Abstract
Streptococcus pneumoniae penicillin-binding protein 2x (PBP2x) is an enzyme involved in the last stages of peptidoglycan assembly and essential for bacterial growth and survival. PBP2x localizes to the division site, a process that depends on its Penicillin-Binding Protein And Serine-Threonine-kinase Associated (PASTA) domains, which was previously demonstrated via GFP-PBP2x in living cells. During this study a mutant strain was isolated in which the GFP-PBP2x fusion protein did not localize at division sites and it contained reduced amounts of the full-length GFP-PBP2x. We now show that this defect is due to a point mutation within the C-terminal PASTA2 domain of PBP2x. The mutant protein was analyzed in detail in terms of beta-lactam binding, functionality, and localization in live cells. We demonstrate that the mutation affects the GFP-tagged PBP2x variant severely and renders it susceptible to the protease/chaperone HtrA.
Introduction
Penicillin-binding proteins (PBPs) are targets for ß-lactam antibiotics. They are membrane-associated enzymes implicated in the last steps of peptidoglycan (PG) biosynthesis, a major component of the bacterial cell wall. Streptococcus pneumoniae contains six PBPs.9 The high molecular weight (hmw) PBPs are subdivided into two classes based on the architecture of their domains (for reviews, see Goffin and Ghuysen,6 Sauvage et al.32; Zapun et al.38). Members of class A (S. pneumoniae PBP1a, PBP1b, and PBP2a) are bifunctional enzymes containing transglycosylase and transpeptidase (TP) domains. The two S. pneumoniae class B PBPs (PBP2x and PBP2b) are monofunctional TPs and individually essential in S. pneumoniae.2,13 Each of the class A hmw PBPs can be deleted in S. pneumoniae, but pbp1apbp2a double mutants are not viable.12,27 The low-molecular-weight PBP3 acts as D,D-carboxypeptidase.8
During cell division, all S. pneumoniae hmw PBPs are localized at mid-cell where cell wall synthesis occurs by a combination of peripheral and septal PG synthesis.2,20,34,39 PBP2x is involved in septal and PBP2b in peripheral synthesis.2,39 The septal localization of PBP2x in S. pneumoniae has been demonstrated by immunofluorescence microscopy and by GFP-tagging in living cells.15,24,25,29
S. pneumoniae PBP2x was the first hmw PBP whose crystal structure was solved.28 The structure of a soluble PBP2x derivative revealed a three-domain organization composed of an elongated, “sugar tongue”-like N-terminal domain, a central TP domain and a C-terminal extension attached to the TP domain via a long flexible linker region. In addition, PBP2x contains an N-terminal cytoplasmic tail of 27 amino acids (aa) and a single membrane spanning segment. The N-terminal domain of unknown function is located adjacent to the TP domain and may serve as a pedestal, placing the catalytic region of the protein away from the cell membrane and toward the PG.17 The TP domain has an active site reminiscent of class A and C β-lactamases and harbors three conserved aa motifs with the active site Ser337 residue, which forms a covalent complex with beta-lactams. Interestingly, the X-ray structure of an acylated PBP2x in complex with cefuroxime shows the presence of two antibiotic molecules, one as expected covalently attached to Ser337, and another sandwiched between the TP domain and the first of two homologous C-terminal noncatalytic domains.7 These domains are named Penicillin-Binding Protein And Serine-Threonine-kinase Associated (PASTA) domains since they exist in single or multiple copies in some hmw PBPs and in bacterial Ser/Thr kinases.37 In S. pneumoniae the only PBP that harbors PASTA domains is PBP2x. Each PASTA domain of ∼70 aa consists of three β-sheets and one α-helix, with a loop region of variable length between the first and second β-sheets. These structural elements are highly conserved among PASTA subunits of different proteins although their sequence identity is only ∼10%.37
Since the structures of β-lactam antibiotics mimic the terminal portion of the PG stem peptide, it has been suggested that PASTA motifs might bind noncross-linked PG, the substrate of PBPs.4,37 Indeed, this has been verified for several Ser/Thr kinases.18,23,33,35 Unfortunately, no specific binding sites have been identified in the PASTA domain of different Ser/Thr kinases. Recent studies with PonA2, a key PBP from Mycobacterium tuberculosis, show that its PASTA domain is not able to bind noncross-linked PG, β-lactams, or polymeric PG.3 These results indicate that the role of PASTA domains cannot be generalized.
Little is known about the PASTA domains of S. pneumoniae PBP2x. Deletion of the last 40 aa of PASTA2 strongly affects beta-lactam binding at the active site, whereas deletion of 30 aa does not.22 In addition, it was recently shown that septal localization of PBP2x is driven by its PASTA domains.29 During this work a mutant was isolated that did not localize at septal sites although it contained a full-length GFP-PBP2x fusion protein albeit at reduced amount. We now show that this defect is due to a mutation localized in the PASTA2 domain of PBP2x. The mutant protein was analyzed in detail in terms of functionality, beta-lactam binding and localization in live cells.
Materials and Methods
Bacterial strains, plasmids, and growth conditions
All strains and plasmids used in this work are listed in Table 1. S. pneumoniae strains were grown at 30°C or 37°C in complex C medium14 supplemented with 0.1% yeast extract (C+Y). D-agar1 supplemented with 3% defibrinated sheep blood was used to grow S. pneumoniae on plates. To induce expression of gfp fusions under the PZn promoter, 0.15 mM ZnCl2 was added to the C+Y medium. Growth in liquid culture was monitored by nephelometry (nephelo units, [N]).
Table 1.
Bacterial Strains and Plasmids Used in This Study
| Relevant genotype or descriptiona | Source or reference | |
|---|---|---|
| Strains | ||
| R6 | Unencapsulated, nonvirulent descendent of D39 | 16,26 |
| DKL03 | R6, ΔbgaA::tetM-PZn-gfp-pbp2x; TetR | 29 |
| DKL04 | R6, ΔbgaA::tetM-PZn-gfp-pbp2xA707D G749V; TetR | This study |
| DKL041 | DKL04, but ΔbgaA::tetM-PZn-gfp-pbp2xG749V; Δpbp2x::aad9; SpcR | This study |
| DKL22 | R6, ΔbgaA::tetM-PZn-gfp-pbp2xA707D; TetR | This study |
| DKL22ΔhtrA | DKL22, but ΔhtrA; KanR | This study |
| Plasmids | ||
| pJWV25 | pPP2 derivative,10 carries PZn-gfp+, Amp,R TetR | 5 |
| pFP098 | pJWV25 derivative, carries PZn-gfp-pbp2xA707D fusion | This study |
| p2xKO | pUC19 derivative carrying an insert composed of Spc resistance cassette and upstream/downstream flanking regions of pbp2x; AmpR | 29 |
Antibiotic resistance markers: Tet, tetracycline; Spc, spectinomycin; Kan, kanamycin; Amp, ampicillin.
Escherichia coli strain DH5α was used as a host for cloning and propagation of plasmids. E. coli strains were grown at 37°C either in luria broth (LB) medium with aeration or on LB agar plates.31 Growth of E. coli was followed by measuring the optical density at 600 nm.
Transformation
Transformation of S. pneumoniae R6 and derivatives was carried out as described previously.19 The transformation efficiency was determined using chromosomal DNA of S. pneumoniae AmiA9 conferring resistance to streptomycin30 and was calculated as the percentage of transformants on selective plates (colony forming units [CFU]) compared to CFU on control plates without selective antibiotic. S. pneumoniae transformants were selected by plating on D-agar supplemented with 3% defibrinated sheep blood and 2.5 μg/ml tetracycline or 20 μg/ml spectinomycin.
E. coli was transformed according to Hanahan11 and transformants were selected in the presence of 100 μg/ml ampicillin.
DNA manipulations and construction of strains and plasmids
Procedures such as DNA isolation, restriction, ligation, and agarose gel electrophoresis were performed as recently depicted.29 DNA modifying enzymes were purchased from New England Biolabs or Fermentas (Thermo Scientific) and used as described by the manufacturer.
To construct the plasmid pFP098, a polymerase chain reaction (PCR) fragment was amplified from strain DKL04 with the primers pbp2x_gfp_f/pbp2x_gfp_r29 using iProof high-fidelity DNA polymerase (Bio-Rad Laboratories). The amplified pbp2x fragment was digested with NotI and SpeI and inserted into the same sites in pJWV25,5 generating the plasmid pFP098.
The plasmid pFP098 was transformed into S. pneumoniae R6 strain using the tetracycline resistance marker tetM for selection, generating strain DKL22. Correct integration into the bgaA region via double-crossover was confirmed by PCR amplification with the oligonucleotides bga_check_F and bga_check_R as well as DNA sequencing.29 To exclude the possibility that any mutation occurred in the pbp2x gene, the gfp-pbp2x fusion was amplified and sequenced.
To delete the PBP2x gene, the plasmid p2xKO29 was transformed into competent DKL04 cells and transformants were selected with spectinomycin in the presence of ZnCl2 (0.15 mM). Correct integration into the genome was verified by PCR and sequencing.
To construct an htrA deletion strain, a PCR cassette replacing htrA with the kanamycin resistance gene aphIII was inserted into the genome of R6 derivatives as described previously.29
Labeling and detection of PBPs
Preparation of samples, PBP labeling of cell lysates with Bocillin™ FL and separation of proteins by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was carried out as described recently.29
Western blot analysis
Cells from exponentially growing cultures were collected at a cell density of N=70–80 by centrifugation. Whole cell lysates were obtained by incubating cells in the presence of 0.2% Triton X-100 for 30 min at 37°C; per sample the cell equivalent of 1.5 ml at N=20 was used. After SDS-PAGE, proteins were transferred onto a PVDF membrane (Roche Diagnostics) and detected with affinity-purified polyclonal PBP2x,21 or anti-GFP antibodies (polyclonal rabbit antibodies; Invitrogen). After incubation with the secondary antibody (alkaline phosphatase-conjugated goat anti rabbit immunoglobulin G; Sigma-Aldrich) staining was conducted with 4-nitrobluetetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate (Roche). Immunoblots were quantified using the programme Image J (NIH).
Fluorescence microscopy
Fluorescence microscopy was performed as previously described.29 Briefly, cells grown in liquid medium at 30°C were analyzed by phase contrast or epi-fluorescence microscopy using an Eclipse E600 (Nikon) microscope and 100×NA 1.4 oil immersion objective; pictures were taken with a DXM1200C camera (Nikon). Fluorescence signals of GFP were visualized with the Epi-FL filterblock B-2E/C (EX: 465–495, DM: 505, BA: 515–555; Nikon) using identical fluorescence intensity. Typical exposure times were between 1 and 2 sec. Image analysis was carried out using Nikon Imaging software Nis-Elements BR.
Results
Identification of mutations in PASTA2 of GFP-PBP2x
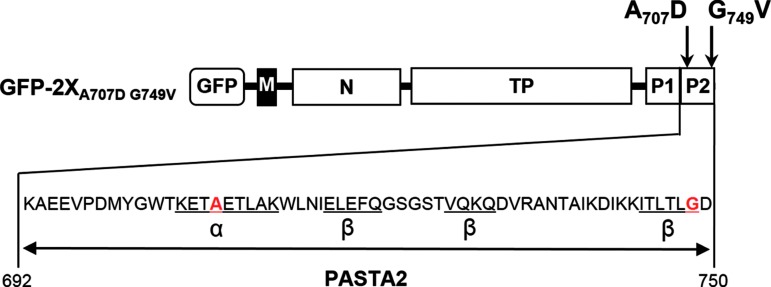
In a recent report we described the construction of an N-terminal GFP-PBP2x fusion protein for localization studies in live cells29 using the vector pJWV25. It harbors the zinc-inducible PZn promotor and integrates in the chromosome of S. pneumoniae by double-crossover at the bgaA locus.5 Such constructs were readily obtained, and most of them showed localization of GFP-PBP2x at septal sites as shown for DKL03.29 However, one transformant was obtained during the screening procedure in which the fluorescence signal was dispersed throughout the cytoplasm. Sequencing of gfp-pbp2x in this particular transformant named DKL04 revealed that it contains two mutations, both of which were located within the PASTA2 domain of GFP-PBP2x named GFP-PBP2xA707D G749V: A707D in the α-helix, and G749V at the very end of β3-sheet (Fig. 1).
FIG. 1.
Schematic outline of the GFP-PBP2xA707D G749V fusion protein. GFP, green fluorescent protein GFP+; M, transmembrane helix; N, N-terminal domain; TP, transpeptidase/penicillin-binding domain; P1 and P2, C-terminal PASTA domains. Structural features of the P2 domain are shown in detail below: α: alpha-helix and β: beta-sheets are underlined according to37; first and last aa of P2 are indicated.7 The mutations in GFP-PBP2xA707D G749V (DKL04) are marked in red. aa, amino acids; PASTA, Penicillin-Binding Protein And Serine-Threonine-kinase Associated; PBP, and Ser/Thr-associated; PBP2x, penicillin-binding protein 2x.
The localization pattern of GFP-PBP2x is dependent on the presence of the two C-terminal PASTA domains of PBP2x.29 Moreover, Maurer et al. have shown that deletion of the last 40 aa in PBP2x resulted in an almost complete loss of beta-lactam binding, that is, the presence of the α-helix of PASTA2 domain appears to be critical for antibiotic binding, whereas deletion of 30 C-terminal aa had no functional consequences.22 Therefore, we analyzed the effect of these two point mutations in GFP-PBP2xA707D G749V on beta-lactam binding and localization in detail.
Localization and beta-lactam binding of GFP-PBP2xA707D G749V
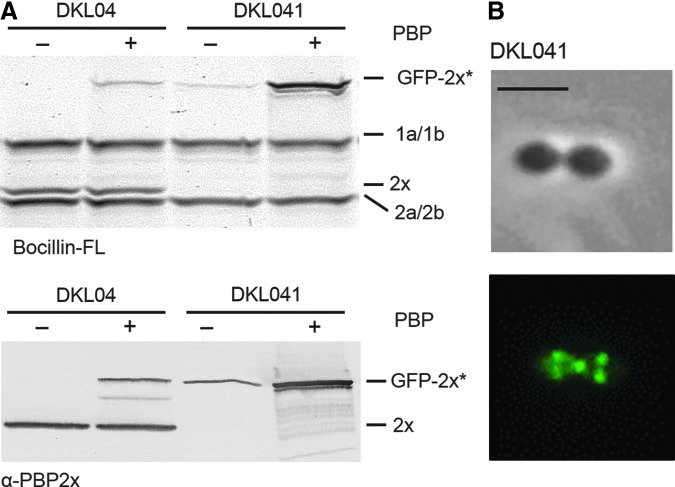
The expression of GFP-PBP2xA707D G749V was induced by addition of ZnCl2 to the growth medium. First, we localized the GFP-PBP2xA707D G749V in DKL04 strain by fluorescence microscopy. The protein did not localize to mid-cell and the fluorescence signal was distributed throughout the cytoplasm. However, a small fraction of the cells (3 out of 480 examined cells) showed septal localization (see arrow in Fig. 2A) and will be discussed below.
FIG. 2.
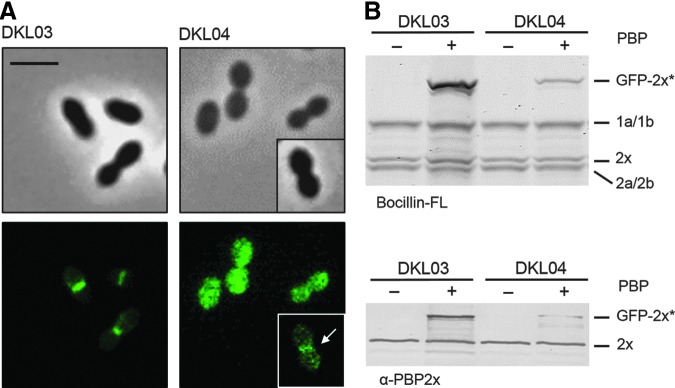
Localization and production of the GFP-PBP2xA707D G749V fusion protein in DKL04. (A) DKL04 (R6, PZn-gfp-pbp2xA707D G749V) and the control strain DKL03 (R6, PZn-gfp-pbp2x) were grown in the presence of ZnCl2. Top: Phase contrast microscopy; Bottom: Fluorescence signal. Micrograph inserts show a cell with septal localization (see arrow), which occurs on rare occasions in DKL04 (3 out of 480 counted cells). Scale bar=2 μm. (B) Top: PBP profiles in strains DKL03 and DKL04. Cell lysates were incubated with Bocillin™ FL and PBPs visualized after separation by SDS-PAGE followed by fluorography. Bottom: Western blot developed with anti-PBP2x (α-PBP2x) antibodies. + and − indicate growth with and without ZnCl2. The positions of PBPs and GFP-2x* (GFP-PBP2x and GFP-PBP2xA707D G749V) are indicated on the right. SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Next, Bocillin FL binding of GFP-PBP2xA707D G749V was examined in cell lysates by fluorography (Fig. 2B, top). Surprisingly, only a thin band with the correct size was detectable in the DKL04 lysate, whereas the GFP-PBP2x band was perfectly present in the lysates of the control strain DKL03. Western blot analyses of cell lysates using anti-PBP2x antibodies showed that the GFP-PBP2xA707D G749V fusion protein is also only present in small amounts in DKL04 (Fig. 2B, bottom). The amount of GFP-PBP2xA707D G749V was more than 10-fold lower when compared with the wild type GFP-PBP2x. In this context it should be noted that the fusion protein GFP-PBP2x in the control strain DKL03 was present in slightly higher amounts compared with native PBP2x.29
Taken together these data indicate that the GFP-PBP2xA707D G749V is not stable and subject to degradation. Alternatively, it is possible that at least one of the mutations is not tolerated by the cell, and that the highly reduced number of cells with septal fluorescence signal is the product of revertants that can easily arise by recombination with the native pbp2x.
The mutation A707 is not tolerated in GFP-PBP2x
To test whether GFP-PBP2xA707D G749V can replace the native PBP2x gene, we tried to delete the native copy of pbp2x in DKL04 using the plasmid p2xKO.29 This is readily possible in the control strain DKL03 where such transformants were obtained at high frequency, confirming that the GFP-PBP2x fusion is fully functional.29 In contrast, the transformation efficiency in DKL04 as recipient was below 1% compared to the DKL03 control (Table 2). DNA sequencing of two such transformants revealed that indeed the native pbp2x was deleted. However, the GFP-PBP2x fusion protein carried only the mutation V749, whereas in both cases the other mutation D707 had reverted to A707. This strongly suggests that D707 severely affects at least the function of the protein, whereas this is not the case for V749.
Table 2.
Transformation Efficiency
Chromosomal donor DNA from Streptococcus pneumoniae AmiA9 was used and StrR transformants were selected.
Spectinomycin-resistant transformants were selected using the plasmid p2xKO.
One transformant, DKL041, was used for further experiments. As can be seen in Fig. 3A, the GFP-PBP2xG749V fusion was produced in clearly higher amounts than in the parental strain DKL04, and was active in Bocillin FL binding. It should be noted that a very thin band corresponding to GFP-PBP2xG749V protein was detected in cell lysates of DKL041 prepared from cultures grown in the absence of ZnCl2 (Fig. 3A). This probably represents some residual GFP-PBP2xG749V originating from the starting culture grown with ZnCl2 since it takes a considerable time before the fusion protein is depleted upon shift to zinc-free medium.29 Moreover, the GFP-PBP2xG749V perfectly localized at the division septum (Fig. 3B).
FIG. 3.
Cellular localization of the GFP-fusion protein in DKL041. (A) Top: PBP profile analysis. PBPs in cell lysates were labeled with Bocillin™ FL and visualized by fluorography. Bottom: Western blot developed with anti-PBP2x antibodies. The positions of PBPs and GFP-2x* (GFP-PBP2xA707D G749V in strain DKL04 and GFP-PBP2xG749V in strain DKL041) are indicated on the right side. + and − indicate growth with and without ZnCl2. Strains used for preparation of cell lysates are indicated on top. (B) Localization of GFP-PBP2xG749V in DKL041 strain. Top: Phase contrast microscopy; Bottom: Fluorescence signal. Scale bar=2 μm.
To exclude the possibility that other mutations are involved in these phenotypes, three independent transformants obtained with a PCR product covering the GFP-PBP2xA707D G749V gene were produced. All three transformants showed the same phenotype as described above. Taken together these data indicate that the mutation V749 is well tolerated by the cells and has no apparent effect on PBP2x properties. In other words, it is the mutation D707 that affects the PBP2x protein, that is, the position A707 in the α-helix of PASTA2 is critical for stability and thus affects localization and function of PBP2x as well. In this context it should be noted that revertants of DKL04 expressing GFP-PBP2xG749V occur at low frequency, probably via recombination with the native pbp2x. Thus, it is possible that the small amount of GFP-PBP2x seen in DKL04 is due to such revertants already present in the culture.
Ala707 is important for PBP2x functionality
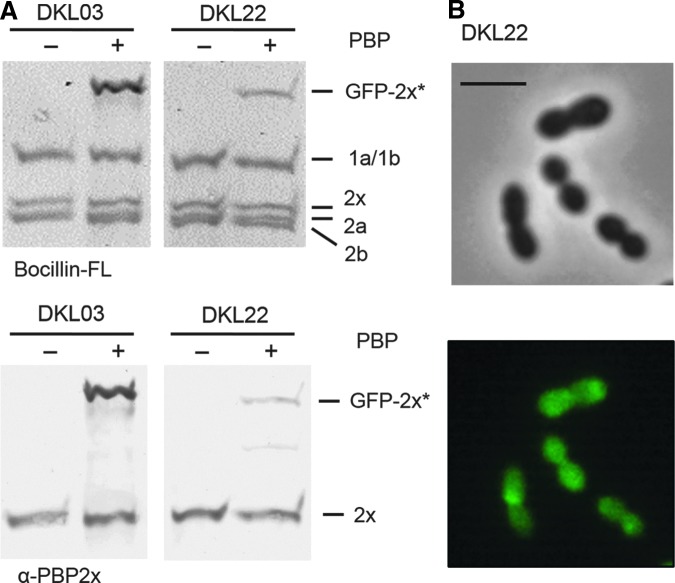
Finally, we constructed the mutant DKL22 that expresses a GFP-PBP2x containing the single mutation D707 to prove that it is indeed this mutation that confers the phenotype observed in DKL04. Upon induction of gfp-pbp2xA707D in DKL22 by ZnCl2, cell lysates were incubated with Bocillin FL for visualization of PBPs and PBP2x was specifically labeled with anti-PBP2x antibodies after western blotting as shown above. Only a thin band was visible in the lysate of strain DKL22 on the fluorogram at the position of the GFP-PBP2x (Fig. 4A, top), and the presence of very low amounts of the GFP-PBP2xA707D fusion protein in DKL22 when compared with DKL03 was confirmed immunologically (Fig. 4A, bottom). The fluorescence signal in DKL22 was dispersed throughout the cells and did not localize at the septum (Fig. 4B), similar to the results obtained with DKL04. Furthermore, we tried to delete the native copy of pbp2x in DKL22 and the transformants were obtained with similar transformation efficiency as in the case of DKL04 (Table 2). The analysis of one transformant revealed that indeed the native pbp2x was deleted but the D707 mutation had reverted to A707. As expected, the GFP-PBP2x fusion in these revertants perfectly localized at the division septum (data not shown). These experiments confirm that the mutation D707 located within the α-helix of PASTA2 severely affects PBP2x functionality.
FIG. 4.
Effect of the mutation D707. (A) Top. PBP in cell lysates of strains DKL03 (R6, PZn-gfp-pbp2x) and DKL22 (R6, PZn-gfp-pbp2xA707D) were labeled with Bocillin FL and visualized by fluorography. Bottom: Western blot of the gel on the top developed with anti-PBP2x antibodies. + and − indicate growth with and without ZnCl2. The positions of the PBPs and GFP-2x* (GFP-PBP2x and GFP-PBP2xA707D) are indicated on the right. Strains used for preparation of cell lysates are indicated on top. (B) Micrographs of strain DKL22 grown in the presence of ZnCl2. Top: Phase contrast microscopy; Bottom: Fluorescence signal. Scale bar=2 μm.
GFP-PBP2xA707D is targeted by HtrA
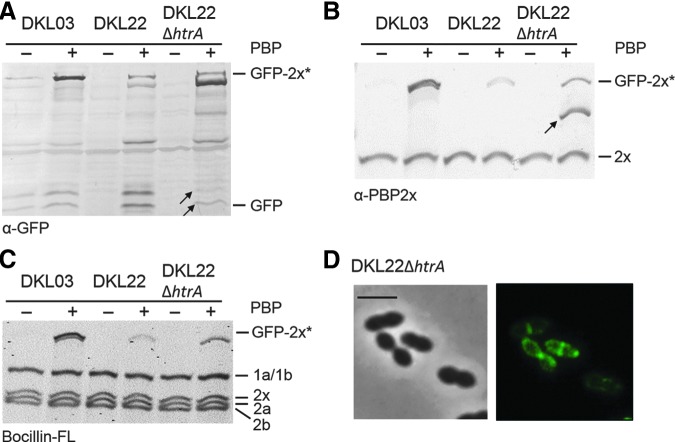
We have recently shown that derivatives of GFP-PBP2x containing various deletions are degraded by the serine protease/chaperon HtrA, and most of them did not localize to septal sites in the presence of HtrA. Therefore, GFP-PBP2xA707D was compared in DKL22ΔhtrA to that in the parental DKL22. Western blot analysis using anti-GFP antibodies demonstrated that indeed the fusion protein GFP-PBP2xA707D was degraded in DKL22 (i.e., in the presence of HtrA; Fig. 5A), whereas the degradation products were barely visible in the HtrA deletion mutant (see arrows in Fig. 5A). With anti-PBP2x antibodies another degradation product of the fusion protein was detected in HtrA deletion strain that was already seen on western blot developed with anti-GFP antibodies (see arrow in Fig. 5B). Moreover, DKL22ΔhtrA contained a GFP-PBP2xA707D protein active in Bocillin™ FL binding, which is hardly visible in DKL22 (Fig. 5C).
FIG. 5.
The fusion protein GFP-PBP2xA707D is a target of HtrA. The protein GFP-PBP2xA707D (DKL22) was analyzed in presence or absence of HtrA. (A) Western blot analysis using anti-GFP (α-GFP) antibodies. The positions of GFP-2x* (GFP-PBP2x and GFP-PBP2xA707D) are indicated on the right. Cells were grown in the presence or absence of ZnCl2 (indicated + and −). Arrows point to degradation products that are barely visible in case of ΔhtrA. (B) Western blot of cell lysates developed with anti-PBP2x antibodies. + and −: growth with and without ZnCl2. The position of GFP-2x* and PBP2x are marked. Arrow: product seen in ΔhtrA background. (C) PBP profile analysis. PBPs in cell lysates were labeled with Bocillin FL and visualized by fluorography after separation by SDS-PAGE. On the right the positions of PBPs are indicated. Strains as indicated on top were grown in the presence (+) or absence (−) of ZnCl2. (D) Localization of GFP-PBP2xA707D in the absence of HtrA. DKL22ΔhtrA was grown in the presence of ZnCl2. Left: Phase contrast microscopy; Right: Fluorescence signal. Scale bar=2 μm.
Fluorescence microscopy showed that deletion of htrA affected the localization of GFP-PBP2xA707D fusion protein. The percentage of cells showing fluorescence signals at septal sites increased from 13% (DKL22) to 35% in DKL22ΔhtrA. In most cells it still localized in the membrane, at cell poles or in irregular patches (Fig. 5D).
Discussion
The present study extends previous observations related to in vivo localization studies of PBP2x, one of the two essential PBPs in S. pneumoniae, which is an important component of the cell division apparatus, and moreover a major determinant for beta-lactam resistance. Using GFP-PBP2x fusion proteins we showed in a previous study that the PASTA domains are required for proper localization at septal sites, and that GFP-PBP2x derivatives with various deletions are targeted and degraded by the serine protease/chaperon HtrA.29 We now show that also a single aa exchange A707D within the PASTA2 domain has severe effects in different respects. First, the GFP-PBP2xA707D mutant protein did not localize at septal sites (Fig. 4B). Second, deletion of the genomic wild-type pbp2x was not possible in strain DKL22, and the amount of GFP-PBP2xA707D was severely reduced. On the other hand, GFP-PBP2xA707D was detectable in cells where htrA was deleted, and degradation products, which were clearly visible in the wild-type genetic background, were hardly detectable any more. Thus, the mutation renders A707D the protein recognizable by HtrA, strongly suggesting a severe structural impact.
This is the first case where a single point mutation in the C-terminal PASTA domain is shown to have a severe effect on PBP2x. So far only mutations within the TP domain have been described that mediate resistance against beta-lactams (for review, see Hakenbeck et al.9), and at least some of them apparently affect the physiology of the cell as well.19,40 It has been discussed that the PASTA domains are mutational hotspots, and that the mutations consistently occur at the same sites, whereas the remaining sites are conserved.4,37 The position A707 is highly conserved in PBP2x of S. pneumoniae and the closely related species Streptococcus mitis including mosaic variants of resistant strains; only in a few Streptococcus oralis strains the alteration V707 occurs, an aa, which contains also a small hydrophobic side chain (own unpublished data). This is in agreement with our hypothesis that the position A707 is of structural and/or functional importance.
The mutant isolated initially contained two mutations: A707D and G749V. PBP2x is 750 aa long, and since deletion of the C-terminal 30 aa are tolerated without any impact on beta-lactam binding,22 it is unlikely that G749V drastically affects function and/or stability of the protein, especially since no large or charged side chains are introduced by this mutation. Moreover, the side chain is oriented toward the surface of the protein and thus has probably little impact on its structure. In fact, in a mutant containing only GFP-PBP2xG749V the protein appears to localize at the septum and to bind Bocillin FL similar to the wild-type PBP2x (Fig. 5B), confirming that there is no detectable effect of this mutation on the functionality of PBP2x. On the other hand, the mutation A707D introduces a large charged side chain, which is directed toward the interior of the C-terminal domain and might well affect its tertiary structure severely and thereby attracting HtrA. On the other hand, GFP-PBP2xG749V was perfectly able to bind Bocillin FL as shown in DKL22ΔhtrA (Fig. 5C). Thus, this mutant protein differs from the deletion derivatives described recently,29 in that its enzymatic function in terms of beta-lactam binding and thus PG synthesis appears not to be affected, whereas localization at the septum and thus its function in the division process is severely hampered. This might indirectly affect the interaction with other components of the divisome complex, which appears to be governed by HtrA, which itself localizes at septal sites.36 Future studies will help to unravel the interactive network of divisome components.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft Ha 1011/11-1.
Disclosure Statement
No competing financial interests exist.
References
- 1.Alloing G., Granadel C., Morrison D.A., and Claverys J.-P. 1996. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21:471–478 [DOI] [PubMed] [Google Scholar]
- 2.Berg K.H., Stamsas G.A., Straume D., and Havarstein L.S. 2013. The effect of low Pbp2b levels on cell morphology and peptidoglycan composition in Streptococcus pneumoniae R6. J. Bacteriol. 195:4342–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvanese L., Falcigno L., Maglione C., Marasco D., Ruggiero A., Squeglia F., Berisio R., and D'Auria G.2013. Structural and binding properties of the PASTA domain of PonA2, a key penicillin binding protein from Mycobacterium tuberculosis. Biopolymers 101:712–719 [DOI] [PubMed] [Google Scholar]
- 4.Dessen A., Mouz N., Gordon E., Hopkins J., and Dideberg O.2001. Crystal structure of PBP2x from a highly penicillin-resistant Streptococcus pneumoniae clinical isolate: a mosaic framework containing 83 mutations. J. Biol. Chem. 276:45106–45112 [DOI] [PubMed] [Google Scholar]
- 5.Eberhardt A., Wu L.J., Errington J., Vollmer W., and Veening J.W.2009. Cellular localization of choline-utilization proteins in Streptococcus pneumoniae using novel fluorescent reporter systems. Mol. Microbiol. 74:395–408 [DOI] [PubMed] [Google Scholar]
- 6.Goffin C., and Ghuysen J.-M.2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66:706–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon E., Mouz N., Duee E., and Dideberg O.2000. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 299:477–485 [DOI] [PubMed] [Google Scholar]
- 8.Hakenbeck R., and Kohiyama M.1982. Purification of penicillin-binding protein 3 from Streptococcus pneumoniae. Eur. J. Biochem. 127:231–236 [DOI] [PubMed] [Google Scholar]
- 9.Hakenbeck R., Brückner R., Denapaite D., and Maurer P.2012. Molecular mechanism of beta-lactam resistance in Streptococcus pneumoniae. Future Microbiol. 7:395–410 [DOI] [PubMed] [Google Scholar]
- 10.Halfmann A., Hakenbeck R., and Brückner R.2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268:217–224 [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 12.Hoskins J., Matsushima P., Mullen D.L., Tang J., Zhao G., Meier T.I., Nicas T.I., and Jaskunas S.R. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552–6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kell C.M., Sharma U.K., Dowson C.G., Town C., Balganesh T.S., and Spratt B.G. 1993. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B and 2X of Streptococcus pneumoniae. FEMS Microbiol. Lett. 106:171–175 [DOI] [PubMed] [Google Scholar]
- 14.Lacks S., and Hotchkiss R.D.1960. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim. Biophys. Acta 39:508–517 [DOI] [PubMed] [Google Scholar]
- 15.Land A.D., Tsui H.C., Kocaoglu O., Vella S.A., Shaw S.L., Keen S.K., Sham L.T., Carlson E.E., and Winkler M.E. 2013. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol. Microbiol. 90:939–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanie J.A., Ng W.L., Kazmierczak K.M., Andrzejewski T.M., Davidsen T.M., Wayne K.J., Tettelin H., Glass J.I., and Winkler M.E. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macheboeuf P., Contreras-Martel C., Job V., Dideberg O. and Dessen A.2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673–691 [DOI] [PubMed] [Google Scholar]
- 18.Maestro B., Novaková L., Hesek D., Lee M., Leyva E., Mobashery S., Sanz J.M., and Branny P. 2011. Recognition of peptidoglycan and b-lactam antibiotics by the extracellular domain of the Ser/Thr protein kinase StkP from Streptococcus pneumoniae. FEBS Lett. 585:357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mascher T., Heintz M., Zähner D., Merai M., and Hakenbeck R.2006. The CiaRH system of Streptococcus pneumoniae prevents lysis during stress induced by treatment with cell wall inhibitors and mutations in pbp2x involved in beta-lactam resistance. J. Bacteriol. 188:1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massidda O., Novakova L., and Vollmer W.2013. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division?. Environ. Microbiol. 15:3133–3157 [DOI] [PubMed] [Google Scholar]
- 21.Maurer P., Koch B., Zerfaß I., Krauß J., van der Linden M., Frère J.-M., Contreras-Martel C., and Hakenbeck R. 2008. Penicillin-Binding Protein 2x of Streptococcus pneumoniae: Three new mutational pathways for remodelling an essential enzyme into a resistance determinant. J. Mol. Biol. 376:1403–1416 [DOI] [PubMed] [Google Scholar]
- 22.Maurer P., Todorova K., Sauerbier J. and Hakenbeck R.2012. Mutations in Streptococcus pneumoniae penicillin-binding protein 2x: importance of the C-terminal penicillin-binding protein and serine/threonine kinase-associated domains for beta-lactam binding. Microb. Drug Resist. 18:314–321 [DOI] [PubMed] [Google Scholar]
- 23.Mir M., Asong J., Li X., Cardot J., Boons G.J., and Husson R.N. 2011. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog. 7:e1002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morlot C., Zapun A., Dideberg O., and Vernet T.2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845–855 [DOI] [PubMed] [Google Scholar]
- 25.Morlot C., Bayle L., Jacq M., Fleurie A., Tourcier G., Galisson F., Vernet T., Grangeasse C., and Di Guilmi A.M.2013. Interaction of Penicillin-Binding Protein 2x and Ser/Thr protein kinase StkP, two key players in Streptococcus pneumoniae R6 morphogenesis. Mol. Microbiol. 90:88–102 [DOI] [PubMed] [Google Scholar]
- 26.Ottolenghi E., and Hotchkiss R.D.1962. Release of genetic transforming agent from pneumococcal cultures during growth and disintegration. J. Exp. Med. 116:491–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paik J., Kern I., Lurz R., and Hakenbeck R.1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pares S., Mouz N., Pétillot Y., Hakenbeck R., and Dideberg O.1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284–289 [DOI] [PubMed] [Google Scholar]
- 29.Peters K., Schweizer I., Beilharz K., Stahlmann C., Veening J.-W., Hakenbeck R., and Denapaite D. 2014. Streptococcus pneumoniae PBP2x mid-cell localization requires the C-terminal PASTA domains and is essential for cell shape maintenance. Mol. Microbiol. [Epub ahead of print]; DOI: 10.1111/mmi.12588 [DOI] [PubMed] [Google Scholar]
- 30.Salles C., Creancier L., Claverys J.P., and Méjean V. 1992. The high level streptomycin resistance gene from Streptococcus pneumoniae is a homologue of the ribosomal protein S12 gene from Escherichia coli. Nucleic Acids Res. 20:6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch E.F., and Maniatis T.1989. MolecularCloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Sauvage E., Kerff F., Terrak M., Ayala J., and Charlier P.2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258 [DOI] [PubMed] [Google Scholar]
- 33.Shah I.M., Laaberki M.H., Popham D.L., and Dworkin J. 2008. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell 135:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sham L.T., Tsui H.C., Land A.D., Barendt S.M., and Winkler M.E. 2012. Recent advances in pneumococcal peptidoglycan biosynthesis suggest new vaccine and antimicrobial targets. Curr. Opin. Microbiol. 15:194–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squeglia F., Marchetti R., Ruggiero A., Lanzetta R., Marasco D., Dworkin J., Petoukhov M., Molinaro A., Berisio R., and Silipo A.2011. Chemical basis of peptidoglycan discrimination by PrkC, a key kinase involved in bacterial resuscitation from dormancy. J. Am. Chem. Soc. 133:20676–20679 [DOI] [PubMed] [Google Scholar]
- 36.Tsui H.C., Keen S.K., Sham L.T., Wayne K.J., and Winkler M.E. 2011. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. MBio 2:e00202–e00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeats C., Finn R.D., and Bateman A.2002. The PASTA domain: a beta-lactam-binding domain. Trends Biochem. Sci. 27:438–440 [DOI] [PubMed] [Google Scholar]
- 38.Zapun A., Contreras-Martel C., and Vernet T.2008a. Penicillin-binding proteins and b-lactam resistance. FEMS Microbiol. Rev. 32:361–385 [DOI] [PubMed] [Google Scholar]
- 39.Zapun A., Vernet T., and Pinho M.G.2008b. The different shapes of cocci. FEMS Microbiol. Rev. 32:345–360 [DOI] [PubMed] [Google Scholar]
- 40.Zerfaß I., Hakenbeck R., and Denapaite D.2009. An important site in PBP2x of penicillin-resistant clinical isolates of Streptococcus pneumoniae: mutational analysis of Thr338. Antimicrob. Agents Chemother. 53:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]