Abstract
The morphogenesis of ovococci has been reviewed extensively. Recent results have provided new insights concerning the mechanisms of elongation in ovoid bacteria. We present here the proteins involved in the elongation (firmly established and more or less hypothetical) and discuss the relationship between elongation and division of ovococci.
Introduction
Bacterial shape has been studied for many years for several reasons. Nature has generated bacteria with a wide variety of appearances, differing in size and morphologies, two features directly observable by optical microscopy. Specific shapes have been shown to confer advantages for survival under various environmental challenges. For example, shape plays a role in nutrient access, predation avoidance, diffusion and motility, and defense against stress (reviewed in Young34). The fact that cells retain their specific shape through generations, but are sometimes able to alter it upon environmental changes are strong indications of the adaptive importance of morphology.
The bacterial peptidoglycan (PG), one of the main constituent of the cell wall, is a giant molecule consisting of glycan chains reticulated by peptide links.4 This structure totally encloses the cell and resists the internal osmotic pressure. The different morphologies observed in bacteria result from distinct mechanisms of PG insertion in the cell wall.
Among a vast diversity,34 three main morphological types have been investigated in some details in bacteria: bacilli that are cylindrical (e.g., Bacillus subtilis, Escherichia coli), cocci that are spherical (e.g., Staphylococcus aureus), and ovococci that are ellipsoid (e.g., Streptococcus pneumoniae, Lactococcus lactis, and Enterococcus faecalis). During growth, bacilli elongate and periodically divide, which contrasts with cocci that only divide. Ovococci exhibit an intermediate behavior with a short peripheral growth phase (or elongation) for each division round. Compared with bacilli, there are few data on the morphogenesis of ovococci, especially concerning their elongation. However, recent studies reviewed herein have brought new insights on this mechanism.
The Two Morphogenetic Machineries Model
Early work by Higgins and Shockman described in detail the growth of ellipsoid bacteria.9 Observation by electronic microscopy of thin sections of Enterococcus hirae ATCC 9790 and careful reconstitution of the cell cycle showed that the cell wall is primarily assembled at midcell, also called equator, where a small amount of crosswall is first incorporated toward the septum. Then, new material is incorporated in the cell wall; while the septum is split at the periphery, the crosswall thus remaining constant in size for some time. Finally, the septal crosswall is completed and the two daughter cells are separated. At this point, PG incorporation at the equators of daughter cells has already begun when cells are growing exponentially.9 The observation that the peripheral wall on each side of the septum is thicker than the crosswall itself ruled out the possibility that the elongation is only due to splitting of the septum. Therefore, they proposed the currently admitted model, including both the septal and peripheral phases of PG assembly.10
This model is supported by a number of functional observations. Inhibition of division by antibiotic treatment or mutations resulted in unchecked elongation in several ovococcal species,14 indicating that peripheral wall incorporation is independent of septum formation. Interestingly, Streptococcus mutans NCTC 10449S was shown to adopt a rod-like or an ellipsoid conformation depending on the salt composition of the medium.24,25 Similarly, planktonic cells of L. lactis IL1403 underwent ovococcus to rod transition in a particular medium. Biofilms grown in this medium had elongated cells at their surface, while bacteria not directly exposed to the medium were only ovococci.19 Two types of cell wall growth, a peripheral elongation in addition to the septation, are thus present in ovococci, the balance between the activities of each being finely tuned by mechanisms that remain unknown.
Although the model ovococcal species S. pneumoniae, L. lactis, and E. faecalis have similar cell cycles, subtle differences were recently described in these bacteria.33 Structured illumination super-resolution microscopy on cells treated with fluorescent vancomycin, which labels the PG precursor and nascent unmatured PG, allowed measurement of the dimensions of PG insertion sites throughout the cell cycle. Briefly, as in E. hirae, S. pneumoniae shows overlapping rounds of cell wall synthesis, with the insertion of new wall material at the equators of daughter cells that are not separated yet, whereas E. faecalis and L. lactis show discrete rounds of division. Whereas crosswall synthesis and splitting are concomitant in S. pneumoniae and L. lactis, E. faecalis cells initiate splitting after the septum is completed. In L. lactis, cell elongation is almost completed when septation begins, in contrast with S. pneumoniae and E. faecalis where septation and elongation are mostly simultaneous.
These observations support a model with two cell wall assembly machineries, although the regulation of these machineries slightly differs among ovococcal species.
Proteins Involved in the Elongation of Ovococci
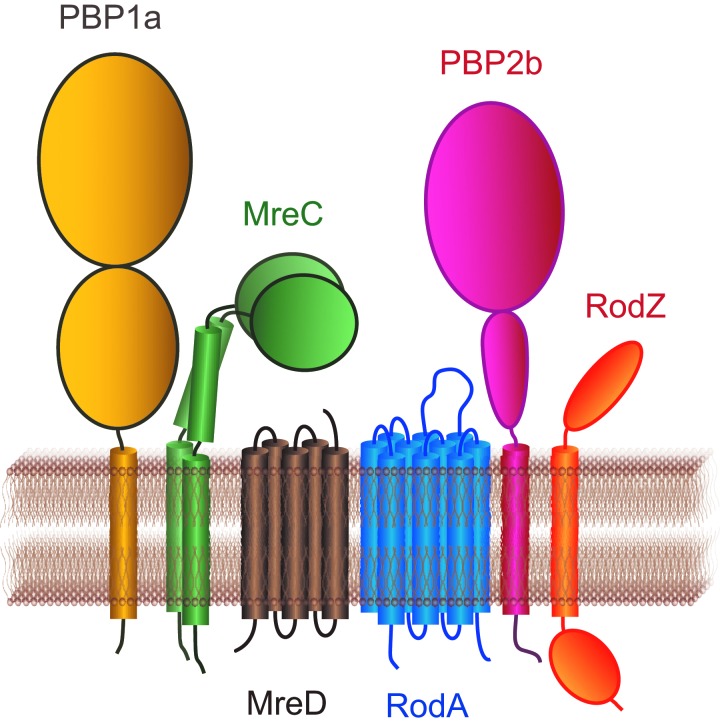
Several proteins of different functions have been assigned to the elongation machinery of rod-like bacteria, such as the Mre proteins, RodA, RodZ, GpsB, bi- and monofunctional class A and class B penicillin-binding proteins (PBPs).5,22,27,38 We will review here those proteins that have also been assigned to the elongation, or peripheral growth, of ovococci (Fig. 1).
FIG. 1.
Topology of the elongation proteins in ovococci. The implication of MreC,13 MreD,13 PBP2b,3,26 and RodA26 in elongation was experimentally shown in ovococci. The implication of PBP1a13 and RodZ1 is hypothetical.
The PBPs assemble the PG from its lipid II precursor, a disaccharide pentapeptide linked to the membrane by a lipid pyrophosphate. Class A PBPs have both the glycosyltransferase (GT) activity, which allows polymerization of the glycan chains, and the transpeptidase (TP) activity that catalyzes the crosslinking of these chains through peptide bonds.36 Class B PBPs are monofunctional and only have the TP activity. Ovococci have three class A PBPs (PBP1a, 1b, and 2a) and two class B PBPs (PBP2b and 2x). Two exceptions are Streptococcus pyogenes that lacks PBP2b (Table 1), but is less elongated than other ovococcal species, and enterococci that have an additional class B PBP, PBP5, with a low affinity for β-lactams.
Table 1.
Proteins Thought to Participate in the Elongation of Ovococci and Bacillus subtilis
| Species/strain | PBP2b | PBP1a | RodA | MreC | MreD | RodZ | GpsB |
|---|---|---|---|---|---|---|---|
| Streptococcus pneumoniae R6 | P0A3M6 | Q8DR59 | Q8DQE8 | Q8DMY2 | Q8DMY3 | Q8DMX7 | Q8DR57 |
| spr1517 | spr0329 | spr0712 | spr2023 | spr2022 | spr2028 | spr0332 | |
| Streptococcus thermophilus CNRZ1066 | Q5M0P5 | Q5M1K8 | Q5LZC8 | Q5M214 | Q5M213 | Q5LXJ1 | Q5M1K5 |
| str0613 | str0230 | str1229 | str0020 | str0021 | str2011 | str0233 | |
| Streptococcus mutans UA159 | Q8DVA0 | Q8DVL4 | Q8CWX3 | Q8DW M4 | I6L914 | Q8DRR7 | Q8DVL1 |
| SMU_597 | SMU_467 | SMU_1279c | SMU_20 | SMU_21 | SMU_2152c | SMU_471 | |
| Streptococcus agalactiae 2603V/R | Q8E0G8 | Q8E1Q5 | Q8E0U9 | A | A | A | Q8E1Q1 |
| SAG0765 | SAG0298 | SAG0621 | SAG0302 | ||||
| Streptococcus dysgalactiae D166B | E8QC63 | E8QD40 | E8Q947 | A | A | E8Q9U7 | C5WIH6 |
| SDE12394_07630 | SDE12394_08565 | SDE12394_03755 | SDE12394_10950 | SDEG_1708 | |||
| Streptococcus pyogenes MGAS10394 | A | Q5XAM7 | A | A | A | Q5X9B3 | Q5XAN0 |
| M6_Spy1401 | M6_Spy1865 | M6_Spy1398 | |||||
| Lactococcus lactis IL1403 | Q9CIL7 | Q9CI23 | Q9CH43 | Q9CDI9 | Q9CDJ0 | Q9CE74 | Q9CF28 |
| LL0339 | LL0543 | LL0896 | LL2231 | LL2230 | LL1971 | LL1653 | |
| Enterococcus faecalis V583 | Q830D1 | Q836G4 | Q820T7 | Q82ZJ4 | Q82ZJ5 | Q82ZB7 | Q836G2 |
| EF_2857 | EF_1148 | EF_2502 | EF_3062 | EF_3061 | EF_3149 | EF_1151 | |
| Enterococcus faecium Aus0004 | H8LF48 | H8LE97 | H8LD04 | H8LAP3 | H8LAP3 | H8LAS4 | H8LE94 |
| EFAU004_01299 | EFAU004_00997 | EFAU004_00554 | EFAU004_02614 | EFAU004_02614 | EFAU004_02645 | EFAU004_00994 | |
| Enterococcus hirae ATCC 9790 | I6T1G1 | I6S3M8 | I6T427 | I6S0D1 | I6T5X3 | I6T5U6 | I6T0I4 |
| EHR_14050 | EHR_12290 | EHR_01910 | EHR_05725 | EHR_05730 | EHR_05540 | EHR_12275 | |
| B. subtilis 168 | P54488 | P39793 | P39604 | Q01466 | Q01467 | O31771 | P0CI74 |
| BSU25000 | BSU22320 | BSU38120 | BSU28020 | BSU28010 | BSU16910 | BSU22180 | |
| (PBP2a YqgF) | (PBP1) | (YmfM) |
For each protein, the UniprotKB number is given with the ordered locus name. The B. subtilis protein name is given in parenthesis if different.
A, absent.
PBP2b of ovococci is the orthologue of B. subtilis PBP2 that is involved in the elongation of this rod-shaped species.21 In an analysis of oxidative stress-resistant mutants of Streptococcus thermophilus, Thibessard et al. identified the depletion of PBP2b.26 S. thermophilus cells depleted of PBP2b grew twice slower than the wild type, and they reduced their ovoid shape to be more spherical. The role of PBP2b or the shape in oxidative stress remains mysterious. A similar morphological effect of PBP2b depletion was also observed in L. lactis.19 More recently, the role of PBP2b has also been described in S. pneumoniae, where it is essential.3 In the absence of an inducer, conditional knocked out pneumococci took a lentil-like shape, confirming a role in peripheral growth. The TP activity of PBP2b from S. pneumoniae has recently been observed in vitro.37
PBP1a has both the GT and TP activities, as demonstrated in vitro with the recombinant enzyme from S. pneumoniae.37 Unencapsulated D39 pneumococci artificially depleted of PBP1a have a smaller diameter, whereas depletion of the other bifunctional PBP2a or PBP1b has no effect on the cell diameter.13 In rod-like bacteria, the elongation machinery was shown to affect the cell diameter,35 supporting a role of PBP1a in the elongation of S. pneumoniae. Another clue that PBP1a participates in the elongation is its genetic relationship with the MreC and MreD proteins. Indeed, the presence of PBP1a, or its sequence, affects the essentiality of these two elongation proteins in S. pneumoniae.13 In the unencapsulated variant of D39, MreC and MreD are essential, but deletion of pbp1a, or a point mutation likely affecting the GT activity of PBP1a, suppresses this essentiality. In R6 S. pneumoniae, the mreCD operon can be deleted, but this appears to depend partly on the specific sequence of PBP1a, which differs at two positions from that in the D39 strain. In B. subtilis, PBP1 (the orthologue of PBP1a) has been proposed to act both in elongation and division,5 in agreement with the possibility that PBP1a plays a role in the elongation of ovococci.
The Mre proteins (for the murein region e) were first discovered in E. coli, where mutation of the mre genes resulted in round-shaped bacteria, indicating a role in elongation.32 Three Mre protein types have been described: the soluble actin homolog MreB (and the likes Mbl and MreBH) that are absent in ovococci, MreC and MreD, two membrane proteins. The exact role of MreC and MreD remains elusive to date. In S. pneumoniae, both are localized at the PG insertion site.13 They were shown to be essential in some S. pneumoniae strains lacking suppressor mutations in pbp1a or other genes of unknown function.13 In this background, MreC and MreD depletion led to cell rounding and lysis, the first experimental evidence for a role of these proteins in the elongation of an ovococcus.13 The mreC and mreD genes are absent in S. pyogenes, which may be consistent with the fact that this species does not elongate and accordingly lacks other components of the elongation system such as PBP2b and RodA. Intriguingly, mreC and mreD are also missing in Streptococcus agalactiae and Streptococcus dysgalactiae (Table 1)1, which is consistent with the fact that these species appear shorter than other ovococci.
SEDS (for shape, elongation, division, and sporulation) proteins are integral membrane proteins with 10 membrane-spanning segments.8 In genomes, their genes are often located in the same operons with genes encoding monofunctional class B PBPs, suggesting that they belong to the same machinery.38 These membrane proteins allow flipping of the lipid II from the inside to the outside of the cell, providing PBPs with their substrate.16 Ovococci have generally two SEDS, RodA and FtsW that are involved in elongation and division, respectively.38 Some exceptions are to be noted: S. pyogenes lacks RodA, whereas Enterococcus faecium has one and E. faecalis and E. hirae have two additional SEDS proteins (Table 1). The absence of RodA in S. pyogenes is consistent with the absence of PBP2b and elongation in this species. The presence of at least one additional SEDS protein in enterococci is consistent with the additional class B PBP5, which has a low affinity for β-lactams. RodA depletion in S. thermophilus results in the same round phenotype as PBP2b depletion26 supporting its role in elongation. Land and Winkler reported preliminary results suggesting the essentiality of RodA in S. pneumoniae.13
More recently, other proteins were proposed to participate in the elongation of ovococci.
In B. subtilis, the small protein GpsB interacts with PBP1 (PBP1a's orthologue) to allow its transition between the division and elongation machineries through the cell cycle.5 Given that GpsB is present in ovococci and that PBP1a probably participates in the elongation of pneumococci, Land and Winkler proposed that GpsB also participates in the peripheral growth of ovococci.13 However, subsequent studies of this essential protein showed that GpsB is rather implicated in the division of S. pneumoniae, as depletion caused cell elongation.12 In B. subtilis, GpsB was proposed to act primarily as a vector to relocate PBP1 from the division to the elongation machinery, with a minor effect on the reverse transition from the elongation to the division machinery. If a similar mechanism operates in ovococci, it could function in the opposite way, explaining that depletion leads to aborted division.
RodZ is a nonessential bitopic membrane protein of E. coli, where its depletion results in shorter cells, and its overexpression causes elongation of the bacteria,23 suggesting a role in elongation. RodZ was proposed to play a role in MreB cytoskeleton polymerization and stability2 and was shown to link MreB filaments to the membrane.28 RodZ is widely conserved in bacteria, but is generally absent in species devoid of MreB with the exception of ovococci where it is present.1 The conserved function of RodZ might therefore be linked to that of the MreC/MreD complex, which is found in ovococci, rather than to MreB. Note that S. agalactiae, which lacks MreC and MreD proteins, also lacks RodZ, but S. pyogenes and S. dysgalactiae that also lack MreC and MreD nevertheless encode RodZ in their genome (Table 1).
To ensure a proper insertion of PG strands in the peripheral wall, additional bifunctional class A PBPs and hydrolases may be required.38
An outstanding observation should be taken into consideration here. In a study of the localization of wall-anchored proteins in S. pyogenes, Raz et al. noted that methicillin treatment at precisely 0.2 μg/ml induced coccus to rod transition.20 This observation is counterintuitive as S. pyogenes does not encode for the elongation proteins PBP2b, RodA, MreC, and MreD (Table 1). Methicillin is known to inhibit specifically PBP2x in ovoid bacteria such as S. pneumoniae12 and L. lactis.19 At 0.2 μg/ml of methicillin, the activity of PBP2x may be inhibited, and the three bifunctional class A PBPs of S. pyogenes may remain functional at a level that allows peripheral PG insertion in the cell wall.
Characterized Interactions Between Elongation Proteins
In E. coli, two-hybrid assays showed that MreC interacts with both MreB and MreD and they were proposed to form a complex with RodA and PBP2, the monofunctional class B PBP assigned to elongation.11 These five proteins have a similar localization in E. coli.31 Surprisingly, however, MreB, MreC, MreD, and RodA are each able to localize correctly in the absence of the four other proteins. Only PBP2 localization appears to depend on the presence of MreC.31 In Helicobacter pylori, MreC and PBP2 (the orthologue of PBP2b) form a complex that is required for the elongation.7 In B. subtilis, two-hybrid experiments have shown that PBP1 (the orthologue of PBP1a) interacts with MreC and GpsB.5
RodZ was shown to interact with MreB in E. coli by a two-hybrid assay, and the crystal structure of a complex between MreB and the cytoplasmic domain of RodZ was solved for proteins from Thermotoga maritima.28 Van den Ent et al. proposed that MreB is brought close to the membrane by interacting with RodZ, which favors its interaction with MreC and links the cytoskeleton to the PG synthesis machinery.28
No such studies have been performed with elongation proteins from ovococci. In these organisms, although some of the same interaction patterns likely occur, significant differences are also expected. For example, the MreB-interacting domain of RodZ is present in the protein from ovococci although MreB is absent.
Recently, some complexes, including up to five proteins of the divisome (a complex, including all the division proteins) of S. pneumoniae (namely, DivIC, DivIB, FtsL, FtsW, and PBP2x), were reconstituted in vitro.18 However, to our knowledge, no characterized protein–protein interactions have demonstrated the presence of an elongasome (a complex, including all the elongation proteins) in ovococcus species. We report in this study, the reconstitution of two complexes of recombinant proteins of S. pneumoniae (Fig. 2).
FIG. 2.
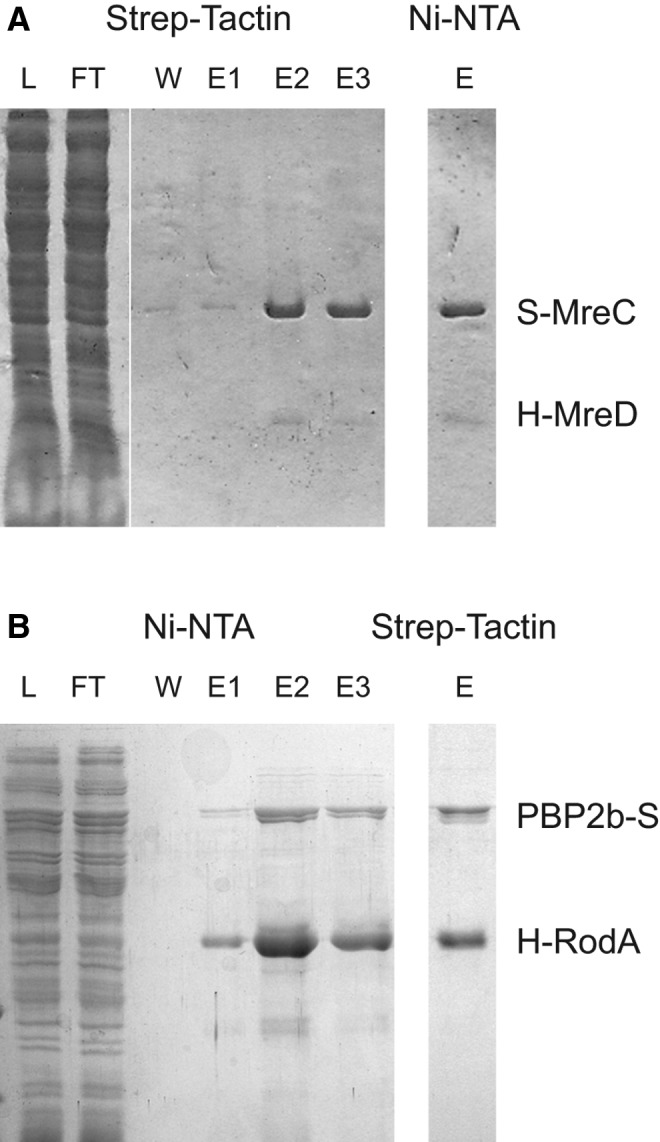
Isolation of recombinant MreC/MreD and PBP2b/RodA complexes from pneumococcus. (A) MreC and MreD were expressed in Escherichia coli from an artificial operon fused with N-terminal Strep- and His8-tags, respectively. Membranes were isolated and solubilized with n-dodecyl-β-d-maltopyranoside. The complex was isolated by two successive affinity chromatography steps on Strep-Tactin® and Ni-NTA. (B) PBP2b with a C-terminal Strep-tag and RodA with an N-terminal His8-tag were similarly expressed and the complex isolated, in that case, first by Ni-NTA chromatography followed by a Strep-Tactin® purification. L, W, and E stand for load, wash, and elution. Samples were analyzed by Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
The bitopic membrane protein MreC and the integral membrane protein MreD (five predicted transmembrane domains) were fused to distinct affinity tags (Strep-MreC and His8-MreD). Coexpression in E. coli followed by two successive purification steps on Strep-Tactin® and Ni-NTA resin allowed us to recover the complex in vitro (Fig. 2). Size-exclusion chromatography–multi-angle laser light scattering analysis of the sample suggested a complex comprising a dimer of MreC and a single MreD unit, although a large portion of the sample was aggregated (55%) affecting the interpretation.
Similarly, a membrane protein complex of recombinant PBP2b and RodA from S. pneumoniae was isolated in the laboratory (Fig. 2). In this case, PBP2b harbored a Strep-tag, whereas RodA was fused to a His8-tag.
Our attempts to isolate a larger elongation protein complex, including the four proteins MreC, MreD, RodA, and PBP2, were not successful so far. The trivial technical explanation cannot be discounted since it is difficult to find appropriate conditions to copurify stable membrane protein complexes as these should solubilize the proteins, while preserving their interaction, two competing goals. Nevertheless, such a complex may indeed not exist. Colocalization does not necessarily imply interaction, and protein may participate in a common function without interacting. If interactions take place, they may occur successively between different partners during the cell cycle, and thus preclude the isolation of one single large assembly. Finally, additional hitherto unknown partners may be required to form stable complexes that can be isolated recombinantly.
Relationship Between the Elongation and the Division Machineries
In 1990, Lleo et al. gave the evidence for the existence of two distinct machineries for the elongation and the division of ovococci. They also hypothesized that these machineries would be independent and mutually exclusive.14 Some experimental observations are in conflict with this latter statement, as several ovococci appear to simultaneously elongate and synthesize a septum, such as E. hirae, S. pneumonia, and E. faecalis.9,33
The link between the two PG synthesis machineries remains unclear in ovococci. In B. subtilis, GpsB was proposed to shuttle between the elongasome and the divisome, forming a link between elongation and division machineries. Proteins from both machineries were shown to colocalize throughout the cell cycle of S. pneumoniae at the resolution of epifluorescence optical microscopy.17,38 However, and most importantly, recent observations with improved resolution using three-dimensional structured illumination microscopy revealed a clear difference in the localization of PBP1a and PBP2x during septum formation of S. pneumoniae.12 The constriction of the PBP1a ring was found to lag behind that of PBP2x. This first observation that different PBPs are in distinct localization at some point during the cell cycle is in strong support of the two-machinery model. PBP1a remaining at the periphery of the closing septum is consistent with the primary role in peripheral growth. Nevertheless, as PBP1a and PBP2x are colocalized at the onset of the cell cycle, it is not excluded that PBP1a plays a role in both machineries, as it was proposed in B. subtilis.5 Such a dual role of PBP1a would also be consistent with intriguing β-lactam resistance phenotypes that hint at an interaction of PBP2x with PBP1a.39 Also, of note, in these high-resolution microscopy experiments is the finding that the rings of the different proteins (PBPs or FtsZ) are discontinuous and constituted of foci of different sizes arranged irregularly in circles.12
In E. coli, FtsZ and PBP2 (the orthologue of PBP2b) can act together in inserting PG in the side wall of cells when MreB is inhibited.30 Also, in these organisms, division is preceded by a short period of PG insertion at the midcell dependent on FtsZ and the elongasome. This phase of the cell cycle is particularly visible when septation is inhibited (e.g., de Pedro et al.6). Consistent with these observations of the PG, it was recently shown by immunofluorescence that elongation proteins are transiently colocalized with those of the division before the septal PG synthesis phases in E. coli.29 Also, fluorescence resonance energy transfer analysis gave evidence of an interaction between the class B PBP2 and PBP3 that belong to the elongation and division, respectively.29 Interestingly, in ovococci, the elongation or peripheral growth occurs while the FtsZ-ring is assembled and possibly even during its constriction. In this respect, peripheral PG synthesis in ovococci is more akin to the localized preseptal PG synthesis observed in rod-shaped bacteria than to their true elongation phase.31
Today, we lack evidence for the existence of two physically separated machineries of PG assembly in ovococci, with the exception of the distinct localization of PBP1a and PBP2x. Nevertheless, a comprehensive model was recently proposed that includes all the morphogenesis proteins in a single large machinery comprising two complexes located at the midcell, the site of insertion of new PG in S. pneumoniae15: the division proteins acting on the leading edge of the closing septum and the elongation proteins on the outer edge of the septal disc. It is expected that various features of this model will be tested in the coming years by various novel multicolor super-resolution microscopy techniques and in vitro reconstitutions.
Acknowledgments
J.P. was funded by a grant from la Région Rhône-Alpes. This work used the platforms PAOL and RoBioMol of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
Disclosure Statement
No conflicts of interest.
References
- 1.Alyahya S.A., Alexander R., Costa T.Henriques A.O.Emonet T., and Jacobs-Wagner C. 2009. RodZ, a component of the bacterial core morphogenic apparatus. Proc. Natl Acad. Sci. U. S. A. 106:1239–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendezu F.O., Hale C.A.Bernhardt T.G. and de Boer P.A. 2009. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 28:193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg K.H., Stamsas G.A., Straume D., and Havarstein L.S. 2013. Effects of low PBP2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J. Bacteriol. 195:4342–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui N.K., Eberhardt A., Vollmer D., Kern T., Bougault C., Tomasz A.Simorre J.P. and Vollmer W. 2012. Isolation and analysis of cell wall components from Streptococcus pneumoniae. Anal. Biochem. 421:657–666 [DOI] [PubMed] [Google Scholar]
- 5.Claessen D., Emmins R.Hamoen L.W.Daniel R.A.Errington J., and Edwards D.H. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68:1029–1046 [DOI] [PubMed] [Google Scholar]
- 6.de Pedro M.A., Quintela J.C.Holtje J.V. and Schwarz H. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El Ghachi M., Mattei P.J., Ecobichon C., Martins A., Hoos S., Schmitt C., Colland F., Ebel C.Prevost M.C.Gabel F., et al. 2011. Characterization of the elongasome core PBP2: MreC complex of Helicobacter pylori. Mol. Microbiol. 82:68–86 [DOI] [PubMed] [Google Scholar]
- 8.Gerard P., Vernet T., and Zapun A.2002. Membrane topology of the Streptococcus pneumoniae FtsW division protein. J. Bacteriol. 184:1925–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins M.L., and Shockman G.D. 1970. Model for cell wall growth of Streptococcus faecalis. J. Bacteriol. 101:643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins M.L., and Shockman G.D. 1976. Study of cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstructions of thin sections of cells. J. Bacteriol. 127:1346–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruse T., Bork-Jensen J., and Gerdes K.2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78–89 [DOI] [PubMed] [Google Scholar]
- 12.Land A.D., Tsui H.C., Kocaoglu O.Vella S.A.Shaw S.L.Keen S.K.Sham L.T.Carlson E.E. and Winkler M.E. 2013. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol. Microbiol. 90:939–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Land A.D., and Winkler M.E. 2011. The requirement for pneumococcal MreC and MreD is relieved by inactivation of the gene encoding PBP1a. J. Bacteriol. 193:4166–4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lleo M.M., Canepari P., and Satta G. 1990. Bacterial cell shape regulation: testing of additional predictions unique to the two-competing-sites model for peptidoglycan assembly and isolation of conditional rod-shaped mutants from some wild-type cocci. J. Bacteriol. 172:3758–3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massidda O., Novakova L., and Vollmer W.2013. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division?. Env. Microbiol. 15:3133–3157 [DOI] [PubMed] [Google Scholar]
- 16.Mohammadi T., van Dam V., Sijbrandi R., Vernet T., Zapun A., Bouhss A., Diepeveen-de Bruin M., Nguyen-Disteche M., de Kruijff B., and Breukink E.2011. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30:1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morlot C., Zapun A., Dideberg O., and Vernet T.2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845–855 [DOI] [PubMed] [Google Scholar]
- 18.Noirclerc-Savoye M., Lantez V., Signor L., Philippe J., Vernet T., and Zapun A.2013. Reconstitution of membrane protein complexes involved in pneumococcal septal cell wall assembly. PLoS One 8:e75522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Nunez D., Briandet R., David B., Gautier C., Renault P., Hallet B., Hols P., Carballido-Lopez R., and Guedon E.2011. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol. Microbiol. 79:759–771 [DOI] [PubMed] [Google Scholar]
- 20.Raz A., Talay S.R., and Fischetti V.A.2012. Cellular aspects of the distinct M protein and SfbI anchoring pathways in Streptococcus pyogenes. Mol. Microbiol. 84:631–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauvage E., Kerff F., Terrak M., Ayala J.A., and Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258 [DOI] [PubMed] [Google Scholar]
- 22.Sham L.T., Tsui H.C., Land A.D., Barendt S.M., and Winkler M.E. 2012. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr. Opin. Microbiol. 15:194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiomi D., Sakai M., and Niki H.2008. Determination of bacterial rod shape by a novel cytoskeletal membrane protein. EMBO J. 27:3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao L., Tanzer J.M., and MacAlister T.J.1987. Bicarbonate and potassium regulation of the shape of Streptococcus mutans NCTC 10449S. J. Bacteriol. 169:2543–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao L., MacAlister T.J., and Tanzer J.M.1988. Factors influencing cell shape in the mutans group of streptococci. J. Bacteriol. 170:3752–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thibessard A., Fernandez A., Gintz B., Leblond-Bourget N., and Decaris B.2002. Effects of rodA and pbp2b disruption on cell morphology and oxidative stress response of Streptococcus thermophilus CNRZ368. J. Bacteriol. 184:2821–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Typas A., Banzhaf M., Gross C.A., and Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10:123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Ent F., Johnson C.M., Persons L., de Boer P., and Lowe J.2010. Bacterial actin MreB assembles in complex with cell shape protein RodZ. EMBO J. 29:1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Ploeg R., Verheul J., Vischer N.O., Alexeeva S., Hoogendoorn E., Postma M., Banzhaf M., Vollmer W., and den Blaauwen T. 2013. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol. Microbiol. 87:1074–1087 [DOI] [PubMed] [Google Scholar]
- 30.Varma A., and Young K.D.2009. In Escherichia coli, MreB and FtsZ direct the synthesis of lateral cell wall via independent pathways that require PBP 2. J. Bacteriol. 191:3526–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vats P., Shih Y.L., and Rothfield L.2009. Assembly of the MreB-associated cytoskeletal ring of Escherichia coli. Mol. Microbiol. 72:170–182 [DOI] [PubMed] [Google Scholar]
- 32.Wachi M., Doi M., Tamaki S., Park W., Nakajima-Iijima S., and Matsuhashi M.1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheeler R., Mesnage S., Boneca I.G., Hobbs J.K., and Foster S.J. 2011. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol. Microbiol. 82:1096–1109 [DOI] [PubMed] [Google Scholar]
- 34.Young K.D.2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70:660–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young K.D.2010. Bacterial shape: two-dimensional questions and possibilities. Annu. Rev. Microbiol. 64:223–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zapun A., Contreras-Martel C., and Vernet T.2008. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]
- 37.Zapun A., Philippe J., Abrahams K.A., Signor L., Roper D.I., Breukink E., and Vernet T. 2013. In vitro reconstitution of peptidoglycan assembly from the gram-positive pathogen Streptococcus pneumoniae. ACS Chem. Biol. 8:2688–2696 [DOI] [PubMed] [Google Scholar]
- 38.Zapun A., Vernet T., and Pinho M.G.2008. The different shapes of cocci. FEMS Microbiol. Rev. 32:345–360 [DOI] [PubMed] [Google Scholar]
- 39.Zerfass I., Hakenbeck R., and Denapaite D.2009. An important site in PBP2x of penicillin-resistant clinical isolates of Streptococcus pneumoniae: mutational analysis of Thr338. Antimicrob. Agents Chemother. 53:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]