTSs, expressing the OTS components in the source strain to create one-pot reactions, and removing RF1 competition.
OTS optimization
The Swartz group has made marked contributions to CFPS development for high yielding NSAA incorporation (Goerke and Swartz, 2009; Bundy and Swartz, 2010). Showcasing the freedom of design in adjusting cell-free system components by direct addition to the reaction, their approach typically adds the NSAA and its purified o-aaRS directly to the reaction, while the o-tRNA is expressed during the cell growth prior to making the extract. As compared to in vivo systems, an advantage of this approach is that the toxicity associated with overexpressing the o-tRNA and o-aaRS is not observed. This is because the OTS elements are sequestered from each other until the protein synthesis reaction. Another advantage is that NSAAs with low solubility or poor transport characteristics can be used. For example, the tyrosine analog p-propargyloxy-L-phenylalanine (pPaF), which can be used in site-specific bioconjugation with the copper-catalyzed azide-alkyne cycloaddition, has low solubility. This is a known limitation in vivo. However, site-specific pPaF incorporation in the CFPS reaction was improved ~27-fold (as based on protein yield) for producing a modified protein when compared to previous in vivo approach (Bundy and Swartz, 2010).
Cell-free systems are not only useful for making protein product but also for assessing the catalytic efficiency of the OTSs. A growing number of studies, for example, have shown that o-aaRSs are poor catalysts, up to 1000 times worse than natural aminoacyl tRNA synthetases, mainly due to the fact that the evolution of the orthogonal pairs occurs under high concentrations of non-standard amino acids (Tanrikulu et al., 2009; Nehring et al., 2012; Umehara et al., 2012; Albayrak and Swartz, 2013b). Future efforts for improving site-specific NSAA incorporation will require the development of o-aaRSs with higher catalytic rates and stronger affinity for the o-tRNAs. One approach to achieve such desired properties is to find strategies to remove fitness and the health of the cell on evolutionary outcomes. Ellington's lab recently published such an approach, compartmentalized partnered replication (Ellefson et al., 2014), but there are other opportunities as well.
In the meantime, NSAA incorporation in cell-free systems is being improved by increasing the amount of o-tRNA and o-aaRS in the CFPS reaction. One approach to achieving increased o-tRNA levels was pioneered by Albayrak and Swartz (2013a) and validated by Hong et al. (2014). Namely, the o-tRNA is co-produced in the CFPS reaction as a transzyme construct. The transzyme construct is a DNA fragment containing hammer-head ribozyme sequence between T7-controlled promoter and o-tRNA sequences. Upon transcription, the hammer-head ribozyme cleaves 5′-end of tRNA liberating active tRNA into the reaction (Fechter et al., 1998) and thereby increased o-tRNA is supplied to the CFPS reaction. With the transzyme technology, up to 0.9–1.7 mg/mL of a modified protein containing NSAA was produced (Albayrak and Swartz, 2013a) and multiple site NSAA incorporation was improved (Hong et al., 2014). As another approach, there are efforts to co-express all the OTS components in the source strain. While there are potential concerns of expressing both the o-tRNA and the o-aaRS in the source strain prior to lysis, Bundy and colleagues recently showed that this was not only possible, but improved CFPS yields of a modified protein (Smith et al., 2014). As an alternative approach, natural amino acids have been depleted from crude extracts to allow for the incorporation of NSAA analogs (Singh-Blom et al., 2014).
Removing RF1 competition
NSAA incorporation using amber codon suppression is limited by RF1 competition (Lajoie et al., 2013). The presence of RF1 causes the production of truncated protein and low yields of protein product in the case of multiple identical site-specific NSAA incorporation (Park et al., 2011; Hong et al., 2014). Deletion of RF1 is lethal in native biological systems. However, this limitation was recently addressed by making a more promiscuous release factor 2 (Johnson et al., 2011, 2012), and genome engineering (Mukai et al., 2010; Heinemann et al., 2012; Ohtake et al., 2012). Most notably, the development of the first genomically recoded E. coli strain was completed; all 321 TAG stop codons were reassigned to synonymous TAA codons allowing the deletion of RF1 without observing growth defects (Lajoie et al., 2013).
With RF1-deficient E. coli strains at hand, efforts are underway to utilize these strains in vivo for improved production of proteins with NSAAs, but also to develop RF1-deficient CFPS systems. In one example, human histone H4 protein was produced with site-specific incorporation of AcK at four amber sites by using a RF1-deficient cell extract (Mukai et al., 2011). In another case, the effect of RF1 deletion was systematically assessed for single and multiple site pPaF incorporation using cell extracts from genomically recoded E. coli with or without RF1 (Hong et al., 2014). The production of modified soluble superfolder green fluorescent protein (sfGFP) containing pPaF was 2.5-fold higher in the RF1-deficient cell extract compared to the RF1-present cell extract. The authors showed that the yield improvement was due to an increase in full-length modified sfGFP synthesis, observing a shift from 20% full-length product (with RF1) to 80% full-length product (without RF1). In a complementary approach, RF1-depleted cell extracts were constructed from selective removal of a RF1 variant tagged with chitin-binding domains (Loscha et al., 2012) or His-tag (Gerrits et al., 2007). Looking forward, we anticipate that RF1-deficient E. coli strains will become an important chassis for NSAA incorporation.
Reconstituted in vitro translation for NSAA incorporation
Although crude extract-based CFPS systems have shown tremendous growth, there are limitations to the number of open coding channels available because one must grow E. coli to obtain cellular lysate. To address this limitation, researchers have turned to purified translation systems, such as the PURE system (protein synthesis using purified recombinant elements) (Shimizu et al., 2001). Since the user defines all of the elements in the PURE system, single or multiple components (e.g., tRNA, aaRS) can be omitted, increased, or decreased according to the experimental purpose (Hirao et al., 2009). This enables highly efficient sense and non-sense suppression and provides unmatched flexibility for genetic code reprogramming to incorporate NSAAs (Shimizu et al., 2005). Efforts using purified translation for NSAA incorporation have mainly centered on the production, screening, and selection of peptidomimetic, or non-standard peptides (Josephson et al., 2005; Tan et al., 2005; Hartman et al., 2007; Passioura and Suga, 2013). As an exemplary illustration, peptidomimetic synthesis was achieved by adding pre-aminoacylated tRNA with NSAAs corresponding to sense codons in the reconstituted translation system lacking aaRS activities (Forster et al., 2003). In an alternative approach, Suga's group has leveraged the highly flexible tRNA acylation Flexizyme technology. Flexizyme is an artificial ribozyme that was developed to charge virtually any amino acid onto any tRNA in vitro, allowing the synthesis of proteins and short peptides containing multiple distinct NSAAs (Murakami et al., 2006; Ohuchi et al., 2007). A drug discovery pipeline has been enabled by combining a modified reconstituted translation system with Flexizyme technology (Goto et al., 2011) for the development of small peptides (Passioura and Suga, 2013), such as macrocyclic peptides (Hayashi et al., 2012; Morimoto et al., 2012). In yet a different approach, Szostak's work has demonstrated the ability to incorporate numerous amino acid analogs using the endogenous machinery. Strikingly, the natural aaRS machinery tolerates many kinds of side chain derivatives, such as α,α disubstituted, N-methyl and α-hydroxy derivatives (Hartman et al., 2007). Even D-amino acids have been shown to be compatible with polypeptide elongation (Fujino et al., 2013).
Although PURE translation is a powerful research tool, the cost of the PURE system is prohibitive for most commercial applications. For example, when compared to crude extract-based CFPS systems, which have been scaled to 100 L (Zawada et al., 2011), the PURE system costs ~1000 times more on a milligram protein produced/$ basis (Hong et al., 2014) and yields lower protein titers than the crude extract-based CFPS system (Lee et al., 2012; Hong et al., 2014). Hence, an important design decision for producing proteins with NSAAs using cell-free systems is choosing between a crude extract and a purified system.
Emerging applications
Marked advancements in productivity, improvements in OTS efficiency, and increases in the ability to incorporate multiple identical NSAAs (in crude extracts) and multiple distinct NSAAs (in the PURE system) are rapidly expanding the possible applications of CFPS systems. In this section, we highlight several emerging applications made possible by these advances. These include the production of protein-based materials and therapeutics.
Protein-based materials
NSAA incorporation is being applied to create new types of sequence-defined polymers for versatile applications in biomaterials synthesis. In an illustrative example, Albayrak and Swartz reported direct polymerization of proteins containing two or three copies of site-specifically incorporated NSAAs that allows copper-catalyzed azide-alkyne cycloaddition to form linear or branched protein polymers (Albayrak and Swartz, in press).
Therapeutics
NSAA incorporation is being applied to (i) clinical scale production of protein therapeutics and vaccines, (ii) discovery of novel biologics through ribosome display methods (Murray and Baliga, 2013), and (iii) structure/function studies to identify protein inhibitors. Swartz and colleagues, for example, have developed a novel pipeline for the production of decorated virus-like particles that could function as potential vaccines and imaging agents (Lu et al., 2013). In another example, Sutro Biopharma has demonstrated the synthesis of site-specific antibody drug conjugates (ADCs) (Zimmerman et al., 2014). Their ADCs, which were synthesized at ~250 μg/mL titers, proved potent in cell cytotoxicity assays. Rather than producing a therapeutic using CFPS, Ugwumba et al. utilized the NSAA 7-(hydroxy-coumarin-4-yl) ethylglycine to structurally probe a protein from the West Nile Virus to identify novel inhibitors (Ugwumba et al., 2011). Collectively, these recent reports highlight the utility of CFPS for producing novel vaccines and therapeutics, as well for serving as a rapid and attractive tool in drug discovery.
Conclusion and outlook
CFPS has emerged as a promising approach to enable site-specific incorporation of NSAAs into proteins and bio-based polymers. With the ability to select peptides and proteins for novel drugs in the PURE system and advent of scalable CFPS from crude extract systems, we anticipate significant growth in the field in years to come. Immediate challenges are (i) the evolution of more efficient OTSs (ii) new codons that can be assigned to NSAAs, and (iii) the development of genomically recoded organisms for preparing highly active cellular extracts. Addressing these challenges and continuing to lower costs will expand the scale and scope of cell-free biology, providing a transformative toolbox that enables new frontiers in synthetic biology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science Foundation (MCB-0943393), the Office of Naval Research (N00014-11-1-0363), the DARPA YFA Program (N66001-11-1-4137), the Army Research Office (W911NF- 11-1-044), the NSF Materials Network Grant (DMR - 1108350), the DARPA Living Foundries Program (N66001-12-C-4211), the David and Lucile Packard Foundation (2011-37152), and the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust for support. We thank Dr. Javin Oza for critical reading of the manuscript.
References
- Albayrak C., Swartz J. R. (2013a). Cell-free co-production of an orthogonal transfer RNA activates efficient site-specific non-natural amino acid incorporation. Nucleic Acids Res. 41, 5949–5963 10.1093/nar/gkt226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albayrak C., Swartz J. R. (2013b). Using E. coli-based cell-free protein synthesis to evaluate the kinetic performance of an orthogonal tRNA and aminoacyl-tRNA synthetase pair. Biochem. Biophys. Res. Commun. 431, 291–295 10.1016/j.bbrc.2012.12.108 [DOI] [PubMed] [Google Scholar]
- Albayrak C., Swartz J. R. (in press). Direct polymerization of proteins. ACS Synth. Biol. 10.1021/sb400116x [DOI] [PubMed] [Google Scholar]
- Axup J. Y., Bajjuri K. M., Ritland M., Hutchins B. M., Kim C. H., Kazane S. A., et al. (2012). Synthesis of site-specific antibody-drug conjugates using unnatural amino acids. Proc. Natl. Acad. Sci. U.S.A. 109, 16101–16106 10.1073/pnas.1211023109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco A., Townsley F. M., Greiss S., Lang K., Chin J. W. (2012). Expanding the genetic code of Drosophila melanogaster. Nat. Chem. Biol. 8, 748–750 10.1038/nchembio.1043 [DOI] [PubMed] [Google Scholar]
- Bröcker M. J., Ho J. M. L., Church G. M., Söll D., O'Donoghue P. (2014). Recoding the genetic code with selenocysteine. Angew. Chem. Int. Ed. Engl. 53, 319–323 10.1002/anie.201308584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundy B. C., Swartz J. R. (2010). Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein-protein click conjugation. Bioconjug. Chem. 21, 255–263 10.1021/bc9002844 [DOI] [PubMed] [Google Scholar]
- Calhoun K. A., Swartz J. R. (2005). Energizing cell-free protein synthesis with glucose metabolism. Biotechnol. Bioeng. 90, 606–613 10.1002/bit.20449 [DOI] [PubMed] [Google Scholar]
- Carlson E. D., Gan R., Hodgman C. E., Jewett M. C. (2012). Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194 10.1016/j.biotechadv.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caschera F., Noireaux V. (2014). Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription–translation system. Biochimie 99, 162–168 10.1016/j.biochi.2013.11.025 [DOI] [PubMed] [Google Scholar]
- Catherine C., Lee K.-H., Oh S.-J., Kim D.-M. (2013). Cell-free platforms for flexible expression and screening of enzymes. Biotechnol. Adv. 31, 797–803 10.1016/j.biotechadv.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Chappell J., Jensen K., Freemont P. S. (2013). Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 41, 3471–3481 10.1093/nar/gkt052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Sun S. B., Furman J. L., Xiao H., Schultz P. G. (2013). A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry 52, 1828–1837 10.1021/bi4000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Xiao H., Schultz P. G. (2012). Evolution of multiple, mutually orthogonal prolyl-tRNA synthetase/tRNA pairs for unnatural amino acid mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 109, 14841–14846 10.1073/pnas.1212454109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Daniel T., Buechler Y. J., Litzinger D. C., Maio Z., Putnam A.-M. H., et al. (2011). Optimized clinical performance of growth hormone with an expanded genetic code. Proc. Natl. Acad. Sci. U.S.A. 108, 9060–9065 10.1073/pnas.1100387108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y., Ohtsuki T., Shimizu Y., Ueda T., Sisido M. (2007). Elongation factor Tu mutants expand amino acid tolerance of protein biosynthesis system. J. Am. Chem. Soc. 129, 14458–14462 10.1021/ja075557u [DOI] [PubMed] [Google Scholar]
- Ellefson J. W., Meyer A. J., Hughes R. A., Cannon J. R., Brodbelt J. S., Ellington A. D. (2014). Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nat. Biotechnol. 32, 97–101 10.1038/nbt.2714 [DOI] [PubMed] [Google Scholar]
- Fechter P., Rudinger J., Giegé R., Théobald-Dietrich A. (1998). Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 436, 99–103 10.1016/S0014-5793(98)01096-5 [DOI] [PubMed] [Google Scholar]
- Forster A. C., Tan Z., Nalam M. N. L., Lin H., Qu H., Cornish V. W., et al. (2003). Programming peptidomimetic syntheses by translating genetic codes designed de novo. Proc. Natl. Acad. Sci. U.S.A. 100, 6353–6357 10.1073/pnas.1132122100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino T., Goto Y., Suga H., Murakami H. (2013). Reevaluation of the d-amino acid compatibility with the elongation event in translation. J. Am. Chem. Soc. 135, 1830–1837 10.1021/ja309570x [DOI] [PubMed] [Google Scholar]
- Gerrits M., Strey J., Claußnitzer I., Groll U. V., Schäfer F., Rimmele M., et al. (2007). “Cell-free synthesis of defined protein conjugates by site-directed cotranslational labeling,” in Cell-Free Protein Expression, eds Kudlicki W., Katzen F., Bennett R. (Austin, TX: Landes Bioscience; ), 166–180 [Google Scholar]
- Goerke A. R., Swartz J. R. (2009). High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol. Bioeng. 102, 400–416 10.1002/bit.22070 [DOI] [PubMed] [Google Scholar]
- Goto Y., Katoh T., Suga H. (2011). Flexizymes for genetic code reprogramming. Nat. Protoc. 6, 779–790 10.1038/nprot.2011.331 [DOI] [PubMed] [Google Scholar]
- Hartman M. C. T., Josephson K., Lin C.-W., Szostak J. W. (2007). An expanded set of amino acid analogs for the ribosomal translation of unnatural peptides. PLoS ONE 2:e972 10.1371/journal.pone.0000972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Morimoto J., Suga H. (2012). In vitro selection of anti-Akt2 thioether-macrocyclic peptides leading to isoform-selective inhibitors. ACS Chem. Biol. 7, 607–613 10.1021/cb200388k [DOI] [PubMed] [Google Scholar]
- Heinemann I. U., Rovner A. J., Aerni H. R., Rogulina S., Cheng L., Olds W., et al. (2012). Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 586, 3716–3722 10.1016/j.febslet.2012.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao I., Kanamori T., Ueda T. (2009). “Cell-free synthesis of proteins with unnatural amino acids. The PURE system and expansion of the genetic code,” in Protein Engineering, eds Köhrer C., Rajbhandary U. (Heidelberg: Springer Berlin; ), 271–290 [Google Scholar]
- Hoesl M. G., Budisa N. (2012). Recent advances in genetic code engineering in Escherichia coli. Curr. Opin. Biotechnol. 23, 751–757 10.1016/j.copbio.2011.12.027 [DOI] [PubMed] [Google Scholar]
- Hong S. H., Ntai I., Haimovich A. D., Kelleher N. L., Isaacs F. J., Jewett M. C. (2014). Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth. Biol. [Epub ahead of print]. 10.1021/sb400140t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R. A., Ellington A. D. (2010). Rational design of an orthogonal tryptophanyl nonsense suppressor tRNA. Nucleic Acids Res. 38, 6813–6830 10.1093/nar/gkq521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett M. C., Calhoun K. A., Voloshin A., Wuu J. J., Swartz J. R. (2008). An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 4, 220 10.1038/msb.2008.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett M. C., Fritz B. R., Timmerman L. E., Church G. M. (2013). In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 9, 678 10.1038/msb.2013.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. B. F., Wang C., Xu J., Schultz M. D., Schmitz R. J., Ecker J. R., et al. (2012). Release factor one is nonessential in Escherichia coli. ACS Chem. Biol. 7, 1337–1344 10.1021/cb300229q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. B. F., Xu J., Shen Z., Takimoto J. K., Schultz M. D., Schmitz R. J., et al. (2011). RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 7, 779–786 10.1038/nchembio.657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson K., Hartman M. C. T., Szostak J. W. (2005). Ribosomal synthesis of unnatural peptides. J. Am. Chem. Soc. 127, 11727–11735 10.1021/ja0515809 [DOI] [PubMed] [Google Scholar]
- Kim C. H., Axup J. Y., Schultz P. G. (2013). Protein conjugation with genetically encoded unnatural amino acids. Curr. Opin. Chem. Biol. 17, 412–419 10.1016/j.cbpa.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. -h., Llopis P. M., Heinritz J., Jacobs-Wagner C., Söll D. (2013). Suppression of amber codons in Caulobacter crescentus by the orthogonal Escherichia coli histidyl-tRNA synthetase/tRNAHis Pair. PLoS ONE 8:e83630 10.1371/journal.pone.0083630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M. J., Rovner A. J., Goodman D. B., Aerni H.-R., Haimovich A. D., Kuznetsov G., et al. (2013). Genomically recoded organisms expand biological functions. Science 342, 357–360 10.1126/science.1241459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. B., Kim H.-C., Kim D.-M., Kang T. J., Suga H. (2012). Comparative evaluation of two cell-free protein synthesis systems derived from Escherichia coli for genetic code reprogramming. J. Biotechnol. 164, 330–335 10.1016/j.jbiotec.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Lee S., Oh S., Yang A., Kim J., Söll D., Lee D., et al. (2013). A facile strategy for selective incorporation of phosphoserine into histones. Angew. Chem. Int. Ed. Engl. 52, 5771–5775 10.1002/anie.201300531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C., Schultz P. G. (2010). Adding new chemistries to the genetic code. Annu. Rev. Biochem. 79, 413–444 10.1146/annurev.biochem.052308.105824 [DOI] [PubMed] [Google Scholar]
- Liu D. R., Magliery T. J., Pastrnak M., Schultz P. G. (1997). Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 94, 10092–10097 10.1073/pnas.94.19.10092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscha K. V., Herlt A. J., Qi R., Huber T., Ozawa K., Otting G. (2012). Multiple-site labeling of proteins with unnatural amino acids. Angew. Chem. Int. Ed. Engl. 51, 2243–2246 10.1002/anie.201108275 [DOI] [PubMed] [Google Scholar]
- Lu Y., Welsh J. P., Chan W., Swartz J. R. (2013). Escherichia coli-based cell free production of flagellin and ordered flagellin display on virus-like particles. Biotechnol. Bioeng. 110, 2073–2085 10.1002/bit.24903 [DOI] [PubMed] [Google Scholar]
- Ma Y., Biava H., Contestabile R., Budisa N., di Salvo M. L. (2014). Coupling bioorthogonal chemistries with artificial metabolism: intracellular biosynthesis of azidohomoalanine and its incorporation into recombinant proteins. Molecules 19, 1004–1022 10.3390/molecules19011004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto J., Hayashi Y., Suga H. (2012). Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew. Chem. Int. Ed. Engl. 51, 3423–2427 10.1002/anie.201108118 [DOI] [PubMed] [Google Scholar]
- Mukai T., Hayashi A., Iraha F., Sato A., Ohtake K., Yokoyama S., et al. (2010). Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 38, 8188–8195 10.1093/nar/gkq707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Yanagisawa T., Ohtake K., Wakamori M., Adachi J., Hino N., et al. (2011). Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem. Biophys. Res. Commun. 411, 757–761 10.1016/j.bbrc.2011.07.020 [DOI] [PubMed] [Google Scholar]
- Murakami H., Ohta A., Ashigai H., Suga H. (2006). A highly flexible tRNA acylation method for non-natural polypeptide synthesis. Nat. Methods 3, 357–359 10.1038/nmeth877 [DOI] [PubMed] [Google Scholar]
- Murray C. J., Baliga R. (2013). Cell-free translation of peptides and proteins: from high throughput screening to clinical production. Curr. Opin. Chem. Biol. 17, 420–426 10.1016/j.cbpa.2013.02.014 [DOI] [PubMed] [Google Scholar]
- Nehring S., Budisa N., Wiltschi B. (2012). Performance analysis of orthogonal pairs designed for an expanded eukaryotic genetic code. PLoS ONE 7:e31992 10.1371/journal.pone.0031992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H., Hancock S. M., Buning R., Routh A., Chapman L., Somers J., et al. (2009). A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell 36, 153–163 10.1016/j.molcel.2009.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J. W. (2010). Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 10.1038/nature08817 [DOI] [PubMed] [Google Scholar]
- Niu W., Schultz P. G., Guo J. (2013). An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem. Biol. 8, 1640–1645 10.1021/cb4001662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donoghue P., Ling J., Wang Y.-S., Söll D. (2013). Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 9, 594–598 10.1038/nchembio.1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake K., Sato A., Mukai T., Hino N., Yokoyama S., Sakamoto K. (2012). Efficient decoding of the UAG triplet as a full-fledged sense codon enhances the growth of a prfA-deficient strain of Escherichia coli. J. Bacteriol. 194, 2606–2613 10.1128/JB.00195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohuchi M., Murakami H., Suga H. (2007). The flexizyme system: a highly flexible tRNA aminoacylation tool for the translation apparatus. Curr. Opin. Chem. Biol. 11, 537–542 10.1016/j.cbpa.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Park H.-S., Hohn M. J., Umehara T., Guo L.-T., Osborne E. M., Benner J., et al. (2011). Expanding the genetic code of Escherichia coli with phosphoserine. Science 333, 1151–1154 10.1126/science.1207203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T., Suga H. (2013). Flexizyme-mediated genetic reprogramming as a tool for noncanonical peptide synthesis and drug discovery. Chem. Eur. J. 19, 6530–6536 10.1002/chem.201300247 [DOI] [PubMed] [Google Scholar]
- Polycarpo C. R., Herring S., Bérubé A., Wood J. L., Söll D., Ambrogelly A. (2006). Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 580, 6695–6700 10.1016/j.febslet.2006.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 10.1038/90802 [DOI] [PubMed] [Google Scholar]
- Shimizu Y., Kanamori T., Ueda T. (2005). Protein synthesis by pure translation systems. Methods 36, 299–304 10.1016/j.ymeth.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Shin J., Noireaux V. (2012). An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 1, 29–41 10.1021/sb200016s [DOI] [PubMed] [Google Scholar]
- Shrestha P., Smith M. T., Bundy B. C. (2014). Cell-free unnatural amino acid incorporation with alternative energy systems and linear expression templates. New Biotechnol. 31, 28–34 10.1016/j.nbt.2013.09.002 [DOI] [PubMed] [Google Scholar]
- Singh-Blom A., Hughes R. A., Ellington A. D. (2014). An amino acid depleted cell-free protein synthesis system for the incorporation of non-canonical amino acid analogs into proteins. J. Biotechnol. 178, 12–22 10.1016/j.jbiotec.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Smith M. T., Hawes A. K., Shrestha P., Rainsdon J. M., Wu J. C., Bundy B. C. (2014). Alternative fermentation conditions for improved Escherichia coli-based cell-free protein synthesis for proteins requiring supplemental components for proper synthesis. Process Biochem. 49, 217–222 10.1016/j.procbio.2013.10.012 [DOI] [Google Scholar]
- Smith M. T., Wu J. C., Varner C. T., Bundy B. C. (2013). Enhanced protein stability through minimally invasive, direct, covalent, and site-specific immobilization. Biotechnol. Prog. 29, 247–254 10.1002/btpr.1671 [DOI] [PubMed] [Google Scholar]
- Swartz J. R. (2012). Transforming biochemical engineering with cell-free biology. AIChE J. 58, 5–13 10.1002/aic.13701 [DOI] [Google Scholar]
- Tan Z., Blacklow S. C., Cornish V. W., Forster A. C. (2005). De novo genetic codes and pure translation display. Methods 36, 279–290 10.1016/j.ymeth.2005.04.011 [DOI] [PubMed] [Google Scholar]
- Tanrikulu I. C., Schmitt E., Mechulam Y., Goddard W. A., Tirrell D. A. (2009). Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc. Natl. Acad. Sci. U.S.A. 106, 15285–15290 10.1073/pnas.0905735106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugwumba I. N., Ozawa K., de la Cruz L., Xu Z.-Q., Herlt A. J., Hadler K. S., et al. (2011). Using a genetically encoded fluorescent amino acid as a site-specific probe to detect binding of low-molecular-weight compounds. Assay Drug Dev. Technol. 9, 50–57 10.1089/adt.2010.0306 [DOI] [PubMed] [Google Scholar]
- Ugwumba I. N., Ozawa K., Xu Z.-Q., Ely F., Foo J.-L., Herlt A. J., et al. (2010). Improving a natural enzyme activity through incorporation of unnatural amino acids. J. Am. Chem. Soc. 133, 326–333 10.1021/ja106416g [DOI] [PubMed] [Google Scholar]
- Umehara T., Kim J., Lee S., Guo L.-T., Söll D., Park H.-S. (2012). N-Acetyl lysyl-tRNA synthetases evolved by a CcdB-based selection possess N-acetyl lysine specificity in vitro and in vivo. FEBS Lett. 586, 729–733 10.1016/j.febslet.2012.01.029 [DOI] [PubMed] [Google Scholar]
- Wan W., Huang Y., Wang Z., Russell W. K., Pai P.-J., Russell D. H., et al. (2010). A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew. Chem. Int. Ed. Engl. 49, 3211–3214 10.1002/anie.201000465 [DOI] [PubMed] [Google Scholar]
- Wang F., Niu W., Guo J., Schultz P. G. (2012a). Unnatural amino acid mutagenesis of fluorescent proteins. Angew. Chem. Int. Ed. Engl. 51, 10132–10135 10.1002/anie.201204668 [DOI] [PubMed] [Google Scholar]
- Wang K., Neumann H., Peak-Chew S. Y., Chin J. W. (2007). Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777 10.1038/nbt1314 [DOI] [PubMed] [Google Scholar]
- Wang L., Brock A., Herberich B., Schultz P. G. (2001). Expanding the genetic code of Escherichia coli. Science 292, 498–500 10.1126/science.1060077 [DOI] [PubMed] [Google Scholar]
- Wang Y.-S., Fang X., Chen H.-Y., Wu B., Wang Z. U., Hilty C., et al. (2012b). Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant. ACS Chem. Biol. 8, 405–415 10.1021/cb300512r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-S., Fang X., Wallace A. L., Wu B., Liu W. R. (2012c). A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J. Am. Chem. Soc. 134, 2950–2953 10.1021/ja211972x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D. D., Young T. S., Jahnz M., Ahmad I., Spraggon G., Schultz P. G. (2011). An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 50, 1894–1900 10.1021/bi101929e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T. S., Ahmad I., Yin J. A., Schultz P. G. (2010). An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 395, 361–374 10.1016/j.jmb.2009.10.030 [DOI] [PubMed] [Google Scholar]
- Zawada J. F., Yin G., Steiner A. R., Yang J., Naresh A., Roy S. M., et al. (2011). Microscale to manufacturing scale-up of cell-free cytokine production—a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 108, 1570–1578 10.1002/bit.23103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E. S., Heibeck T. H., Gill A., Li X., Murray C. J., Madlansacay M. R., et al. (2014). Production of site-specific antibody–drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug. Chem. 25, 351–361 10.1021/bc400490z [DOI] [PubMed] [Google Scholar]