Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is a common nephropathy caused by mutations in either PKD1 or PKD2. Mutations in PKD1 account for ~85% of cases and cause more severe disease than mutations in PKD2. Diagnosis of ADPKD before the onset of symptoms is usually performed using renal imaging by either ultrasonography, CT or MRI. In general, these modalities are reliable for the diagnosis of ADPKD in older individuals. However, molecular testing can be valuable when a definite diagnosis is required in young individuals, in individuals with a negative family history of ADPKD, and to facilitate preimplantation genetic diagnosis. Although linkage-based diagnostic approaches are feasible in large families, direct mutation screening is generally more applicable. As ADPKD displays a high level of allelic heterogeneity, complete screening of both genes is required. Consequently, such screening approaches are expensive. Screening of individuals with ADPKD detects mutations in up to 91% of cases. However, only ~65% of patients have definite mutations with ~26% having nondefinite changes that require further evaluation. Collation of known variants in the ADPKD mutation database and systematic scoring of nondefinite variants is increasing the diagnostic value of molecular screening. Genic information can be of prognostic value and recent investigation of hypomorphic PKD1 alleles suggests that allelic information may also be valuable in some atypical cases. In the future, when effective therapies are developed for ADPKD, molecular testing may become increasingly widespread. Rapid developments in DNA sequencing may also revolutionize testing.
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is characterized by the progressive development of cysts and is an important cause of end-stage renal disease (ESRD).1,2 Individuals at risk of ADPKD due to a positive family history are normally diagnosed by imaging of the kidneys by ultrasonography, CT or MRI. Identification of the genes that cause ADPKD, PKD1 and PKD2, in addition to extensive mutation screening in affected families, has provided the framework for the clinical molecular tests that are currently available. Although not necessary in every case, mutation-based diag nostics are increasingly used in individuals who are at risk of ADPKD and are especially helpful in cases where imaging studies are equivocal and a definite diagnosis is required. In this review, we discuss the role of molecular diagnostics in ADPKD, including when it should be considered and the value of the results. The advantages of molecular diagnostics are balanced against the potential pitfalls of this approach. Recently identified genetic complexities in ADPKD will also be discussed and the potential for obtaining prognostic data considered. Finally, the future role of molecular diagnostics in ADPKD will be evaluated.
ADPKD: clinical and genetic details
ADPKD is one of the most common human monogenic diseases, with an incidence between one in 400 and one in 1,000, and accounts for 4–5% of patients who require renal transplantation or dialysis.3–5 The disease is characterized by the progressive development and expansion of cysts in the kidney. Severe polycystic liver disease and an increased risk of intracranial aneurysms are other causes of morbidity and mortality in a minority of cases.1 Familial clustering of intracranial aneurysms is found in patients with ADPKD, suggesting that genetic factors may influence the development of this entity.6,7 The management of intracranial aneurysms in patients with ADPKD is beyond the scope of this Review and is discussed in detail elsewhere.8 The progressive nature of the kidney disease in ADPKD and the variability in kidney and cyst volumes between affected individuals was demonstrated by the CRISP study, which performed magnetic resonance analyses over a 3-year period and revealed an average annual increase in kidney volume of 5.27%.9 The full range of disease severity extends from patients with adequate renal function in their tenth decade to in utero diagnosis with greatly enlarged and echogenic kidneys.10,11 Very early-onset cases are rare, but interestingly an increased risk of recurrence exists in siblings.12
ADPKD is genetically heterogeneous, being caused by mutations in either PKD1 (located on chromosome 16p13.3), which account for ~85% of cases in clinically defined populations, or mutations in PKD2 (located on chromosome 4p21), which account for ~15% of cases.13–15 The particular gene that is involved is a major determinant of disease severity—the average age of ESRD onset is 54.3 years for individuals with mutations in PKD1 and 74 years for those with mutations in PKD2.10 Age and gender-corrected renal volume is two-thirds larger in individuals with mutations in PKD1 than in those with PKD2 mutations.16 Consistent with this difference in disease severity, PKD2 mutations are more common in elderly patients with ADPKD,17 and studies of populations that included the whole spectrum of disease indicate that mutations in PKD2 may be more prevalent in the total population than is suggested by data from renal clinics.18–20 However, even within a genic group (that is, a group associated with a mutation to just one of the genes) or within affected families, considerable variability exists in the severity of renal and extrarenal disease.21–23
The ADPKD genes and proteins
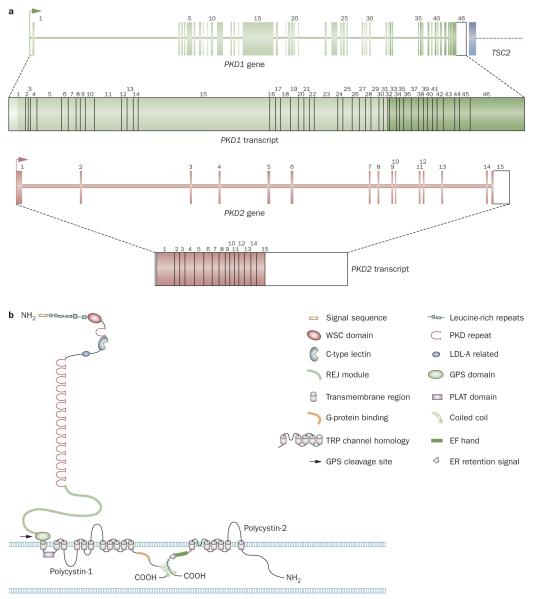
PKD1 contains 46 exons, has a genomic region spanning 50 kb, and is compact, with a large, ~14 kb, messenger (m)RNA containing a 12,909 bp coding sequence (Figure 1).24,25 Analysis of PKD1 is unusually complex because exons 1–33 are encoded by a genomic region that has undergone an intrachromosomal duplication with six copies of this region present as pseudogenes (PKD1P1–P6) located ~13–16 Mb proximal to PKD1 on the short arm of chromosome 16.13,26,27 These pseudogenes are generally expressed but have early stop codons so probably do not generate large protein products. Compared with PKD1, the PKD1 pseudogenes have deletions and other rearrangements but are 98–99% identical to PKD1 in homologous areas. Therefore, anchored and locus-specific long-range PCR protocols that take advantage of the rare sequence differences between PKD1 and the pseudogenes have been developed to specifi cally amplify PKD1 for screening for mutations.15,28,29 PKD2 contains 15 exons, has a genomic span of 68 kb, and a coding sequence of 2,904 bp. PKD2 is a less compact gene than PKD1.14,30
Figure 1.
Gene structure of PKD1 and PKD2 and protein structure of polycystin-1 and polycystin-2. a ∣ Gene and messenger RNA structure of PKD1 and PKD2. The 5′ end of PKD1 from exon 1 to 33 lies in a duplicated genomic region (light green) and the 3′ end of the gene is immediately adjacent (within ~60 bp) to the 3′ end of the tuberous sclerosis gene, TSC2. b ∣ Protein structures of polycystin-1 and polycystin-2. Details of the known domain structures of the polycystins are shown in the figure. Abbreviations: ER, endoplasmic reticulum; GPS, GPCR proteolytic site; LDL, low-density lipoprotein; PKD, polycystic kidney disease; TRP, transient receptor potential.
PKD1 encodes a large, multidomain integral membrane protein, polycystin-1, whereas PKD2 encodes a calcium ion channel of the transient receptor potential (TRP) family, polycystin-2 (also known as TRPP2) (Figure 1).14,24,31 PKD is a ciliopathy and a large body of evidence implicates defective primary cilia in cyst development.32 The polycystin proteins, which are thought to form a complex, have been localized to the cilium.33,34 Roles in the sensing of flow,35 pressure,36 modulation of centrosome duplication and/or cell cycle regulation,37,38 and involvement in planar cell polarity39 are potential mechanisms by which the polycystin proteins may contribute to cyst formation, but the precise mechanism of cystogenesis in ADPKD is unknown.
Diagnosing ADPKD: renal imaging
Since ADPKD is a dominant disease with a high degree of penetrance, 50% of siblings and children of an affected individual will, on average, also develop the disease. As no proven therapy for ADPKD is available, asymptomatic family members who are at-risk of disease are typically not tested until they are 18 years of age, so as not to negate the autonomy of childhood. Furthermore, despite the fact that the Genetic Information Nondiscrimination Act (GINA) legislation was passed in the US in 2008 to prevent the use of genetic information in employment and insurance decisions, discrimination in terms of an individual’s life, disability and long-term care insurability can still result from a positive test result.40
Renal imaging by ultrasonography, CT or MRI is the most common means by which family members who are at risk of ADPKD are diagnosed. In ~10% of affected families no prior history of polycystic kidney disease (PKD) exists.41,42 In these instances, asymptomatic individuals who have ADPKD are often diagnosed with the disease incidentally, during abdominal imaging for other purposes.
Healthy individuals often develop a small number of renal cysts with age (known as simple cysts); therefore, specific criteria with regard to the number of cysts at an individual’s age have been developed for the diagnosis of ADPKD by ultrasonography. These criteria have recently been revised.43,44 Although these criteria have a high positive predictive value, they are not very effective at excluding a diagnosis of ADPKD in young individuals and in those with mild disease caused by mutations in PKD2. CT and MRI are more expensive than ultrasonography but have greater sensitivity so that smaller cysts (~2 mm compared with ~10 mm for ultrasonography) can be detected.45 Consequently, a greater number of cysts are detected by use of these techniques in both affected and normal individuals. Although some data about the frequency of simple cysts detected by CT and MRI are available,45,46 specific criteria to differentiate individuals with ADPKD from those with simple cysts have not been developed.
Diagnosing ADPKD: molecular testing
Molecular diagnostics: who to test
The identification and characterization of PKD1 and PKD2 provided an opportunity for mutation-based molecular diagnostics to be used for ADPKD. These diagnostic tools are now available for clinical use in the US and elsewhere in the world. The development of sensitive molecular methods for the diagnosis of ADPKD can be critical to identify potential living related donors in families affected by ADPKD, especially when imaging tests are equivocal. In 2009 an algorithm was proposed to determine when molecular testing may be useful for evaluating potential kidney donors in families affected by ADPKD.47 However, these criteria did not differentiate between analyses made by ultrasonography and those made by the more sensitive CT and MRI methods, which could result in ~50% of potential donors requiring genetic testing at a considerable cost.40,48 In our opinion, this algorithm also seems too conservative; for instance, it recommends that all potential donors from a family affected by ADPKD who are less than 40 years of age and are found to have three cysts or fewer by ultrasonography, CT, or MRI, should undergo genetic testing. Whether potential donors in their thirties who have no cysts detected by sensitive CT or MR also require genetic testing, especially if the disease in their family is usually severe (which suggests that they carry a PKD1 mutation) is questionable. Nevertheless, there is a place for molecular testing; for example, for the potential donor in whom a single cyst or a small number of cysts (below the diagnostic threshold) are detected, or for young individuals <25 years of age in whom no cysts are detected by ultrasonography, CT or MRI, especially if the disease in the family is typically mild (see Box 1 for other examples).
Box 1. Situations in which to consider molecular testing in ADPKD.
Potential living, related donor in an affected family with equivocal imaging data. Patient usually has one or a small number of renal cysts detected but less than the number required for an ADPKD diagnosis by published criteria47
Factors to consider:
▪ MRI and CT are more sensitive than ultrasonography and can detect smaller cysts (≤0.2 cm compared to ~1 cm)45,46
▪ Severity of disease in family (PKD1 vs PKD2)
Examples:
▪ In a 20-year-old with no cysts detected by CT or MRI and with PKD1-like disease in the family: ADPKD is unlikely and molecular testing is not necessary
▪ In a 20-year-old with no cysts detected by ultrasonography and with PKD2-like disease in the family: ADPKD is still a possibility and molecular testing is appropriate
▪ In a 30-year-old with one cyst detected by CT or MRI and PKD1-like disease in family: ADPKD is not likely but molecular testing may provide reassurance
▪ In a 40-year-old with three cysts detected by CT or MRI and PKD2-like disease in family: ADPKD is a possibility and molecular testing is appropriate
▪ In a 40-year-old with no renal cysts detected by CT or MRI and PKD2-like disease in family: ADPKD is unlikely and molecular testing is not necessary
▪ In a 60-year-old with two renal cysts detected by CT or MRI and PKD1-like disease in family: ADPKD is unlikely and molecular testing is not necessary
Individuals with a negative family history of ADPKD
Factors that influence whether a molecular test would be particularly valuable:
▪ An atypical radiological presentation: e.g. a patient with much more severe disease in one kidney than the other, or a patient manifesting with multiple very small cysts
▪ In patients with mild renal disease
▪ In patients with extrarenal manifestations atypical of ADPKD
▪ To provide prognostic information where no guidance from other family members is available
In families affected by early-onset PKD
▪ In a family with otherwise typical ADPKD to identify variants that may be associated with severe disease
▪ In individuals with a negative family history of ADPKD, but who have negative PKHD1 mutation test results and/or who have ADPKD radiological features
▪ For PGD in families with a history of early-onset disease
In patients requesting a definite diagnosis
▪ For prognostic value
▪ To aid informed family planning choices
Abbreviations: ADPKD, autosomal dominant polycystic kidney disease; PGD, preimplantation genetic diagnostics; PKD, polycystic kidney disease.
Another situation in which molecular testing can be valuable is for obtaining a definite diagnosis for individuals with a negative family history of ADPKD, because of potential phenotypic overlap of ADPKD with several other disorders.1 Data from the past couple of years suggest that bilateral renal cysts, similar to those found in individuals with ADPKD, are a common finding in patients with maturity-onset diabetes of the young (MODY) type 5 (caused by a mutation in HNF1β), even in the absence of features that are typically associated with this disorder, such as genital malformations and early-onset diabetes.49–51 In individuals with severe polycystic liver disease and a few renal cysts, phenotypic overlap exists with autosomal dominant polycystic liver disease, which is caused by mutations in PRKCSH or SEC63.52–54 Autosomal recessive polycystic kidney disease (ARPKD), which is associated with mutations in PKHD1, can also sometimes manifest late in life with an ADPKD-like renal phenotype.55 In addition, misdiagnosis can occur in the assessment of patients with other syndromic forms of PKD, including diseases such as tuberous sclerosis.1 Differentiation of mild ADPKD from simple cysts, and early-onset ADPKD from ARPKD, plus the identification of risk factors for severe disease (as is discussed later), are other instances in which molecular testing may be useful (Box 1). Molecular testing of children less than 18 years of age should in general be avoided, even if requested by the parents, unless the child or a young sibling has symptomatic disease, indicating an increased risk of early-onset disease in the family.
Molecular testing can be performed by genetic linkage analysis with markers that flank the affected genes or by direct mutation analysis of the genes themselves.
Linkage analysis
Linkage analysis for the diagnosis of ADPKD has been possible since PKD1 and PKD2 were mapped to chromosomal regions in the early 1990s. Multiple flanking and intragenic markers are now available for the analysis of these two genes.56,57 However, because of the genetic heterogeneity of ADPKD, characterization of several affected members of a family is generally required to identify which gene is affected, and so only a minority of families are informative for linkage-based diag nostics. Other factors, such as genetic heterogeneity, the presence of de novo mutations, mosaicism, and other facets to genetic complexity (discussed in further detail later) mean that linkage analyses need to be applied with caution and are best used in families in whom the mutation has been characterized.57 However, situations in which linkage analysis is particularly helpful do exist, such as for screening embryos before implantation (preimplantation genetic diagnostics), where the typing of several markers is required to ensure against problems associated with screening a very small amount of DNA, such as allele dropout.58
Screening for mutations
Although various indirect methods have been used for screening for mutations associated with ADPKD,29,59 direct sequencing of exonic and flanking intronic regions from genomic DNA is the method now generally used for diagnostic purposes.15,60 Numerous studies have demon strated a high degree of allelic heterogeneity in PKD1 and PKD2 with no single mutation found to account for more than 2% of affected families; in the majority of families, ADPKD is caused by a unique mutation. Consequently, diagnostic screening of a new family requires sequencing of all PKD1 and PKD2 exons, which is relatively expensive.
Mutations that are known to cause ADPKD have now been collated in a central database.61 These mutations are found throughout the length of both PKD1 and PKD2 with no obvious clustering, although the distribution of the mutations may not be entirely uniform.15,62 A wide range of different mutation types cause ADPKD; any mutation that inactivates one of the two gene alleles is probably pathogenic. The types of mutation that are known to cause ADPKD are summarized in Box 2. Although truncating mutations can generally be considered pathogenic, further evaluation of in-frame changes is required (Box 3). The evaluation of in-frame variants is particularly necessary for PKD1 because even unaffected individuals harbor an average of ~10 neutral variants.12 Publications over the past few years have used a range of bioinformatic tools to evaluate the likely pathogenicity of variants15,59,60,63 and similar methods have been used to score all nondefinite mutations in the ADPKD mutation database.61 The evaluation criteria used to assess the pathogenicity of the different mutations is described in Box 3.
Box 2. Mutations in PKD1 and PKD2 that cause ADPKD.
Definite mutations: many types of mutations are predicted to truncate the protein and are generally considered pathogenic
▪ Nonsense: a substitution that results in a stop codon
▪ Frameshift: a deletion or insertion that changes the reading frame of the mRNA
▪ Typical splicing: a variant in the canonical dinucleotides flanking the exon that alters the way an intron is excised and the exons are assembled into a mature mRNA
▪ Large rearrangements: deletion or insertion of a considerable region of the gene, usually encompassing more than one exon
Nondefinite mutations: mutations that do not alter the reading frame of the gene or are of uncertain consequence. Their pathogenicity is less clear and requires further analysis
▪ Missense: substitution of one amino acid residue for another
▪ In-frame deletions and insertions: removal or gain of a nucleotide number divisible by three so that the reading frame is maintained
▪ Atypical splicing: a variant that may alter splicing but is not in the canonical dinucleotides that flank the exon
De novo mutation: a new mutation that has occurred in the family
Mosaicism: a mutation that occurs in the embryo at an early stage but does not affect all cells (the affected patient is a chimera). Mosaicism can also affect gonadal cells (the patient generates germ cells with and without the mutation)
Hypomorphic allele (also termed an incompletely penetrant allele): a mutation that alone does not result in the full ADPKD phenotype because some partially functional protein is still generated. Patients with two such mutations inherited from different parents (in trans) are viable and develop typical to severe PKD. A hypomorphic allele in trans with an inactivating allele may cause early-onset ADPKD
Abbreviations: ADPKD, autosomal dominant polycystic kidney disease; mRNA, messenger RNA, PKD, polycystic kidney disease.
Box 3. Evaluation of nondefinite mutations.
Information about the position and the nature of a substitution (or other nondefinite variant) and the context in which it is found can be used to score its likely pathogenicity
Substitution score
Assesses both the chemical difference caused by the substitution (the Grantham difference [GD]; based on the Grantham matrix)95 and the level of conservation of the residue in a multisequence alignment of orthologous proteins (the Grantham variation [GV]):
▪ Programs such as PolyPhen,96 SIFT,97 and Align-GVGD98 score the significance of substitutions by use of various algorithms based on GV and GD values
▪ Use of an accurate and well-validated multisequence alignment that contains orthologs but not other homologous proteins increases the reliability of these predictions
Determines the importance (conservation) of the substituted residue within defined domains (Figure 1):
▪ Especially valuable if a three-dimensional structure of the domain has been determined
Atypical splicing variants are ideally evaluated by reverse-transcriptase PCR, but changes can be scored for their likely effect on splicing by use of programs such as Human Splicing Finder99
Context score
Assesses whether the variant has previously been described as a pathogenic mutation or as a neutral polymorphism
Assesses whether the mutation is the only variant that is likely to be pathogenic found on complete screening of PKD1 and PKD2
Assesses whether the variant segregates with the disease:
▪ Lack of segregation in two affected members of a family demonstrates that a variant is not a fully penetrant mutation
▪ In a family known to have mutations in either PKD1 or PKD2 by linkage analysis, segregation of a variant in two affected individuals from different generations supports the pathogenicity of the variant and shows that it is at a minimum, a marker of the disease allele
▪ In a family for which the mutated gene is unknown, appropriate segregation of a variant in six or more family members (or fewer if the members are distantly related) with a known disease status supports the pathogenicity of the variant and shows that at a minimum, it is a marker of the disease allele
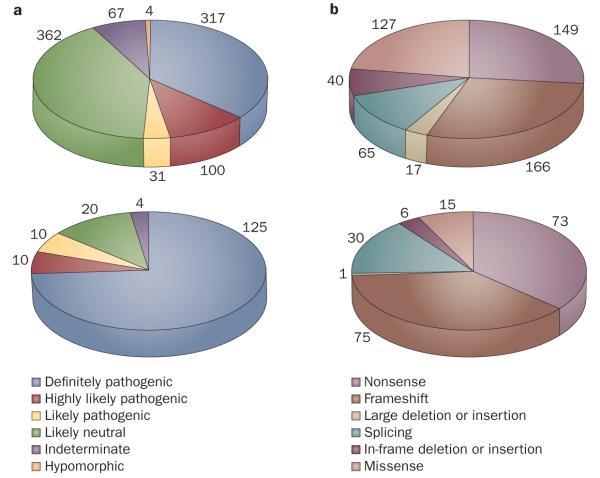
Collection and collation of all published mutations and other variants in the ADPKD mutation database enables the array of ADPKD mutations to be analyzed.61 436 different pathogenic PKD1 mutations exist, which are known to cause ADPKD in 564 families, and 115 different PKD2 mutations have been found in 200 different pedigrees. Figure 2 shows a breakdown of the types of mutations found in each gene (see also Box 2). Comprehensive screens of well-characterized ADPKD populations have revealed definite (truncating) base-pair mutations in up to 61% of affected families. Another ~26% of families have in-frame changes that are scored as pathogenic.15,59,60 Screening for larger rearrangements, such as multi-exon deletions or duplications using methods such as multiplex ligation-dependent probe amplification, detects mutations in a further ~4% of families.64 This group includes PKD1 deletions that can extend 100 kb 5′ to the gene, removing up to 10 additional genes, without apparent additional phenotypic consequences. Deletion mutations that extend 3′ to PKD1 disrupt the TSC2 gene (Figure 1), and result in the PKD1/TSC2 contiguous gene deletion syndrome.64–66 Individuals with this syndrome have a distinctive phenotype consisting of variable signs of tuberous sclerosis, plus severe PKD that is usually evident during childhood.
Figure 2.
Summary of data from the ADPKD mutation database. a ∣ Classification and numbers of different variants found in PKD1 (top) and PKD2 (bottom) in the ADPKD mutation database. Changes that are probably pathogenic are classified into three groups depending on the likelihood that they are associated with disease. Many more neutral variants are found in PKD1 than in PKD2.
b ∣ Classification and numbers of families affected by changes in PKD1 and PKD2 that are probably pathogenic. A greater number of missense changes are found in PKD1 than in PKD2, and a higher proportion of splicing variants are found in PKD2.
Note that the numbers in panels a and b differ because panel a concerns the number of different variants and panel b concerns the number of affected families. Some mutations are found in more than one family.
In total, mutations are detected in up to 91% of patients with ADPKD—65% of patients have truncating mutations that can readily be used for diagnostics.15 Nondefinite mutations are found in 26% of patients and need careful consideration before use in the clinical setting. However, changes at highly conserved sites that have been described multiple times (for example, Glu2771Lys in PKD1 and Arg322Trp and Arg322Gln in PKD261) can increasingly be used diagnostically, especially if supported by segregation with the disease in a family (Box 2). Similarly, novel changes at highly conserved sites with high pathogenicity scores (Box 3), can also be used at a minimum as a marker of the mutant allele when supported by segregation data. As further data on ADPKD mutations are compiled in the ADPKD mutation database, the use of nondefinite mutations that are predicted to be pathogenic for diagnostic purposes, in the absence of linkage data, will probably become increasingly common.
Functional analysis of ADPKD mutations
Whether mutations are causative of a disease is ideally examined by functional assays. This approach has been successfully used in the study of mutations associated with ADPKD, especially those in PKD2, where, for instance, Asp511Val was shown by electrophysiological studies to prevent channel function.67 In addition, some missense changes in the REJ domain of PKD1 (Figure 1) have been shown to prevent cleavage of polycystin-1 at its GPS domain,68 while substitutions in the PKD repeats of PKD1 can alter its mechanical properties when stretched by atomic force microscopy.69–71 However, these functional tests usually require exogenous expression of the mutant protein in cells, are technically challenging, and have a complex functional readout that may in itself not completely predict the pathogenicity of the mutation. Although functional tests are ideal, at present it is not possible to test multiple variants in a diagnostic setting and it seems that improving bioinformatic prediction tools will probably have a greater influence than functional tests in diagnostics.
Complexities in molecular diagnostics
ADPKD in individuals with no detectable mutation
Analysis of molecular diagnostic tests results for ADPKD are further complicated by the ~9% of individuals for whom no mutation in either PKD1 or PKD2 is detected.15 Studies of such patients found that on average they have milder disease and are more likely than other patients to have a negative family history for ADPKD. Therefore, the inability to detect mutations in these individuals is probably not simply due to missed mutations in the complex PKD1 gene.41,72 Deep intronic changes that influence splicing, or gene promoter changes, which are not detected by the exon-based screening approaches, probably account for some of these cases.73 In addition, some variants that might be detected but score poorly from the substitution and contextual analysis (Box 3) may exert pathogenic effects by altering the protein structure in unexpected ways or by inducing subtle influences on splicing—for example, by changing a splice enhancer site.74 Additional genetic heterogeneity is also possible as several ADPKD families apparently unlinked to PKD1 and PKD2 have been described,75,76 although careful evaluation of at least one of these families questions the existence of an additional locus.77 ADPKD phenocopies resulting from mutations in other known cystogenes, such as HNF1β, PRKCSH, SEC63 or PKHD1, may also account for the disease in some individuals.
Mosaicism in ADPKD
Another complexity that adds to the difficulty in obtaining reliable diagnostics in ADPKD is mosaicism (Box 2), which has been described in two ADPKD families but is probably more prevalent in de novo cases.64,78 Mosaicism may explain the etiology of some affected families who have a very different expressivity of disease between generations. Determining whether mosaicism is present in a family is important because this feature radically alters the risk of disease in siblings of the individual with apparently de novo disease. Mosaicism can also confuse the findings from linkage analyses with affected and unaffected individuals having the same haplotype but differing in whether or not they have the mutation.78 Furthermore, as mosaicism can result in very different levels of the mutant allele in different tissues, the level of the mutant allele in leukocytes is a poor indicator of its frequency in the kidney and, hence, of disease severity.64
The role of hypomorphic alleles in ADPKD
In contrast to the evidence that genic effects have considerable impact on the severity of disease in ADPKD, the data supporting a role for allelic effects in disease severity are much less clear. No clear correlation between the disease phenotype and the type or position of mutation has been reported for PKD2.62 In PKD1, no correlation with mutation type and disease severity has been described but modest associations between mutation position and age at onset of ESRD and occurrence of intracranial aneurysms has been reported, although the mechanisms of these associations are unclear.7,79 These data are currently of insufficient predictive value to direct the testing of families at risk of aneurysmal rupture.
Despite findings that suggest that most missense changes completely inactivate the gene, data from a 2009 study suggest that some PKD1 alleles may be hypomorphic or incompletely penetrant.63 In fact, in ARPKD, clear evidence that some missense alleles are not fully penetrant exists. Individuals who have two truncating mutations in PKHD1 die soon after birth, whereas many individuals who have one or two missense mutations can live beyond childhood.80 Evidence of hypomorphic alleles is also found in mouse models of ADPKD. While homozygous truncating mutations of either Pkd1 or Pkd2 results in an embryonic lethal phenotype, mice that have down regulated levels of polycystin-1 or polycystin-2 (to 15–20% of normal levels) are viable as homozygotes but develop progressive cystic disease.81–83
Studies of human pedigrees with typical to severe PKD have revealed individuals who are homozygous or compound heterozygous for PKD1 variants that are probably hypomorphic mutations.63 Patients with one such allele often have just a few cysts, whereas if this allele is inherited in trans with an inactivating allele, early-onset PKD can result. Although such in trans inheritance of two PKD1 variants is probably not the only cause of early-onset ADPKD, this mode of inheritance seems to be important. As the penetrance of disease associated with a single hypomorphic allele is low, families segregating such alleles often seem to have either a negative history of disease, recessive inheritance, or marked heterogeneity in severity between generations. Hypomorphic alleles provide a new diagnostic challenge, but also present an opportunity as these alleles are potentially of prognostic importance. In a family with early-onset disease it may be possible to predict the likelihood of a sibling developing severe disease, facilitating preimplantation genetic diagnostics. Hypomorphic alleles may also more generally underlie some of the phenotypic variability seen in ‘typical’ families affected by ADPKD.84 However, the reliable differentiation of such changes from neutral or fully penetrant mutations is a particular challenge. At present, their identification relies on bioinformatic tools that highlight them as likely pathogenic alleles in situations in which the family data indicate that the mutation is not fully penetrant, but rather has a modifying role.
From family studies, 43–50% of the variability in the time to ESRD onset in patients with ADPKD is estimated to be caused by genetic modifier effects.85,86 Although some of these effects may be accounted for by the presence of the ‘normal’ ADPKD allele, many other loci probably also have an influence. Identifying these loci by use of genome-wide association studies and re-sequencing methods (discussed later) may identify further factors of prognostic significance in the near future.
Present status of molecular testing
Molecular testing for ADPKD is not recommended in patients in whom a firm positive or negative diagnosis can be obtained by imaging analysis alone, because molecular diagnostics is a relatively expensive approach and does not provide usable data in every case. However, once a mutation has been identified in a family, a presymptomatic diagnosis in other at-risk family members can be made relatively inexpensively by just screening for the known mutation. As described earlier, the value of molecular testing is especially high for patients with equivocal imaging results, especially in instances that involve potential living, related kidney donors (Box 1). A kidney from a potential donor can generally be used if the individual is negative for the mutation characterized in the family, even if they have one cyst or a small number of cysts.57 One caveat is the parent of a patient with an apparent de novo mutation, where low-level mosaicism in the parent is possible.64,78 Other situations in which a molecular diagnosis is helpful is to establish a firm diagnosis in patients who have a negative family history and in those with mild disease where other disorders may be phenotypically similar. Mosaicism or more complex inheritance may be revealed that influences the risk status of other family members. Although the family history of disease severity can to some extent suggest which gene is involved,18 molecular data are more reliable. Determining whether a family has mutations in PKD1 or PKD2 is of prognostic value, especially if no known family history of ADPKD exists. In families that have a member with early-onset disease, the detection of hypomorphic alleles may allow the identification of siblings who are at risk of early-onset disease and enable preimplantation genetic diagnostics to be performed.
Impediments to the further use of molecular testing at this time include the costs of the tests and our ability to interpret the results for cases in which only a nondefinite mutation is detected. As further missense changes are characterized and bioinformatics analyses of the likely consequences of specific substitutions are improved, further use of these data in a diagnostic setting will be possible, especially when used in combination with analyses of entire families. At present, no effective therapies for ADPKD exist; however, several potential therapies are now in phase III trials so treatments may become available in the next few years. If an effective therapy was available, treatment would probably be initiated at a young age, when renal function is normal and diagnosis by imaging is less reliable. In this scenario, the role of molecular testing may be greatly expanded to identify affected individuals from at-risk families.
Prospects for improved screening
Technology for DNA-based molecular diagnostics has evolved enormously over the past decade. The new genomic revolution triggered by the introduction of high-throughput sequencing approaches (so-called ‘next-generation’ sequencing87) holds promise to markedly change the way molecular diagnostics in PKD and the other genetic diseases are performed.88,89 The next-generation sequencing technologies that are available,87,90 and those under development (‘third-generation sequencing’91–93), may enable a qualitatively different approach to molecular testing than current methods. In fact, the capability to generate massive datasets by parallel sequencing will enable an analysis of the complete genomic structure of the PKD genes, and complete haplotype information to be obtained. The clinical applicability of these new technologies are reliant on decreasing the associated costs, the development of more efficient workflows for sample preparation, sequencing and data analysis, multiplexing of samples, and the simplification of validation tools.
Conclusions
Molecular testing for ADPKD seems poised to move from its present niche, for example, the assessment of potential living, related donors, to mainstream diagnostics for this disease. Factors that are propelling this movement include an improved understanding of the mutation data increasing the level of diagnostically informative tests, the potential to obtain prognostic as well as diagnostic information, facilitation of preimplantation genetic diagnostics for early-onset ADPKD, the advent of effective therapies for ADPKD, and improved technology to simplify and reduce the cost of the tests. As these technologies develop, it will be important that providers of clinical tests indicate clearly the results obtained and interpret them in a way that the nonspecialized nephrologist can understand. In addition, limitations and caveats associ ated with the test should be stressed. Further collection of mutation data in the ADPKD mutation database will not only improve our interpretation of future tests but will also help our understanding of the functional significance of key residues in polycystin-1 and polycystin-2. The developing concept of the hypomorphic allele in ADPKD is important as it also influences how the process of cystogenesis is understood.
Key points.
▪ Molecular diagnostics is available and increasingly informative in autosomal dominant polycystic kidney disease (ADPKD)
▪ Determining the disease status of potential living, related donors is where molecular diagnostics is most valuable at present
▪ A molecular diagnosis can clarify the disease status in patients with a negative family history and/or unusually mild or severe polycystic kidney disease
▪ Determining whether a family carries mutations in PKD1 or PKD2 is of prognostic value
▪ Hypomorphic PKD1 alleles can significantly modify the ADPKD phenotype and the identification of specific alleles may be of prognostic value, especially in early-onset ADPKD
▪ As therapies for ADPKD are developed, molecular testing will likely become increasingly valuable
Review criteria.
A literature search was performed in PubMed using the key words: “ADPKD”, “PKD1”, “PKD2”, “polycystic kidney disease”, “mutation classification”, “amino acid substitution classification”, “mosaicism”, “hypomorphic allele”, and “diagnostics”. Recently published abstracts were quoted if no suitable article was found. Papers were selected based on the authors’ judgment to make a comprehensive and readable review.
Footnotes
Competing interests The authors declare no competing interests.
References
- 1.Harris PC, Torres VE. GeneReviews at GeneTests: Medical Genetics Information Resource [database online] Copyright, University of Washginton; Seattle: 2008. 1997–2008 [online], http://www.genetests.org. [Google Scholar]
- 2.Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 3.Dalgaard OZ. Bilateral polycystic disease of the kidneys: A follow-up of two hundred and eighty-four patients and their families. Acta Med. Scand. Suppl. 1957;328:1–255. [PubMed] [Google Scholar]
- 4.Iglesias CG, et al. Epidemiology of adult polycystic kidney disease, Olmsted County, Minnesota. Am. J. Kidney Dis. 1983;2:630–639. doi: 10.1016/s0272-6386(83)80044-4. [DOI] [PubMed] [Google Scholar]
- 5.US Renal Data System . National Institutes of Health. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2007. [Google Scholar]
- 6.Belz MM, et al. Familial clustering of ruptured intracranial aneurysms in autosomal dominant polycystic kidney disease. Am. J. Kidney Dis. 2001;38:770–776. doi: 10.1053/ajkd.2001.27694. [DOI] [PubMed] [Google Scholar]
- 7.Rossetti S, et al. Association of mutation position in polycystic kidney disease 1 (PKD1) gene and development of a vascular phenotype. Lancet. 2003;361:2196–2201. doi: 10.1016/S0140-6736(03)13773-7. [DOI] [PubMed] [Google Scholar]
- 8.Pirson Y, Chauveau D, Torres v. Management of cerebral aneurysms in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2002;13:269–276. doi: 10.1681/ASN.V131269. [DOI] [PubMed] [Google Scholar]
- 9.Grantham JJ, et al. Volume progression in polycystic kidney disease. N. Engl. J. Med. 2006;354:2122–2130. doi: 10.1056/NEJMoa054341. [DOI] [PubMed] [Google Scholar]
- 10.Hateboer N, et al. Comparison of phenotypes of polycystic kidney disease types 1 and 2. Lancet. 1999;353:103–107. doi: 10.1016/s0140-6736(98)03495-3. [DOI] [PubMed] [Google Scholar]
- 11.Shamshirsaz A, et al. Autosomal-dominant polycystic kidney disease in infancy and childhood: progression and outcome. Kidney Int. 2005;68:2218–2224. doi: 10.1111/j.1523-1755.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- 12.Zerres K, Rudnik-Schöneborn S, Deget F, German Working Group on Paediatric Nephrology Childhood onset autosomal dominant polycystic kidney disease in sibs: clinical picture and recurrence risk. J. Med. Genet. 1993;30:583–588. doi: 10.1136/jmg.30.7.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Polycystic Kidney Disease Consortium The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell. 1994;77:881–894. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki T, et al. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science. 1996;272:1339–1342. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 15.Rossetti S, et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2007;18:2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 16.Harris PC, et al. Cyst number but not the rate of cystic growth is associated with the mutated gene in ADPKD. J. Am. Soc. Nephrol. 2006;17:3013–3019. doi: 10.1681/ASN.2006080835. [DOI] [PubMed] [Google Scholar]
- 17.Torra R, et al. Increased prevalence of polycystic kidney disease type 2 among elderly polycystic patients. Am. J. Kidney Dis. 2000;36:728–734. doi: 10.1053/ajkd.2000.17619. [DOI] [PubMed] [Google Scholar]
- 18.Barua M, et al. Family history of renal disease severity predicts the mutated gene in ADPKD. J. Am. Soc. Nephrol. 2009;20:1833–1838. doi: 10.1681/ASN.2009020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dicks E, et al. Incident renal events and risk factors in autosomal dominant polycystic kidney disease: a population and family-based cohort followed for 22 years. Clin. J. Am. Soc. Nephrol. 2006;1:710–717. doi: 10.2215/CJN.01581105. [DOI] [PubMed] [Google Scholar]
- 20.Rossetti S, et al. An Olmsted County population-based study indicates that PKD2 is more common than previously described [abstract] J. Am. Soc. Nephrol. 2007;18:365A. [Google Scholar]
- 21.Geberth S, Ritz E, Zeier M, Stier E. Anticipation of age at renal death in autosomal dominant polycystic kidney disease (ADPKD)? Nephrol. Dial. Transplant. 1995;10:1603–1606. [PubMed] [Google Scholar]
- 22.Gabow PA. Autosomal dominant polycystic kidney disease - more than a renal disease. Am. J. Kidney Dis. 1990;16:403–413. doi: 10.1016/s0272-6386(12)80051-5. [DOI] [PubMed] [Google Scholar]
- 23.Reed BY, et al. Variation in age at ESRD in autosomal dominant polycystic kidney disease. Am. J. Kidney Dis. 2008;51:173–183. doi: 10.1053/j.ajkd.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes J, et al. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat. Genet. 1995;10:151–160. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 25.International Polycystic Kidney Disease Consortium Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell. 1995;81:289–298. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 26.Loftus BJ, et al. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60:295–308. doi: 10.1006/geno.1999.5927. [DOI] [PubMed] [Google Scholar]
- 27.Martin J, et al. The sequence and analysis of duplication-rich human chromosome 16. Nature. 2004;432:988–994. doi: 10.1038/nature03187. [DOI] [PubMed] [Google Scholar]
- 28.Phakdeekitcharoen B, Watnick TJ, Germino GG. Mutation analysis of the entire replicated portion of PKD1 using genomic DNA samples. J. Am. Soc. Nephrol. 2001;12:955–963. doi: 10.1681/ASN.V125955. [DOI] [PubMed] [Google Scholar]
- 29.Rossetti S, et al. A complete mutation screen of the ADPKD genes by DHPLC. Kidney Int. 2002;61:1588–1599. doi: 10.1046/j.1523-1755.2002.00326.x. [DOI] [PubMed] [Google Scholar]
- 30.Veldhuisen B, et al. A spectrum of mutations in the second gene for autosomal dominant polycystic kidney disease (PKD2) Am. J. Hum. Genet. 1997;61:547–555. doi: 10.1086/515497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandford R, et al. Comparative analysis of the polycystic kidney disease 1 (PKD1) gene reveals an integral membrane glycoprotein with multiple evolutionary conserved domains. Hum. Mol. Genet. 1997;6:1483–1489. doi: 10.1093/hmg/6.9.1483. [DOI] [PubMed] [Google Scholar]
- 32.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell. Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 33.Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr. Biol. 2002;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- 34.Yoder BK, Hou X, Guay-Woodford LM. The polycystic kidney disease proteins, polycystin-1, polycystin-2, polaris, and cystin, are co-localized in renal cilia. J. Am. Soc. Nephrol. 2002;13:2508–2516. doi: 10.1097/01.asn.0000029587.47950.25. [DOI] [PubMed] [Google Scholar]
- 35.Nauli SM, et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 36.Sharif-Naeini R, et al. Polycystin-1 and -2 dosage regulates pressure sensing. Cell. 2009;139:587–596. doi: 10.1016/j.cell.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 37.Battini L, et al. Loss of polycystin-1 causes centrosome amplification and genomic instability. Hum. Mol. Genet. 2008;17:2819–2833. doi: 10.1093/hmg/ddn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, et al. Polycystin-1 and polycystin-2 regulate the cell cycle through the helix-loop-helix inhibitor Id2. Nat. Cell Biol. 2005;7:1202–1212. doi: 10.1038/ncb1326. [DOI] [PubMed] [Google Scholar]
- 39.Happé H, et al. Toxic tubular injury in kidneys from Pkd1-deletion mice accelerates cystogenesis accompanied by dysregulated planar cell polarity and canonical Wnt signaling pathways. Hum. Mol. Genet. 2009;18:2532–2542. doi: 10.1093/hmg/ddp190. [DOI] [PubMed] [Google Scholar]
- 40.Torres VE, Harris PC. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 2009;76:149–168. doi: 10.1038/ki.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed B, et al. Presence of de novo mutations in autosomal dominant polycystic kidney disease patients without family history. Am. J. Kidney Dis. 2008;52:1042–1050. doi: 10.1053/j.ajkd.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossetti S, et al. Mutation analysis of the entire PKD1 gene: genetic and diagnostic implications. Am. J. Hum. Genet. 2001;68:46–63. doi: 10.1086/316939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pei Y, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. J. Am. Soc. Nephrol. 2009;20:205–212. doi: 10.1681/ASN.2008050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravine D, et al. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet. 1994;343:824–827. doi: 10.1016/s0140-6736(94)92026-5. [DOI] [PubMed] [Google Scholar]
- 45.Nascimento AB, et al. Rapid MR imaging detection of renal cysts: age-based standards. Radiology. 2001;221:628–632. doi: 10.1148/radiol.2213010178. [DOI] [PubMed] [Google Scholar]
- 46.Carrim ZI, Murchison JT. The prevalence of simple renal and hepatic cysts detected by spiral computed tomography. Clin. Radiol. 2003;58:626–629. doi: 10.1016/s0009-9260(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 47.Huang E, et al. DNA testing for live kidney donors at risk for autosomal dominant polycystic kidney disease. Transplantation. 2009;87:133–137. doi: 10.1097/TP.0b013e318191e729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blumenfeld JD. Pretransplant genetic testing of live kidney donors at risk for autosomal dominant polycystic kidney disease. Transplantation. 2009;87:6–7. doi: 10.1097/TP.0b013e318191965d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edghill EL, Bingham C, Ellard S, Hattersley AT. Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J. Med. Genet. 2006;43:84–90. doi: 10.1136/jmg.2005.032854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Faguer S, et al. Massively enlarged polycystic kidneys in monozygotic twins with TCF2/HNF-1beta (hepatocyte nuclear factor-1beta) heterozygous whole-gene deletion. Am. J. Kidney Dis. 2007;50:1023–1027. doi: 10.1053/j.ajkd.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 51.Heidet L, et al. Spectrum of HNF1B mutations in a cohort of patients harboring renal diseases [abstract] J. Am. Soc. Nephrol. 2009;20:773A. doi: 10.2215/CJN.06810909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davila S, et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat. Genet. 2004;36:575–577. doi: 10.1038/ng1357. [DOI] [PubMed] [Google Scholar]
- 53.Drenth JP, te Morsche RH, Smink R, Bonifacino JS, Jansen JB. Germline mutations in PRKCSH are associated with autosomal dominant polycystic liver disease. Nat. Genet. 2003;33:345–347. doi: 10.1038/ng1104. [DOI] [PubMed] [Google Scholar]
- 54.Li A, et al. Mutations in PRKCSH cause isolated autosomal dominant polycystic liver disease. Am. J. Hum. Genet. 2003;72:691–703. doi: 10.1086/368295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adeva M, et al. Clinical and molecular characterization defines a broadened spectrum of autosomal recessive polycystic kidney disease (ARPKD) Medicine. 2006;85:1–21. doi: 10.1097/01.md.0000200165.90373.9a. [DOI] [PubMed] [Google Scholar]
- 56.Peral B, et al. Evidence of linkage disequilibrium in the Spanish polycystic kidney disease 1 population. Am. J. Hum. Genet. 1994;54:899–908. [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao X, et al. Molecular diagnostics in autosomal dominant polycystic kidney disease: utility and limitations. Clin. J. Am. Soc. Nephrol. 2008;8:146–152. doi: 10.2215/CJN.03430807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Rycke M, et al. PGD for autosomal dominant polycystic kidney disease type 1. Mol. Hum. Reprod. 2005;11:65–71. doi: 10.1093/molehr/gah128. [DOI] [PubMed] [Google Scholar]
- 59.Tan YC, et al. Novel method for genomic analysis of PKD1 and PKD2 mutations in autosomal dominant polycystic kidney disease. Hum. Mutat. 2009;30:264–273. doi: 10.1002/humu.20842. [DOI] [PubMed] [Google Scholar]
- 60.Garcia-Gonzalez MA, et al. Evaluating the clinical utility of a molecular genetic test for polycystic kidney disease. Mol. Genet. Metab. 2007;92:160–167. doi: 10.1016/j.ymgme.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.PKD Foundation Autosomal Dominant Polycystic Kidney Disease: Mutation Database. 2010 [online], http://pkdb.mayo.edu.
- 62.Magistroni R, et al. Genotype-renal function correlation in type 2 autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2003;14:1164–1174. doi: 10.1097/01.asn.0000061774.90975.25. [DOI] [PubMed] [Google Scholar]
- 63.Rossetti S, et al. Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int. 2009;75:848–855. doi: 10.1038/ki.2008.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Consugar MB, et al. Characterization of large rearrangements in autosomal dominant polycystic kidney disease and the PKD1/TSC2 contiguous gene syndrome. Kidney Int. 2008;74:1468–1479. doi: 10.1038/ki.2008.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brook-Carter PT, et al. Deletion of the TSC2 and PKD1 genes associated with severe infantile polycystic kidney disease—a contiguous gene syndrome. Nat. Genet. 1994;8:328–332. doi: 10.1038/ng1294-328. [DOI] [PubMed] [Google Scholar]
- 66.Sampson JR, et al. Renal cystic disease in tuberous sclerosis: role of the polycystic kidney disease 1 gene. Am. J. Hum. Genet. 1997;61:843–851. doi: 10.1086/514888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koulen P, et al. Polycystin-2 is an intracellular calcium release channel. Nat. Cell Biol. 2002;4:191–197. doi: 10.1038/ncb754. [DOI] [PubMed] [Google Scholar]
- 68.Qian F, et al. Cleavage of polycystin-1 requires the receptor for egg jelly domain and is disrupted by human autosomal-dominant polycystic kidney disease 1-associated mutations. Proc. Natl Acad. Sci. USA. 2002;99:16981–16986. doi: 10.1073/pnas.252484899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Forman JR, Qamar S, Paci E, Sandford RN, Clarke J. The remarkable mechanical strength of polycystin-1 supports a direct role in mechanotransduction. J. Mol. Biol. 2005;349:861–871. doi: 10.1016/j.jmb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 70.Ma L, Xu M, Forman JR, Clarke J, Oberhauser AF. Naturally occurring mutations alter the stability of polycystin-1 PKD domains. J. Biol. Chem. 2009;284:32942–32949. doi: 10.1074/jbc.M109.021832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qian F, Wei W, Germino G, Oberhauser A. The nanomechanics of polycystin-1 extracellular region. J. Biol. Chem. 2005;280:40723–40730. doi: 10.1074/jbc.M509650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Consugar MB, et al. Characteristics of CRISP ADPKD patients with no detected base-pair mutations [abstract] J. Am. Soc. Nephrol. 2008;19:125A. [Google Scholar]
- 73.King K, Flinter FA, Nihalani V, Green PM. Unusual deep intronic mutations in the COL4A5 gene cause X linked Alport syndrome. Hum. Genet. 2002;111:548–554. doi: 10.1007/s00439-002-0830-3. [DOI] [PubMed] [Google Scholar]
- 74.Gorlov IP, Gorlova OY, Frazier ML, Amos CI. Missense mutations in hMLH1 and hMSH2 are associated with exonic splicing enhancers. Am. J. Hum. Genet. 2003;73:1157–1161. doi: 10.1086/378819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daoust MC, Reynolds DM, Bichet DG, Somlo S. Evidence for a third genetic locus for autosomal dominant polycystic kidney disease. Genomics. 1995;25:733–736. doi: 10.1016/0888-7543(95)80020-m. [DOI] [PubMed] [Google Scholar]
- 76.de Almeida S, et al. Autosomal dominant polycystic kidney disease: evidence for the existence of a third locus in a Portuguese family. Hum. Genet. 1995;96:83–88. doi: 10.1007/BF00214191. [DOI] [PubMed] [Google Scholar]
- 77.Consugar M, et al. PKD3 revisited with improved PKD1 and PKD2 haplotyping and mutation screening [abstract] J. Am. Soc. Nephrol. 2005;16:358A. [Google Scholar]
- 78.Connor A, et al. Mosaicism in autosomal dominant polycystic kidney disease revealed by genetic testing to enable living related renal transplantation. Am. J. Transplant. 2008;8:232–237. doi: 10.1111/j.1600-6143.2007.02030.x. [DOI] [PubMed] [Google Scholar]
- 79.Rossetti S, et al. The position of the polycystic kidney disease 1 (PKD1) gene mutation correlates with the severity of renal disease. J. Am. Soc. Nephrol. 2002;13:1230–1237. doi: 10.1097/01.asn.0000013300.11876.37. [DOI] [PubMed] [Google Scholar]
- 80.Bergmann C, et al. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1) J. Am. Soc. Nephrol. 2003;14:76–89. doi: 10.1097/01.asn.0000039578.55705.6e. [DOI] [PubMed] [Google Scholar]
- 81.Jiang ST, et al. Defining a link with autosomal-dominant polycystic kidney disease in mice with congenitally low expression of Pkd1. Am. J. Pathol. 2006;168:205–220. doi: 10.2353/ajpath.2006.050342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim I, et al. Polycystin-2 expression is regulated by a PC2-binding domain in the intracellular portion of fibrocystin. J. Biol. Chem. 2008;283:31559–31566. doi: 10.1074/jbc.M805452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lantinga-van Leeuwen IS, et al. Lowering of Pkd1 expression is sufficient to cause polycystic kidney disease. Hum. Mol. Genet. 2004;13:3069–3077. doi: 10.1093/hmg/ddh336. [DOI] [PubMed] [Google Scholar]
- 84.Sandford RN. The diversity of PKD1 alleles: implications for disease pathogenesis and genetic counseling. Kidney Int. 2009;75:765–767. doi: 10.1038/ki.2009.17. [DOI] [PubMed] [Google Scholar]
- 85.Fain PR, et al. Modifier genes play a significant role in the phenotypic expression of PKD1. Kidney Int. 2005;67:1256–1267. doi: 10.1111/j.1523-1755.2005.00203.x. [DOI] [PubMed] [Google Scholar]
- 86.Paterson AD, et al. Progressive loss of renal function is an age-dependent heritable trait in type 1 autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2005;16:755–762. doi: 10.1681/ASN.2004090758. [DOI] [PubMed] [Google Scholar]
- 87.Tucker T, Marra M, Friedman JM. Massively parallel sequencing: the next big thing in genetic medicine. Am. J. Hum. Genet. 2009;85:142–154. doi: 10.1016/j.ajhg.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 89.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin. Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 90.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 91.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 92.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 93.Branton D, et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ravine D, Gibson RN, Donlan J, Sheffield LJ. An ultrasound renal cyst prevalence survey: specificity data for inherited renal cystic diseases. Am. J. Kidney Dis. 1993;22:803–807. doi: 10.1016/s0272-6386(12)70338-4. [DOI] [PubMed] [Google Scholar]
- 95.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 96.Bork Group and Sunyaev Lab PolyPhen: prediction of functional effect on human nsSNPs. 2009 [online], http://genetics.bwh.harvard.edu/pph/
- 97.J. Craig venter Institute SIFT. 2009 [online], http://sift.jcvi.org/
- 98.International Agency for Research on Cancer Align GVGD. 2009 [online], http://agvgd.iarc.fr/agvgd_input.php.
- 99.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acid Research. 2009 doi: 10.1093/nar/gkp215. [online], http://www.umd.be\HSF\. [DOI] [PMC free article] [PubMed] [Google Scholar]