Abstract
Bacillus anthracis has a complex cell wall structure composed of a peptidoglycan (PG) layer to which major structures are anchored such as a neutral polysaccharide, an S-layer, and a poly-γ-D-glutamate (PDGA) capsule. Many of these structures have central roles in the biology of B. anthracis, particularly, in virulence. However, little attention has been devoted to structurally study the PG and how it is modified in the presence of these secondary cell wall components. We present here the fine structure of the PG of the encapsulated RPG1 strain harboring both pXO1 and pXO2 virulence plasmids. We show that B. anthracis has a high degree of cross-linking and its GlcNAc residues are highly modified by N-deacetylation. The PG composition is not dependent on the presence of either LPXTG proteins or the capsule. Using NMR analysis of the PG-PDGA complex, we provide evidence for the anchoring of the PDGA to the glucosamine residues. We show that anchoring of the PDGA capsule is impaired in two PG N-deacetylase mutants, Ba1961 and Ba3679. Thus, these multiple N-deactylase activities would constitute excellent drug targets in B. anthracis by simultaneously affecting its resistance to lysozyme and to phagocytosis impairing B. anthracis survival in the host.
Introduction
Bacillus cereus group comprises, among others, three well-known species, Bacillus anthracis, Bacillus thuringiensis, and Bacillus cereus sensu stricto. B. anthracis is pathogenic for mammals, including man, eliciting anthrax, B. thuringiensis is an entomopathogen, and B. cereus is a ubiquitous soil bacterium, some strains being opportunistic pathogens, causing gastrointestinal and nongastrointestinal infections.25,31 On genetic basis, all three are the same species.24 Some, but not all strains, harbor virulence plasmids, which specify the pathogenic phenotypes.19,26 B. thuringiensis strains possess plasmids responsible for the toxin Cry production, B. cereus sensu sricto may harbor toxin synthetic operon or pXO1- and pXO2-like plasmids, and B. anthracis possesses pXO1 and pXO2.19 The B. anthracis plasmids carry the genetic components responsible, respectively, for the production of a tripartite toxin (composed of protective antigen, edematogen factor, and lethal factor) and of a poly-γ-D-glutamate (PDGA) capsule (synthesized by CapB, CapC, CapA, and CapE and anchored by CapD).7,8 Tripartite toxin and the capsule are the two major B. anthracis virulence factors. Another difference between these Bacillus species lies in the regulatory networks. Indeed, a pleiotropic regulator PlcR activates many B. cereus and B. thuringiensis genes encoding surface or extracellular proteins (including virulence factors), whereas this regulator is inactivated in B. anthracis.1,26,30 Instead, another pleiotropic regulator AtxA (atxA is carried on pXO1) is found to regulate more than 70 genes comprising the major virulence factors of B. anthracis,4,17,23,40 (for review,15,18). Finally, bicarbonate was shown to modulate the expression of an important number of genes.9,17,38
B. anthracis surface is central to host defenses and to bacterial survival. It is composed of, outside-in, a PDGA capsule, an S-layer, that is a bidimensional proteinic crystal, a peptidoglycan (PG) layer, and the membrane.14,16 The S-layers and all the SLH-harboring proteins are noncovalently anchored to the PG,28,29 whereas LPXTG proteins are covalently anchored by three sortases (encoded by srt genes) to the PG.2,11,20 These proteins are potentially involved in virulence.42 Finally, the PDGA filaments (capsule constituents) are directly and covalently linked to the PG.7 In fact, a preparation of PDGA covalently linked to PG affords protection against anthrax.6 PG is therefore a key element of B. anthracis surface and virulence.
Despite the central role of the PG layer in organizing the surface structure of B. anthracis, there are few studies addressing its fine structure. The B. cereus RSVF1 was considered by Severin et al. to be a B. anthracis cured of its virulence plasmids and, therefore, the B. cereus RSVF1 PG composition was analyzed as a surrogate.37 The Bacillus PG belongs to the A1γ, meaning that peptidic parts are directly linked between the positions 3 and 4 containing the meso diaminopemilic acid (mesoDAP).36,37 However, in the absence of the covalently anchored capsule, the study does not reflect the exact fine structure of B. anthracis PG. In fact, the PDGA capsule is directly anchored by the gamma-glutamyl transpeptidase (GGT) CapD to the PG.5,7 It has been shown in vitro that CapD can anchor the PDGA to mesoDAP residues with a free e-amino group,35 although direct in vivo evidence is still missing.
In addition, polysaccharide polymers covalently linked through phosphodiester linkage to B. anthracis PG consist of →6)-α-GlcNAc-(1 →4)-β-ManNAc-(1 →4)-β-GlcNAc-(1→in which the α-GlcNAc residue is substituted with α-galactose and β-galactose at O-3 and O-4, respectively, and the β-GlcNAc substituted with α-galactose at O-3.10 This neutral polysaccharide is the anchoring site of SLH-containing proteins.28,29 Finally, it has been proposed that glucosamine residues are in majority N-deacetylated.37,43 This suggestion was reinforced by the fact that B. cereus and B. anthracis possess ten GlcNAc N-deacetylases of which five belong to the GlcNAc PG N-deacetylase subfamily.34 Recently, Balomenou et al. have shown that of these five GlcNAc N-deacetylases, three have the PG as substrate and two act on the neutral polysaccharide.3 Among the GlcNAc N-deacetylases modifying the PG, BA1977 is involved in resistance to host lysozyme, whereas BA1961 and BA3679 contribute to the PG biogenesis.
Here, we report the analysis of the fine structure of the B. anthracis PG in an encapsulated strain and show that it is similar to that in an isogenic mutant where the PDGA is not synthesized and to that in all sortase mutant strains. We studied the anchoring site of the capsule and we provide evidence for the role of two GlcNAc N-deacetylases in modulating the anchoring of the capsule to the surface of B. anthracis. Our data suggest that targeting these activities would not only affect bacterial cell shape,3 but also impair capsule anchoring or possibly synthesis, affecting its anti-phagocytic role.
Results
PG composition and structure
Despite the central role in virulence of the PDGA capsule, there is no direct evidence on its precise site of in vivo anchoring to the PG. To address this issue, we attempted to characterize the fine structure of the PG of B. anthracis in the presence and in the absence of the capsule. First, to get rid of capsule and yet grow cells in physiological conditions, a capB mutant (RTC10) was used to analyze the PG composition and structure. B. anthracis RTC10 was grown in the presence of bicarbonate at 37°C to mimic the environmental conditions found in the host and PG was purified thereafter.
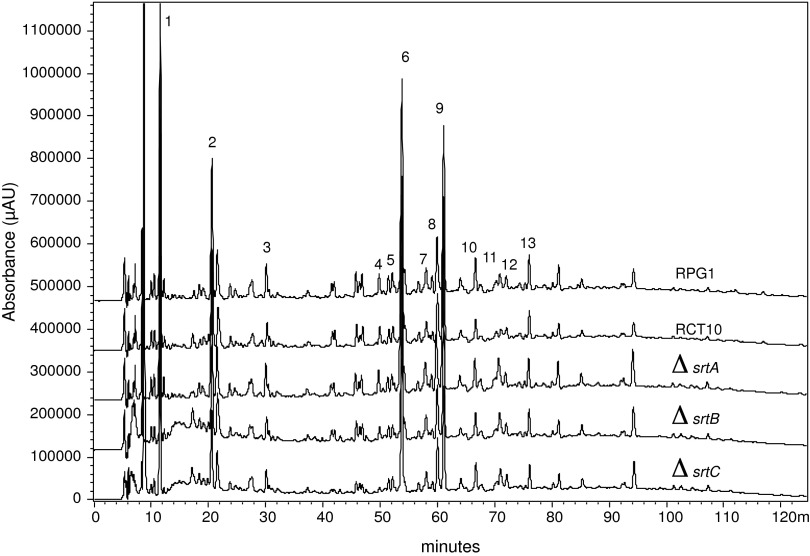
After PG purification, a first global amino acid analysis was done to verify the amino acids contained in B. anthracis PG. Only alanine, glutamate, and mesoDAP were found confirming the global composition of amino acid classically found in Bacillus PG. PG was then hydrolyzed with mutanolysin; muropeptides were purified and analyzed by mass spectrometry. An average of 35% of muropeptides were in monomer form (Fig. 1, peaks 1 to 3). Other muropeptides were dimers (55%) and trimers (10%). These percentages defined the cross-linkage of the wall at approximately 32%. Although this cross-linkage is high, no tetramers or higher multimers were found. All peaks corresponded to one muropeptide, except for the peak 9 that contained two muropeptides separated after desalting (Fig. 1 and Table 1). Monomers were composed of tripeptides (peak 1, 55%), tetrapeptides (peak 2, 34%), and pentapeptides (peak 3, 11%). No dipeptides were detected. Dimers were composed of tetra-tripeptides (peaks 4 to 6, 24.5%), tetra-tetrapeptides (peaks 7 to 9, 18.5%), and penta-tetrapeptide (peak 7 and 10, 3.5%). Two unusual muropeptides were found in minority in peaks 9 and 12. After mass spectrometry analysis, the corresponding mass was found to be respectively a penta-tetrapeptide and a tetra-tetrapeptide minus 14 daltons. It has been described in some bacteria that glycine can replace the terminal alanine.36 The difference between alanine and glycine is 14 daltons. These peaks were therefore assumed to be a tetra-glycine-tetrapeptide (peak 9, 2%) and tri-glycine-tetrapeptide (peak 12, 2.5%). This result was confirmed by MS/MS spectrometry (data not shown). The peptidic portion of PG was not modified by amidation. In contrast, the GlcNAc was vastly deacetylated (92%) as described previously by Zipperle et al.43 Indeed, all monomers were deacetylated (Table 1). All dimers were di-deacetylated except for a minority of tri-tetrapeptide, tetra-tetrapeptide, and penta-tetrapeptide that were found under a mono-acetylated form.
FIG. 1.
Muropeptide composition of Bacillus anthracis RCT10 (ΔcapB) mutant strain. The PG was digested with mutanolysin, reduced with sodium borohydride and separated by reverse-phase high-performance liquid chromatography using a hypersyl ODS18 column (A). The muropeptides were collected, desalted, and analyzed by MALDI-TOF and MALDI-MS/MS (Table 1). The assignment of the different peaks structures are presented in (B). G and M stand for GlcNAc (or glucosamine) and MurNAc, respectively. Ala, Gly A2pm stand for alanine, glutamate, and mesoDAP, respectively. Note that 1 volt corresponds to 1 absorbance unit at 206 nm.
Table 1.
Composition and Degree of N-Deacetylation of the Muropeptides of Bacillus anthracis
| Molecular mass | |||||
|---|---|---|---|---|---|
| Peak | Percentage of total UV | Muropeptide structure | Theoretical | Experimental | Degree of acetylation |
| 1 | 19.59 | Tripeptide | 851.35 | 851.37 | Deacetylate |
| 2 | 12.05 | Tetrapeptide | 922.39 | 922.35 | Deacetylate |
| 3 | 3.81 | Pentapeptide | 993.42 | 993.43 | Deacetylate |
| 4 | 2.95 | Tetra-tripeptide | 1732.74 | 1732.73 | Di-deacetylate |
| 5 | 1.33 | Tetra-tripeptide | 1774.74 | 1774.73 | Mono-deacetylate |
| 6 | 20.39 | Tetra-tripeptide | 1732.74 | 1732.75 | Di-deacetylate |
| 7 | 6.73 | Tetra-tetrapeptide | 1845.78 | 1845.73 | Mono-deacetylate |
| Traces | Penta-tetrapeptide | 1916.82 | 1917.01 | Mono-deacetylate | |
| 8 | 3.55 | Tetra-tetrapeptide | 1803.77 | 1803.79 | Di-deacetylate |
| 9 | 11.47 | Tetra-tetrapeptide | 1803.77 | 1803.73 | Di-deacetylate |
| 2.27 | Tetra-Gly-tetrapeptide | 1860.82 | 1860.74 | Di-deacetylate | |
| 10 | 3.60 | Penta-tetrapeptide | 1874.81 | 1874.81 | Di-deacetylate |
| 11 | 6.53 | Tetra-tetra-tripeptide | 2614.12 | 2615.15 | Tri-deacetylate |
| 12 | 2.49 | Tetra-tri-Gly-peptide | 1789.79 | 1790.78 | Di-deacetylate |
| 13 | 3.24 | Tetra-tetra-tetrapeptide | 2685.16 | 2686.17 | Tri-deacetylate |
As differences of regulation occur with or without bicarbonate, we decided to analyze B. anthracis PG from RTC10 grown in the absence of bicarbonate. The muropeptide profiles of PG purified after culture with or without bicarbonate were identical (data not shown).
LPXTG proteins, for the most, have no known function, but, because of their localization, they may have, directly or not, an impact on PG maturation. Consequently, we assessed their role in PG composition by analyzing it in each of the three sortase mutant strains grown in the presence of bicarbonate. All muropeptide profiles analyzed were identical (Fig. 2). This result suggests that LPXTG proteins have no impact on PG composition.
FIG. 2.
Muropeptide profile of different B. anthracis mutants. The muropeptide profile of the parental strain RPG1 was compared to its isogenic mutants RTC10 (ΔcapB), RSBON30 (ΔsrtA), RSBTR30 (ΔsrtB), and RSBTO30 (ΔsrtC). The mutants produced muropeptide profiles similar to those of the parental strain. The peak numbers correspond to the peak nomenclature of Figure 1. Note that 1 volt corresponds to 1 absorbance unit at 206 nm.
PG composition and structure in the presence of anchored PDGA
To analyze whether the presence of PDGA disturbs the PG composition and structure, we extracted PG from encapsulated RPG1 strain grown in the presence of bicarbonate. As shown previously, PDGA was found at this step linked to PG.7 PDGA presence was therefore tested after elution of each peak by dot-blots with antibodies specifically directed against PDGA, as previously described.7 PDGA was found to elute before 10 minutes (data not shown) with salts and highly charged molecules. Analysis of the muropeptide profile indicated that there were no differences between this PG and that extracted from the capB mutant strain (Fig. 2), indicating that PDGA anchoring did not modify the global PG composition and structure. Due to technical difficulties for PG analysis by mass spectrometry in the presence of salts, we were unable to determine the exact PDGA anchoring site to PG.
PDGA anchoring site
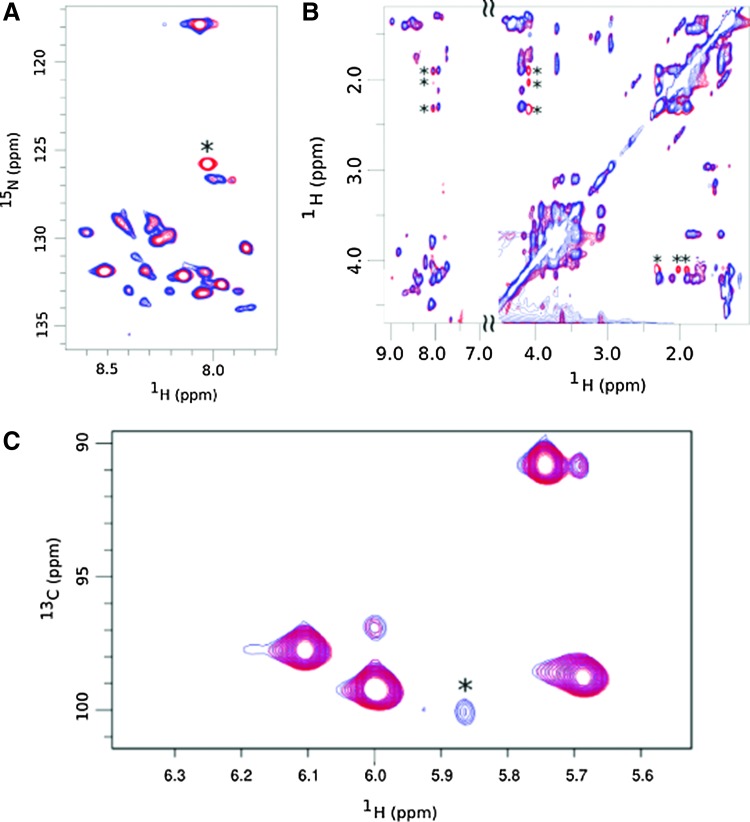
Since the fine structure analysis of the PG by high-performance liquid chromatography (HPLC) did not yield information on the nature of the anchoring site of the capsule to the PG, we decided to study the structural environment of the PG by NMR. The direct and covalent anchoring of the capsule to the PG is predicted to affect the chemical shift of the neighboring residues. For example, based on the work of Richter et al.,35 we anticipated observing changes in the environment of mesoDAP residues in the presence or absence of the capsule. Bacteria were grown in conditions such that macromolecules, including PG and PDGA, were labeled with 13C and 15N and the PG was purified from the parental RPG1 strain and RTC10, its isogenic capB derived strain. We analyzed the NMR spectra using 2D-TOCSY. In the proton–nitrogen and proton–proton correlation spectra (Fig. 3A, B, respectively), comparison of the capsulated and noncapsulated strain was readily observed for the PGDA (asterisk). In the NH region, the two peaks corresponding to backbone amide and the lateral amide can be assigned to the mesoDAP residues. No difference could be observed between the capsulated and noncapsulated strain. In the proton–carbon correlation spectra (Fig. 3C), we observed in the region corresponding to the anomeric protons of the sugars a clear change in the chemical shift between capsulated and noncapsulated strain, suggesting that the presence of the PDGA was changing the chemical environment of the sugar backbone. Anchoring of the PGDA to the PG layer is performed by CapD, a GGT.7,35 This requires a free amine group on the PG layer. Our NMR data suggest that the capsule is anchored to the glycan backbone of the PG layer. The glycan backbone is composed of repeating units of the N-GlcNAc ß-1,4 linked to N-acetylmuramic acid (MurNAc). The glycan backbone has been shown by others and this work to be extensively N-deacetylated (Table 1) at the GlcNAc residues freeing the amine group at C2 of the GlcNAc.
FIG. 3.
Comparison of peptidoglycan and polyglutamate linked peptidoglycan using NMR (A) 15N-1H correlation recorded on muropeptide prepared from digestion of the cell wall of the RPG1 strain (red) and the RTC10 mutant (blue). The amide correlation of the polyglutamate is indicated by a star. (B) 1H-1H TOCSY experiments carried out with the PG from the RPG1 strain (red) and the RTC10 mutant (blue) in the same conditions. Extra correlation peaks obtained for the RPG1 strain can be assigned to the HN, CHα, CH2β, and CH2γ glutamate resonances and are indicated by stars. (C) Difference observed for the sugar in the 13C-1H correlation recorded on the RPG1 and RTC10 peptidoglycans. Region of the 13C-1H correlation recorded on muropeptide prepared from digestion of the cell wall of the RPG1 strain (red) and the RTC10 mutant (blue) in the same conditions. 1H-13C anomeric correlation only present in the RTC10 mutant is indicated by an asterisk.
Previously, we have shown that N-deacetylation of the PG of B. anthracis is performed by at least three distinct N-deacetylases, BA1961, BA1977, and BA3679.3 Given our NMR data, we tested whether the N-deacetylase mutants could be involved in the anchoring of the PDGA capsule to the PG. We grew the different strains (parental and the single mutant strains) in the presence of bicarbonate at 37°C to mimic the environmental conditions that induce the synthesis of the capsule. The presence of the capsule was visualized by staining the bacteria with India ink (Fig. 4). Both the parental RPG1 strain and the RΔba2944 mutant strain, which was shown to be involved in modification of the neutral polysaccharide and used as a control, displayed capsule surrounding homogeneously the bacilli (Fig. 4A, B). RΔba1977, RΔba1961, and RΔba3679, all three harboring a modified PG, also produced a capsule. However, RΔba1961 and RΔba3679, both known to be impaired in the biogenesis of the PG, produced cells with a heterogeneous distribution of PDGA on the surface (Fig. 4D, E), while RΔba1977, which is sensitive to lysozyme, behaved like the parental strain (Fig. 4C). These results suggest that both RΔba1961 and RΔba3679 are impaired in PDGA anchoring to the cell surface of B. anthracis and/or in its synthesis.
FIG. 4.
Visualization of the PDGA capsule by India ink staining. B. anthracis bacilli grown in the presence of bicarbonate were observed in the presence of India ink at the same magnification. RPG1 (A), RΔba2944 (B), and RΔba1977 (C) harbored a capsule, whereas RΔba1961 (D) and RΔba3679 (E) strains display heterogeneous and discontinuous capsule.
Discussion
Despite the importance of B. cereus sensu lato species, and, in particular, of B. anthracis as a pathogen, few studies have addressed the fine structure of the major cell wall component of this bacterium, the PG. A pioneering study by Zipperle43 showed that the global composition was similar to the one found in the Bacillus group. The PG of the Sterne strain (also known as strain 7702) was characterized by a high degree of N-deacetylation at both the GlcNAc (88%) and the MurNAc (34%) and by a low degree of cross-linking. However, this study used crude and indirect methods to probe the level of cross-linking and N-deacetylation. Furthermore, the Sterne strain does not carry the pXO2 virulence plasmid encoding the capsule biosynthetic operon.
More recently, Severin et al. reported the fine structure of the PG of B. cereus RSVF137 arguing that this strain could serve as a surrogate for B. anthracis. The HPLC and mass spectrometry analysis of the muropeptides of strain RSVF1 confirmed the high level of N-deacetylation of the PG. Furthermore, the authors observed a high degree of amidation of the stem peptide. A major limitation of this work was the choice of the strain. Indeed, although closely related to the clonal B. anthracis, strain RSVF1 does not belong to the same lineage and does not carry the two virulence plasmids pXO1 and pXO2.
Thus, we determined the structure of the PG of the virulent B. anthracis strain RPG1. Our results showed significant differences with both previous studies. First, we observed N-deacetylation exclusively of the GlcNAc residues (and none of the MurNAc) and the levels were not identical to those previously described. Second, we did not observe any amidation of the stem peptide in contrast to the Severin study.37 Third, we observed a high degree of cross-linking compared with the Zipperle study.43 Fourth, we isolated muropeptides indicative of glycine presence into the stem peptide. These three studies provide common features of the PG of B. anthracis and B. cereus such as the high degree of N-deacetylation of the GlcNAc conferring high resistance to lysozyme. They also highlight that despite the phylogenetic relationship between these species, there are strain-dependent variations in the PG composition in the B. cereus group, warranting a wider study of the PG composition of strains representing the diversity of the B. cereus group. For example, we have recently published the muropeptide profile of strain 7702.3 Comparison between 77023 and RPG1 (Fig. 2 and data not shown for growth without bicarbonate) indicates that the global composition is similar (similar cross-linking and similar N-deacetylation). However, strain 7702 has more muropeptides with tetrapeptides while RPG1 has more of the tripeptides. These differences could be attributed to the presence of pXO2 in RPG1. Interestingly, the monomer GM-tripeptide is the preferred Nod1 ligand.22 Thus, many of these variations could be under selective pressure such as the immune system of their natural hosts (insects for B. thuringiensis and mammals for B. anthracis).
Next, we attempted to structurally characterize the anchoring site of PDGA to B. anthracis PG. The previous work by Richter et al. proposed PDGA to be anchored to the mesoDAP residues of the stem peptides.35 However, these experiments were done in vitro using small PDGA filaments, which may change the substrate preference. Currently, there is no direct evidence on the preferred anchoring site of PGDA in vivo. Serological detection of the PDGA along the entire HPLC profile of B. anthracis muropeptides indicated it eluted with the flow-through fraction containing the salts precluding any possible MALDI-TOF and MS/MS analysis. CapD as a GGT cleaves the PDGA and transfers it, either to H2O leading to hydrolysis, or to an acceptor molecule leading to a transpeptidation. According to Richter and collaborators, CapD is able to act as an endopeptidase cleaving PDGA into two PDGA filaments.35 It is noteworthy that classical GGTs transfer only a glutamyl while CapD transfers a filament of PDGA.32 During this reaction CapD forms an acyl-intermediate with PDGA as already described for GGTs. Richter et al. showed that in vitro CapD can transfer a cleaved PDGA molecule onto the mesoDAP of B. anthracis PG.35 However, the acceptor group may also be the free amine of N-deacetylated GlcNAc residues. Thus, we attempted to characterize PDGA anchoring to the PG by directly purifying PDGA-harboring PG from the bacilli and using NMR analysis. Surprisingly, no impact on the mesoDAP due to the PDGA presence was noticed. Instead, chemical shifts corresponding to the sugar backbone were observed. Our results suggest that in vivo CapD covalently anchors the PDGA filaments to the glucosamine of the B. anthracis PG rather than on the mesoDAP as observed in vitro. The transfer on mesoDAP may also exist in vivo albeit with a frequency such that it was not observed in our analysis. To further support this conclusion, we show that two of the PG N-deacetylases, BA1961 and BA3679, are involved in the anchoring of PGDA to the B. anthracis cell surface. Thus, BA1961 and BA3679 would generate free amino groups by N-deacetylating the GlcNAc residues creating the site on the PG on which CapD anchors PDGA. This is reminiscent of their role in modifying the PG of B. anthracis spores.3 In the absence of both activities, the PG of the spores is no longer GlcNAc N-deacetylated precluding the sequential action of the amidase CwlD and of the MurNAc N-deacetylase PdaA required for the formation of the ∂-lactam muramic acid. Alternatively, our results cannot exclude that due to the cell shape defect in BA1691 and BA3679 mutants, nonmodified PG might impact on the whole cell wall machinery including the PDGA synthetic machinery through sensing mechanisms such as the WalKR two-component system.13 We observed a heterogeneous distribution of the PDGA in the N-deacetylase mutants clearly distinct from a capD phenotype7 suggesting that PG N-deacetylation probably interferes with the site of PDGA anchoring and eventually of its synthesis.
In conclusion, N-deacetylases are widely distributed in bacteria and contribute to pathogens virulence by escaping the host innate immunity. Thus, strategies aiming these activities would have a dramatic impact on B. anthracis survival by simultaneously targeting mechanisms of resistance to lysozyme and to phagocytosis.
Materials and Methods
Strains and growth conditions
Escherichia coli HB101 harboring pRK24 was used for mating experiments.39 B. anthracis RPG1 strain was used for the study.21 This strain possesses both virulence plasmids pXO1 and pXO2; however, because of point mutations in cya and lef, encoding the edematogen and lethal factors, respectively, nonfunctional toxins are produced. The capB mutant strain (RTC10) derives from the RPG1 strain.8 The strains RΔba1961, RΔba1977, RΔba2929, and RΔba3679 are deleted in ba1961, ba1977, ba2929, and ba3679 encoding N-deacetylases.3
The srt mutants were constructed in the RPG1 background using recombinant plasmids previously described, namely pSON30, pSTR30, and pSTO30,11 to yield RSBON30 (RPG1 ΔsrtA), RSBTR30 (RPG1 ΔsrtB), and RSBTO30 (RPG1 ΔsrtC). Recombinant plasmids were transferred from E. coli to B. anthracis by heterogramic conjugation.39 Allelic exchange was carried out as described previously.33
B. anthracis was grown in BHI (Difco) medium without or with addition of sodium bicarbonate (0.6%).
Labeling PG with15N and 13C was carried out as described previously.27 We grew E. coli up to stationary phase in minimal M9 medium in which we replaced the normal glucose and NH4Cl by 13C-glucose and 15NH4Cl, respectively. Bacteria were harvested by centrifugation, resuspended in 15 ml of Tris-HCl pH 7.5 25 mM and NaCl 150 mM, and sonicated. The lysate was treated with DNAseI and RNAseA for 1 hour at 37°C followed by proteinase K overnight at 37°C. Remaining cellular debris were removed by centrifugation, the soluble lysate was autoclaved 15 minutes at 105°C and lyophilized. B. anthracis was then grown in a R-bic-like medium composed of 50 mM potassium phosphate buffer, 15NH4Cl (0.6 g/L), 3C-glucose (0.25%), thiamine (1 g/L), MgSO4 (17.5 mg/L), MnSO4 (0.9 mg/L), CaCl2 (7.5 mg/L), E. coli lysate extracts (0.5%), and sodium bicarbonate (0.6%). Bacterial cells grown under these conditions were harvested to prepare the labeled PG and PGDA used for the NMR analysis.
Capsule visualization
B. anthracis capsule was visualized using India ink as previously described.7
PG extraction and analysis
PG was extracted as described previously from 200 ml stationary phase cultures.7 PG was digested with mutanolysin treatment (50 μg/ml) and further reduced with sodium borohydride. Muropeptide were then separated on phase reverse Hypersil ODS18 column (250 by 4.6 mm, 3 μm particular size) with 0–15% methanol gradient and 4.3–5 pH sodium phosphate gradient. Chromatograms were obtained at λ=206 nm. Separated muropeptides were desalted on the same column, but with 0.05% trifluoric acid with a 0–25% acetonitrile gradient. They were finally analyzed by mass spectrometry with a Voyager DE-STR MALDI-TOF from the Proteomics platform at Institut Pasteur. Amino acid analysis was performed by “unité de Chimie Organique” at the Institut Pasteur, using the ninhydrin technique and a Beckman 6300 analyzer.
All NMR data were collected at 25°C in the same conditions on two muropeptide samples obtained by digestion of the 13C,15N-labeled cell wall of the RPG1 and the RTC10 B. anthracis strains. The sample was dissolved in 100 mM sodium phosphate buffer, pH 6.5 containing 10% D2O. Spectra were recorded on Agilent spectrometers equipped with cryogenic triple 1H, 13C, 15N resonance probes operating at 600 MHz1H NMR frequencies. TOCSY experiments were performed with a DIPSI mixing time of 70 ms and an experimental time of 6 hours. 15N-1H and 13C-1H Sensitivity enhanced HSQC were recorded with an experimental time of 15 minutes and 2 hours, respectively. NMR data were processed with NMRPipe12 and analyzed using the CcpNmr Analysis 2.2 software.41
Acknowledgments
The Proteomics Plateform and Unité de Chimie Organique from Institut Pasteur are thanked for MS and amino acids analysis. TC was funded by Caisse Nationale d'Assurance Maladie des nonsalariés (CANAM). SB was supported by a short-term EMBO fellowship. Work in IGB laboratory was supported by an ERC starting Grant (PGNfromSHAPEtoVIR 202283). The work of SB and VB was co-financed by EU (ERDF) and Greek funds through the “THALIS” program. We thank Michèle Mock and Agnès Labigne, in whose laboratories part of this work was carried out. Catherine Chaput is also thanked for her help with the HPLC manipulation.
Disclosure Statement
No competing financial interests exist.
References
- 1.Agaisse H., Gominet M., Okstad O.A., Kolsto A.B., and Lereclus D. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043–1053 [DOI] [PubMed] [Google Scholar]
- 2.Aucher W., Davison S., and Fouet A.2011. Characterization of the Sortase Repertoire in Bacillus anthracis. PLoS One 6:e27411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balomenou S., Fouet A., Tzanodaskalaki M., Couture-Tosi E., Bouriotis V., and Boneca I.G.2013. Distinct functions of polysaccharide deacetylases in cell shape, neutral polysaccharide synthesis and virulence of Bacillus anthracis. Mol. Microbiol. 87:867–883 [DOI] [PubMed] [Google Scholar]
- 4.Bourgogne A., Drysdale M., Hilsenbeck S.G., Peterson S.N., and Koehler T.M. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Candela T.2012. CapD Poly-γ-glutamate Hydrolase/Transpeptidase. In: Rawlings and Salvesen (ed.), Handbook of Proteolytic Enzymes, 3rd Edition. Elsevier, United Kingdom, pp. 3719–3722 [Google Scholar]
- 6.Candela T., Dumetz F., Tosi-Couture E., Mock M., Goossens P.L., and Fouet A. 2012. Cell-wall preparation containing poly-gamma-D-glutamate covalently linked to peptidoglycan, a straightforward extractable molecule, protects mice against experimental anthrax infection. Vaccine 31:171–175 [DOI] [PubMed] [Google Scholar]
- 7.Candela T., and Fouet A.2005. Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol. Microbiol. 57:717–726 [DOI] [PubMed] [Google Scholar]
- 8.Candela T., Mock M., and Fouet A.2005. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J. Bacteriol. 187:7765–7772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chitlaru T., Gat O., Gozlan Y., Ariel N., and Shafferman A.2006. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J. Bacteriol. 188:3551–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choudhury B., Leoff C., Saile E., Wilkins P., Quinn C.P., Kannenberg E.L., and Carlson R.W. 2006. The structure of the major cell wall polysaccharide of Bacillus anthracis is species-specific. J. Biol. Chem. 281:27932–27941 [DOI] [PubMed] [Google Scholar]
- 11.Davison S., Couture-Tosi E., Candela T., Mock M., and Fouet A.2005. Identification of the Bacillus anthracis (gamma) phage receptor. J. Bacteriol. 187:6742–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., and Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 13.Dubrac S., Bisicchia P., Devine K.M., and Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70:1307–1322 [DOI] [PubMed] [Google Scholar]
- 14.Fouet A. 2009. The surface of Bacillus anthracis. Mol. Aspects Med. 30:374–385 [DOI] [PubMed] [Google Scholar]
- 15.Fouet A. 2010. AtxA, a Bacillus anthracis global virulence regulator. Res. Microbiol. 161:735–742 [DOI] [PubMed] [Google Scholar]
- 16.Fouet A., and Mesnage S.2002. Bacillus anthracis cell envelope components. Curr. Top. Microbiol. Immunol. 271:87–113 [DOI] [PubMed] [Google Scholar]
- 17.Fouet A., and Mock M.1996. Differential influence of the two Bacillus anthracis plasmids on regulation of virulence gene expression. Infect. Immun. 64:4928–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouet A., and Mock M.2006. Regulatory networks for virulence and persistence of Bacillus anthracis. Curr. Opin. Microbiol. 9:160–166 [DOI] [PubMed] [Google Scholar]
- 19.Fouet A., and Moya M. 2008. Virulence Megaplasmids in Bacillus anthracis and their relatives in the Bacillus cereus group. In: Schwartz (ed.), Microbial Megaplasmids. Springer, Münster, pp. 187–206 [Google Scholar]
- 20.Gaspar A.H., Marraffini L.A., Glass E.M., Debord K.L., Ton-That H., and Schneewind O. 2005. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J. Bacteriol. 187:4646–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimenez A.P., Wu Y.Z., Paya M., Delclaux C., Touqui L., and Goossens P.L. 2004. High bactericidal efficiency of type iia phospholipase A2 against Bacillus anthracis and inhibition of its secretion by the lethal toxin. J. Immunol. 173:521–530 [DOI] [PubMed] [Google Scholar]
- 22.Girardin S.E., Boneca I.G., Carneiro L.A., Antignac A., Jehanno M., Viala J., Tedin K., Taha M.K., Labigne A., Zahringer U., and others. 2003. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science 300:1584–1587 [DOI] [PubMed] [Google Scholar]
- 23.Guignot J., Mock M., and Fouet A.1997. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiol. Lett. 147:203–207 [DOI] [PubMed] [Google Scholar]
- 24.Helgason E., Okstad O.A., Caugant D.A., Johansen H.A., Fouet A., Mock M., Hegna I., and Kolsto A.B. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen G.B., Hansen B.M., Eilenberg J., and Mahillon J. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631–640 [DOI] [PubMed] [Google Scholar]
- 26.Kolsto A.B., Tourasse N.J., and Okstad O.A. 2009. What sets Bacillus anthracis apart from other Bacillus species?. Annu. Rev. Microbiol. 63:451–476 [DOI] [PubMed] [Google Scholar]
- 27.Lecoq L., Bougault C., Kern T., Hugonnet J.E., Veckerle C., Pessey O., Arthur M., and Simorre J.P. 2012. Backbone and side-chain 1H, 15N and 13C assignment of apo- and imipenem-acylated L,D-transpeptidase from Bacillus subtilis. Biomol. NMR Assign. 6:205–208 [DOI] [PubMed] [Google Scholar]
- 28.Mesnage S., Fontaine T., Mignot T., Delepierre M., Mock M., and Fouet A.2000. Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19:4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesnage S., Tosi-Couture E., and Fouet A.1999. Production and cell surface anchoring of functional fusions between the SLH motifs of the Bacillus anthracis S-layer proteins and the Bacillus subtilis levansucrase. Mol. Microbiol. 31:927–936 [DOI] [PubMed] [Google Scholar]
- 30.Mignot T., Mock M., Robichon D., Landier A., Lereclus D., and Fouet A.2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189–1198 [DOI] [PubMed] [Google Scholar]
- 31.Mock M., and Fouet A.2001. Anthrax. Annu Rev Microbiol 55:647–671 [DOI] [PubMed] [Google Scholar]
- 32.Okada T., Suzuki H., Wada K., Kumagai H., and Fukuyama K.2006. Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc. Natl. Acad. Sci. U. S. A. 103:6471–6476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pezard C., Berche P., and Mock M.1991. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 59:3472–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Psylinakis E., Boneca I.G., Mavromatis K., Deli A., Hayhurst E., Foster S.J., Varum K.M., and Bouriotis V. 2005. Peptidoglycan N-acetylglucosamine deacetylases from Bacillus cereus, highly conserved proteins in Bacillus anthracis. J. Biol. Chem. 280:30856–30863 [DOI] [PubMed] [Google Scholar]
- 35.Richter S., Anderson V.J., Garufi G., Lu L., Budzik J.M., Joachimiak A., He C., Schneewind O., and Missiakas D. 2009. Capsule anchoring in Bacillus anthracis occurs by a transpeptidation reaction that is inhibited by capsidin. Mol. Microbiol. 71:404–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schleifer K.H., and Kandler O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Severin A., Tabei K., and Tomasz A.2004. The structure of the cell wall peptidoglycan of Bacillus cereus RSVF1, a strain closely related to Bacillus anthracis. Microb. Drug Resist. 10:77–82 [DOI] [PubMed] [Google Scholar]
- 38.Sirard J.C., Mock M., and Fouet A. 1994. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J. Bacteriol. 176:5188–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trieu-Cuot P., Carlier C., Martin P., and Courvalin P.1987. Plasmid transfer by conjugation from Escherichia coli to Gram-positive bacteria. FEMS Microbiol. Lett. 48:289–294 [Google Scholar]
- 40.Uchida I., Makino S., Sekizaki T., and Terakado N.1997. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Mol. Microbiol. 23:1229–1240 [DOI] [PubMed] [Google Scholar]
- 41.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., Ulrich E.L., Markley J.L., Ionides J., and Laue E.D. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696 [DOI] [PubMed] [Google Scholar]
- 42.Xu Y., Liang X., Chen Y., Koehler T.M., and Hook M. 2004. Identification and biochemical characterization of two novel collagen binding MSCRAMMs of Bacillus anthracis. J. Biol. Chem. 279:51760–51768 [DOI] [PubMed] [Google Scholar]
- 43.Zipperle G.F., Jr., Ezzell J.W., Jr., and Doyle R.J. 1984. Glucosamine substitution and muramidase susceptibility in Bacillus anthracis. Can. J. Microbiol. 30:553–559 [DOI] [PubMed] [Google Scholar]