Summary
Despite over a decade of intense research, the identity and differentiation potential of human adult cardiac progenitor cells (aCPC) remains controversial. Cardiospheres have been proposed as a means to expand aCPCs in vitro, but the identity of the progenitor cell within these 3D structures is unknown. We show that clones derived from cardiospheres could be subdivided based on expression of thymocyte differentiation antigen 1 (THY-1/CD90) into two distinct populations that exhibit divergent cardiac differentiation potential. One population, which is CD90+, expressed markers consistent with a mesenchymal/myofibroblast cell. The second clone type was CD90− and could form mature, functional myocytes with sarcomeres albeit at a very low rate. These two populations of cardiogenic clones displayed distinct cell surface markers and unique transcriptomes. Our study suggests that a rare aCPC exists in cardiospheres along with a mesenchymal/myofibroblast cell, which demonstrates incomplete cardiac myocyte differentiation.
Highlights
-
•
The majority of human adult CS-derived cardiomyocytes do not form sarcomeres
-
•
There is little overlap between the C-KIT and NKX2.5 cell populations in human CS
-
•
CD90 distinguishes two populations that exhibit divergent cardiogenic potential
-
•
CD90neg CS cells can differentiate into mature cardiomyocytes at a very low rate
Cardiospheres (CSs) have been reported to contain an adult cardiac progenitor cell (aCPC) with multilineage differentiation potential. MacLellan and colleagues show that CS-derived cells differentiate robustly into endothelial and smooth muscle cells but predominantly form immature cardiomyocytes. CD90 expression distinguishes CS-derived cells with a mesenchymal cell phenotype with incomplete cardiomyocyte differentiation (CD90+) and a CD90− population that could give rise to mature cardiomyocytes, although infrequently.
Introduction
Adult cardiac progenitor cells (aCPCs) are a rare subpopulation of cells first reported in the adult rat heart that are self-renewing, clonogenic, and multipotent with the capacity to give rise to cardiac myocytes (CMs), smooth muscle cells (SMCs), and endothelial cells (ECs) in vitro and in vivo (Beltrami et al., 2003). Their biology, particularly human aCPCs, remains poorly understood because techniques to study human aCPCs in vivo are very limited, and in vitro studies are challenging due to the rarity of this cell type and lack of reliable methods to propagate and expand these cells in their undifferentiated state. Cardiosphere (CS) formation, a 3D culture system, has been proposed as one method to expand aCPCs in vitro (Messina et al., 2004; Davis et al., 2009). Sphere systems have been used to culture many stem cell types, including neural and skin stem cells (Reynolds and Weiss, 1992; Fernandes et al., 2004; Gago et al., 2009). Rat CS-derived clones are multipotent (Davis et al., 2009), but whether these cells exist in human CS or their identity is unknown. CSs are a heterogeneous mix of nonmyocyte cells derived from the mononuclear fraction of dissociated heart tissue, which includes mesenchymal stem cells, SMCs, ECs, and cardiac fibroblasts. Although CSs have been reported to contain an aCPC, this has been questioned, and its identity or cell surface markers that can be used to isolate these cells are unknown (Masuda et al., 2012; Smith et al., 2007). Interestingly, there is no panel of cell surface markers that can directly identify endogenous CPCs. Numerous putative aCPCs have been reported in adult mouse heart, which were identified based on the expression of C-KIT, SCA1, or the ability to exclude Hoechst dye (Beltrami et al., 2003; Oh et al., 2004; Tomita et al., 2005). Much less data exist in human hearts, although a rare cardiac progenitor cell population of C-KIT and NKX2.5+ cells has been described (Mishra et al., 2011; Goumans et al., 2007; Bearzi et al., 2007; Smits et al., 2009). However, C-KIT by itself is not specific and cannot be used by itself to identify aCPCs (Bearzi et al., 2009; Sandstedt et al., 2010). A significant proportion of the C-KIT cells in human hearts coexpresses CD45, suggesting that they may be hematopoietic in origin (Kubo et al., 2008). The early cardiac transcription factor, NK2 homeobox 5 (NKX2.5), is often used in conjunction with c-KIT to identify aCPCs (Wu et al., 2006, 2008; Mishra et al., 2011), but because it is an intracellular marker, its use is restricted to fixed cells and cannot be used for purifying human live cells. At least a subset of cardiac C-KIT cells expressed NKX2.5, but whether this C-KIT+/NKX2.5+ subpopulation in adult human heart is an authentic aCPC and whether this C-KIT+/NKX2.5+ cell also represents the aCPCs within CS is unknown. No systematic analysis of cell surface markers of cloned human aCPCs has been performed, and no panel of cell surface markers allowing direct isolation of aCPCs is available.
Another major limitation of the adult cardiac stem cell field is the inability to get full differentiation from putative stem/progenitor cells in vitro or in vivo. In fact, although expression of cardiac genes and proteins is demonstrated frequently, the levels, relative to authentic CMs, and their ability to form sarcomeres and display calcium transients are rarely quantified (Barile et al., 2007; Bearzi et al., 2007; He et al., 2011). In vivo engraftment studies have often demonstrated low-level retention of cells that, whereas expressing sarcomeric proteins, do not morphologically appear to resemble adult CMs (Li et al., 2010). This is also true of the CMs differentiated from human CS (Davis et al., 2009). The basis for this paradox is unknown but is a critical issue to resolve if these cells are to be used therapeutically. Despite these limitations, the therapeutic efficacy of autologous CS-derived cells after myocardial infarction was tested in a recent clinical trial by Makkar et al. (2012). The authors reported that the therapy appeared safe, reduced myocardial scar, and increased viable myocardium, but there was no global improvement in patients’ cardiac function.
In the present study, we sought to determine the identity of the cell(s) with cardiogenic potential in CS generated from human hearts. We describe two distinct populations of CS-derived cells based on their expression of CD90 and their cardiomyogenic potential. The first cell population is enriched for the cardiac transcription factor NKX2.5 and ISL1 but negative for mesenchymal stem cell/fibroblast surface marker CD90 and negative for cardiac stem cell marker C-KIT. These CD90− cells can differentiate into SMCs, ECs, and CMs, which form complete sarcomeres and beat spontaneously but at a very low frequency. The second cell population appears to be a cardiac mesenchymal/fibroblast cell that is positive for CD90 and Periostin but is NKX2.5low and ISL1low. This cell demonstrated endothelial and SMC differentiation but only incomplete cardiac differentiation into CM-like cells without sarcomere formation or calcium cycling. These results may account for previous reports of multiple stem cells in the heart (Barile et al., 2007; Beltrami et al., 2003; Davis et al., 2010; Deb et al., 2003; Laflamme et al., 2002; Murry et al., 2004; Toma et al., 2002) and suggest that a cell with a phenotype consistent with a mesenchymal stem cell or myofibroblast exists in the adult heart that is capable of differentiating into CM-like cells that are not authentic mature CMs.
Results
Differentiation of Adult Human CS-Derived Cells Primarily Gives Rise to Immature CM-like Cells
Heart-derived cells (HDCs) obtained from first plating of acutely dissociated mononuclear human cells were plated in adherent culture (ADH) or used to create CSs (Figure 1A). Undifferentiated HDCs, ADHs, and CSs were negative for the CM marker Troponin I (cTNI). ADHs but not CSs contained significant numbers of cells for SMC marker calponin (CNN; 17.8% ± 3%), or EC marker von Willebrand factor (vWF; 45% ± 15%) (Figures 1B and 1C; Figure S1A available online). After 15 days of differentiation, ADH and CS cells demonstrated similar percentages of CNN+ cells (51% ± 5% versus 42% ± 8%, respectively; p < 0.05), but ADH demonstrated higher rates of EC differentiation in comparison with CS (vWF; 49% ± 2% versus 12% ± 7%, respectively; p < 0.05). Compared to ADH cells, differentiated CS-derived cells demonstrated higher percentages of cTNI+ cells (6.5% ± 3% versus 27% ± 5%, respectively; p < 0.05). However, these CM-like cells, whereas expressing low levels of cardiac proteins, did not morphologically resemble mature myocytes (Figures 1D, 1E, and S1B).
Figure 1.
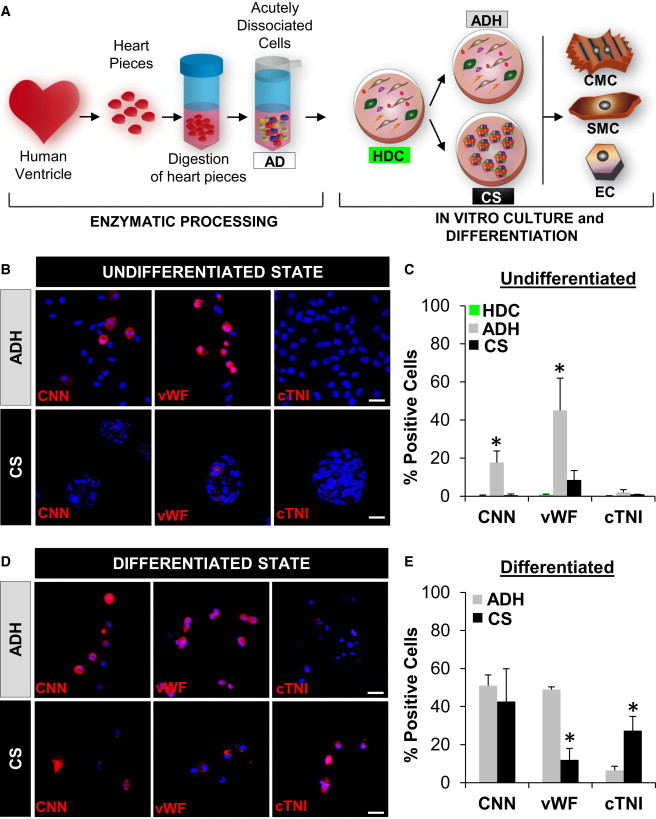
Cardiovascular Differentiation Potential of Adult Human CS-Derived Cells
(A) Experimental design. Human heart samples were minced, digested, and dissociated to single ADs. ADs were grown in primary adhesion culture that we call HDCs. After HDCs reached confluence, they were harvested and then cultured in either adhesion cultures (ADHs) or CSs. After 7 days, ADHs and CSs were induced to differentiate into CMs, SMCs, and ECs.
(B and D) Confocal imaging of ADH and CS cells immunostained with CNN, vWF, and cTNI pre- (B) and post- (D) differentiation. Nuclei were stained with DAPI (blue). Scale bars, 40 μm.
(C and E) Quantification of the number of cells positive for CNN, vWF, and cTNI from ADH and CS pre- (C) and post- (E) differentiation. Characterization of HDCs was performed as an additional control of undifferentiated progenitor/precursor cells in the primary culture (C, green bars). All experiments were performed with a minimum of three independent human heart samples. ∗p < 0.05.
To determine if this incomplete differentiation was related to our culture conditions or the cells themselves, we generated CSs from both fetal and adult human hearts (Figures 2Aa and 2Ai). Differentiated fetal CS-derived cells exhibited typical CM morphology with organized sarcomeres that coexpressed the cardiac-specific markers cTNT and cTNI (Figures 2Ae–2Ah). Although cells from adult CS actually exhibited higher rates of cells expressing cTNT and cTNI (27% ± 5% versus 13.3% ± 7%, respectively; p < 0.05), the absolute expression levels were low, and sarcomere formation was rare (Figures 2Am–2Ap; Figure 2B, right). In contrast, the majority of CMs from fetal CSs demonstrated spontaneous contractions and calcium transients (Movies S1 and S2). No differences were observed in the morphology of fetal or adult CS-derived ECs, but adult CS-derived cells exhibited lower rates of cells expressing vWF (12% ± 6% versus 28% ± 4%, respectively; p < 0.05). Both fetal and adult CS-derived ECs formed typical microtubular capillary networks in Matrigel (Figures 2Ab, 2Ac, 2Aj, and 2Ak; Figure 1B, left). Likewise, SMC differentiation was similar for fetal and adult CS-derived cells (CNN; 64% ± 2% versus 43% ± 16%, respectively; p < 0.05) (Figures 2Ad and 2Al; Figure 2B, middle).
Figure 2.
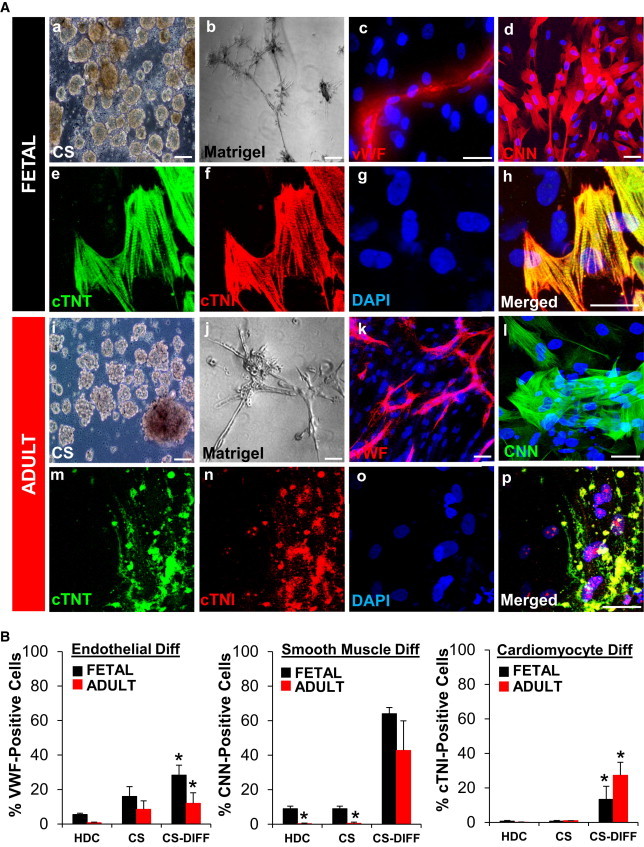
Adult Human CS-Derived Cells Differentiate into Immature Cardiomyocytes
(A) Fetal and adult HDCs formed CSs after 1 week in suspension culture (a and i). CS-derived cells formed microtubules after 24–48 hr in Matrigel angiogenesis assay (b and j). CS-derived cells were plated on coverslips treated with laminin for 15 days to induce cardiovascular differentiation. Confocal images showed expression of vWF (c and k), CNN (d and l), and cardiac proteins cTNT (green) and cTNI (red) (e–h and m–p). Sarcomeres were rare in adult CS-derived CM-like cells (m–p). Nuclei were stained with DAPI (blue). Scale bars, 40 μm.
(B) Cardiovascular differentiation (Diff) of HDCs and CS-derived cells pre- and postdifferentiation was quantitated by FACS. All experiments were performed with a minimum of three independent human heart samples. ∗p < 0.05.
Human CS Contains a Heterogeneous Population of Cells
To determine if the difference in differentiation potential between CS-derived cells from fetal and adult hearts was due to divergent cell populations, we characterized CS cells by flow cytometry for the expression of a panel of surface markers that have been linked to progenitor/stem cells, including C-KIT, VEGF receptors (FLK1, FLT1, and FLT4), CD31, CD144, CD34, CD90, CD114, CD184, and CD271 (Baum et al., 1992; Coultas et al., 2005; Nelson et al., 2008; Shakhova and Sommer, 2010; Shimoji et al., 2010). We compared the expression of these surface markers on acutely dissociated mononuclear cells (ADs), HDCs, and CSs from adult and fetal human hearts (Figure S2A). The most significant differences were observed in the percentage of cells expressing C-KIT, FLT4, CD34, CD90, and CD184 between HDCs and CS cells. The only significant difference between fetal versus adult CSs was the expression of CD184 (Figure S2).
C-KIT has been the cell surface marker most commonly used to identify human aCPCs (Bearzi et al., 2007). The C-KIT+ resident population is very rare in adult compared to fetal human cells (0.3% ± 0.1% versus 5.5% ± 0.6%, respectively; p < 0.05). Interestingly, there was a 5- and 2.5-fold increase in C-KIT+ cells in HDCs from adult and fetal hearts, respectively. Because C-KIT is expressed in aCPCs, hematopoietic, mast cells, endothelial, and certain fibroblasts (Arsalan et al., 2012; Loffredo et al., 2011; Sandstedt et al., 2010), we further phenotyped these C-KIT+ cardiac cells to delineate their potential origin (Figures 3A and 3B). The majority of cardiac C-KIT+ cells coexpressed markers consistent with hematopoietic/EC lineages. Of the fetal and adult C-KIT+ cells, 68%–85% and 61%–77%, respectively, were positive for CD34, CD90, CD31, or CD144. We also analyzed HDCs and CS cells for the coexpression of C-KIT and NKX2.5. Only 10% of C-KIT+ cells coexpressed NKX2.5 (Figures 3B and 3C). Conversely, 5%–20% of the NKX2.5+ population was C-KIT+ (Figure 3D). Fetal and adult NKX2.5+ cells were negative for endothelial surface markers CD31 and CD144, but 50% coexpressed CD271, a neural crest marker (Figures 3D and S3). There were dramatic differences in the expression of CD90, CD34, FLT1, FLT4, and CD184 on NKX2.5+ cells in fetal compared to adult CS cells (Figure 3D). Thus, C-KIT identifies a limited subset of NKX2.5+ cells, and CD271 appears to capture the greatest proportion of the NKX2.5+ subpopulation in both fetal and adult cells.
Figure 3.
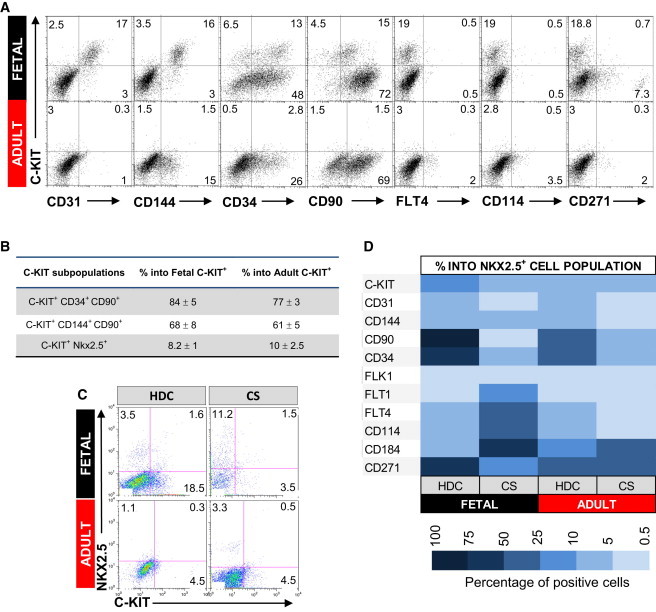
Characterization of Fetal and Adult Human HDCs
(A) HDCs were analyzed for a panel of surface markers (x axis) and C-KIT (y axis) by FACS. Percentage of total positive cells for each marker is shown in the representative dot plot graph.
(B) Percentage of C-KIT cells positive for CD34 and CD90 (hematopoietic markers), CD144 and CD90 (endothelial markers), and NKX2.5 (cardiac progenitor); n = 3.
(C) Costaining for C-KIT (x axis) and NKX2.5 (y axis) in HDCs and CS cells from fetal and adult heart. Percentage of total positive cells for each marker is shown in the representative dot plot graph.
(D) Comparative table showing the percentage of cells positive for C-KIT, CD31, CD144, CD90, CD34, FLK1, FLT1, FLT4, CD114, CD184, and CD271 within the NKX2.5 cell population from fetal and adult HDCs and CSs. All experiments were repeated with a minimum of three independent samples.
Clonal Analysis of Adult CS-Derived Cells Identifies a Cell with Multilineage Potential
To determine if CS contained cells capable of complete cardiac differentiation, we dilutionally cloned adult human CS-derived cells and analyzed their phenotype (Figure 4A). A total of 16 single CS cells were clonally expanded on fibronectin-coated plates leading to homogeneous cultures with elongated, fibroblast-like morphology (Figure S4A). Seven of the clones stopped proliferating, becoming senescent in the first or second passage and were not analyzed in further assays (Figure S4B). Six of the long-term proliferative clones (ID# 2R, 5L, 7L, 5R, 4L, and 8L) were chosen for further characterization of phenotype, differentiation potential, and gene expression profile. Flow cytometry and quantitative PCR (qPCR) demonstrated that all six clones expressed CD105 (endoglin), chondroitin sulfate proteoglycan NG2, as well as weak expression for vascular growth factor receptor Flt4 (Figures 4B and 4C). In contrast, all clones were negative for endothelial markers (CD31, CD144, and FLT1), cardiac stem cell markers (FLK1 and C-KIT), neural crest stem cell marker CD271, and stem cell mobilization marker CD114. Some clones had weak expression for CD106 (7L), CD184 (7L and 8L), and CD34 (5R). CD90 separated the clones into two distinct subpopulations with differing differentiation potential. However, there was no obvious difference in morphology between clones negative for CD90 (2R, 5L, and 7L) or clones positive for CD90 (5R, 4L, and 8L) (Figure S4A).
Figure 4.
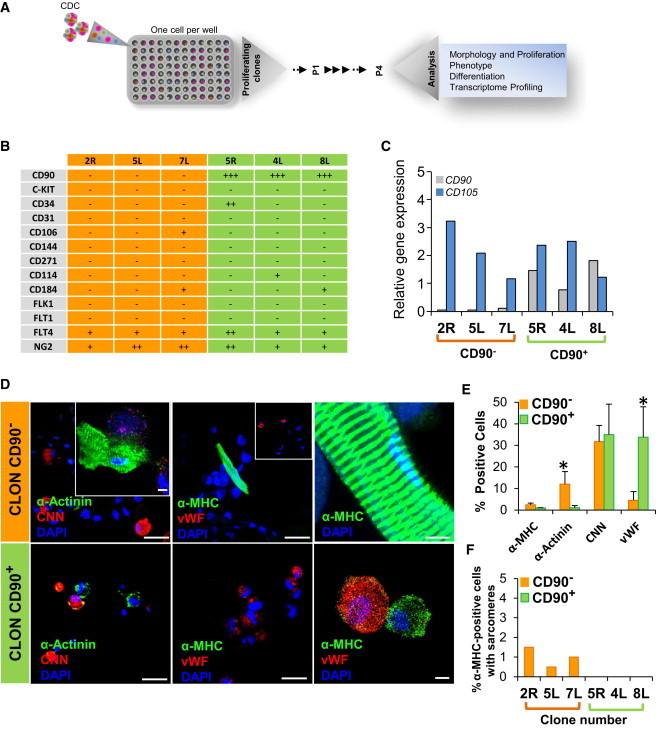
CD90− CS-Derived Clones Are Multipotent, Giving Rise to Mature CMs
(A) Schematic diagram outlining the strategy for cloning and analysis of CS-derived cells.
(B) Characterization for cell surface markers on long-term proliferative CS-derived clones by FACS (n = 6 clones).
(C) Quantitative RT-PCR analysis of CD90 and CD105 expression in long-term proliferative CS-derived clones (n = 6). ∗p < 0.05.
(D) CS-derived clones were differentiated and immunostained for α-actinin (α-SA) and CNN or α-MHC, and vWF and imaged by confocal microscopy. Nuclei were stained with DAPI (blue). Scale bars, 40 μm and 5 μm (inset).
(E) FACS quantification of α-MHC, α-SA, CNN, and vWF in CD90− and CD90+ clones (n = 6). ∗p < 0.05.
(F) Percentage of cardiomyocytes with mature sarcomere formation from CD90− (orange bars) and CD90+ (green bars) clones (n = 6).
To determine if these proliferating clones possess properties of stem or progenitor cells, we analyzed their ability to differentiate into the three cell lineages that constitute the major heart cells. All CD90− clones could differentiate into cells expressing cardiac-specific genes (α-sarcomeric actinin [α-actinin], 12% ± 3.7%; α-myosin heavy chain [α-MHC], 2.5% ± 0.7%), with 60% of the α-MHC-expressing cells demonstrating sarcomeres (1.5% ± 1% of total cells; Figures 4D–4F and S4C). CD90− clones could also differentiate into SMCs (CNN, 30% ± 7.5%) and ECs (vWF, 5% ± 4%). In contrast, although some CD90+ clones could express low levels of cardiac-specific proteins (α-actinin, 1% ± 0.5%, and α-MHC, 0.75% ± 0.1% of total cells), they did not form mature CMs with complete sarcomeres (Figures 4D–4F). CD90+ clones were at least as efficient as CD90− clones in differentiating into SMCs (CNN, 35% ± 14%) and ECs (vWF, 33% ± 14%). These data suggest that the CD90− subpopulation contains a cell with cardiogenic potential.
Adult Human CS-Derived CD90− and CD90+ Clones Display Distinct Transcriptional Profiles
To characterize gene expression in CD90− versus CD90+ clones, we assayed the expression of aCPC candidate genes. ISL-1 was 10.5-fold elevated in CD90− versus CD90+ clones (Figure 5A; p < 0.05). In contrast, expression of fibroblast/myofibroblast gene Periostin was 3.9-fold higher, respectively, in CD90+ versus CD90− clones (Figure 5A; p < 0.05), suggesting a fundamentally different phenotype between the two type of clones. To examine genome-wide expression patterns, we performed next-generation RNA sequencing (RNA-seq). The gene expression profiles of 22,449 genes were examined after removal of duplicate genes following transcript aligning. The low number of CD90 transcripts in the negative versus positive clones confirmed phenotypes of samples (Figure S5). Hierarchical clustering analysis further revealed the molecular difference between the CD90− and CD90+ clones (Figure 5B). A total of 272 genes were differentially expressed in CD90− versus CD90+ clones, of which 133 genes were upregulated and 139 downregulated in CD90− as compared to CD90+ (Table S1). The fold change of major genes differentially upregulated (red) and downregulated (green) in CD90− versus CD90+ clones is shown in Figure 5C.
Figure 5.
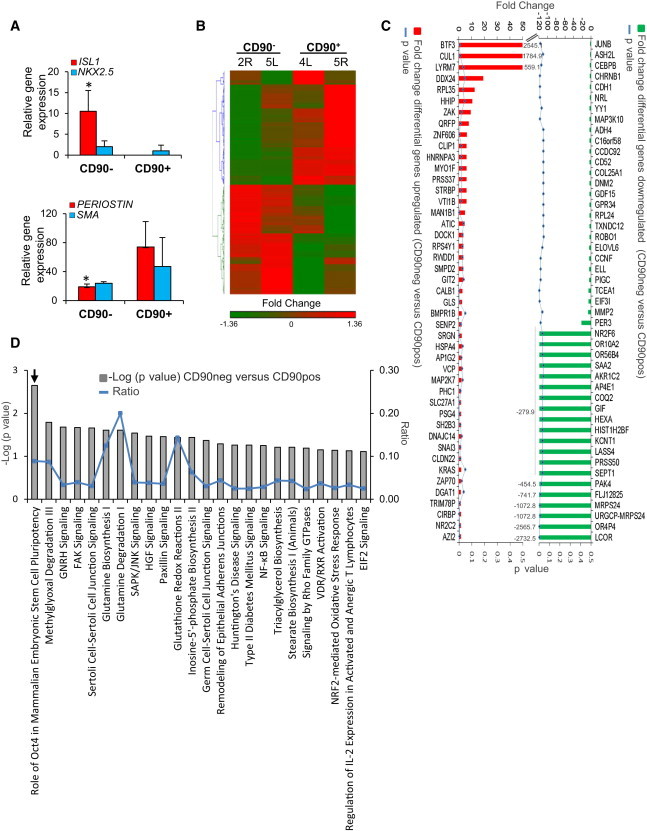
Transcriptome Profile of CS-Derived Clones
(A) Quantitative real-time PCR analysis for ISL-1, NKX2.5, Periostin, and SMA in CD90− and CD90+ CS-derived clones.
(B) Hierarchical clustering analysis of the four CS-derived clones. Differential gene expression was determined using the mean for each sample of CD90− clones versus CD90+ clones. Hierarchical clustering analysis was performed using Partek Genomic Suite software.
(C) Differential expression of genes upregulated (red) and downregulated (green). Blue line represents p value of <0.05.
(D) Pathway analysis using ingenuity pathway analysis (IPA). Significant enrichment of canonical pathways in CD90− samples versus CD90+ samples (gray bars) is shown. Blue line represents ratio.
n = 6; ∗p < 0.05.
To better characterize the molecular basis of the cellular phenotypes and differentiation potential of CD90− and CD90+ clones, we used Gene Ontology (GO) enrichment analysis and canonical pathway analysis to functionally annotate and predict the biological roles of these genes (Figure 5D; Tables S2 and S3). Biological processes that were preferentially represented in the CD90− clones were those associated with embryonic morphogenesis, muscle cell development, proliferation, and telomere maintenance consistent with progenitor-like behavior (Table S2). The most enriched pathway in CD90− clones was the Oct4 and mammalian embryonic stem cell pluripotency pathway (CCNF, PHC1, NR2F6, and ASH2L) (Campbell et al., 2007). In contrast, upregulated genes in CD90+ clones point to networks and pathways of filament-forming cytoskeletal and motility and migration of activated fibroblasts (PAK4, SEPT1, MMP2, and FAP) (Abo et al., 1998; Stawowy et al., 2004; Howard et al., 2012). Transcriptome profiling suggests that human CD90− CS-derived clones are enriched in expression of genes related to stemness, whereas CD90+ clones were enriched with fibroblast-associated genes.
Adult CD90− CS-Derived Cells Have Limited Capacity to Differentiate into More Mature CMs
To determine how specific the lack of CD90 expression may be for the identification of aCPCs in CSs, we isolated and cloned directly by fluorescence-activated cell sorting (FACS) CD90− and CD90+ adult human CS-derived cells (Figure S6A). Purity of individual clones was confirmed, and 21 CD90− clones and 24 CD90+ clones were recovered. Of CD90+ clones, 79.1% stopped proliferating within 30 days in culture in contrast to only the 47.6% of CD90− clones (Figures S6B and S6C). Long-term proliferative CD90− and CD90+ clones grew with a doubling time between 36 and 72 hr (Figure S6C, right). A total of 21 CD90− and 11 CD90+ clones were chosen for further characterization of differentiation potential and gene expression profile before and after differentiation (scheme in Figure 6A). Of CD90− clones, 28.5% (6 of 21 total CD90− clones: #5, #10, #14, #16, #17, and #19) were tripotent for the 3 cell lineages (Figures 6Ba–6Bc, 6C, and 6D), 47.6% of CD90− clones were bipotent for myogenic lineages (CM and SMC), and the remaining 23.8% of CD90− clones were unipotent for SMCs (Figures 6C and 6D). Of 21 CD90− clones analyzed, 16 were cardiomyogenic, expressing α-actinin (Figures 6C and S6D), whereas only 1 CD90+ clone of 11 clones was tripotent (clone number #36), with less than 5% α-actinin-positive cells, 55% of CD90+ clones differentiated into vascular lineages (SMC and EC), and 36% of CD90+ clones were unipotent (3 clones for SMCs [#22, #35, and #45] and 1 clone for ECs [#41]) (Figures 6Bf–6Bh and 6C). After 90 days in culture, some CD90− clones went on to form more mature CMs with complete sarcomeres but at a low rate (<1%) (Figures 6Bd and 6Be). Sarcomere formation was never seen with CD90+ clones.
Figure 6.
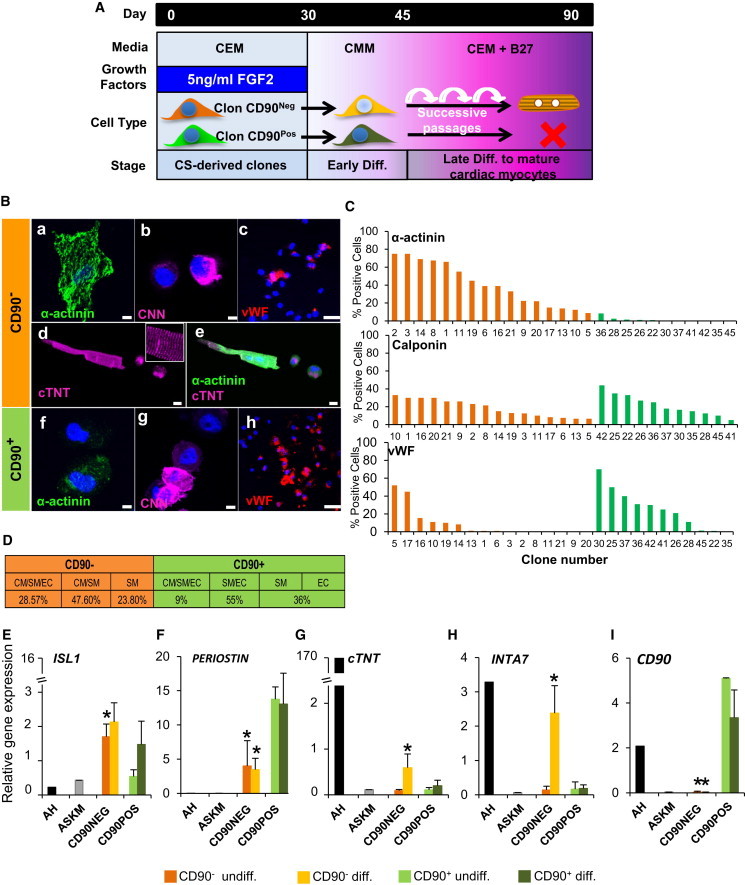
Absence of CD90 Expression Identifies a Population of Cells Containing an aCPC-like Cell
(A) Schematic diagram outlining culture conditions.
(B) CS-derived clones were differentiated and immunostained for α-actinin (a and f), CNN (b and g), and vWF (c and h) and imaged by confocal microscopy. Only CD90− CS-derived clones give rise to myocytes with sarcomeres (d and e) costained with α-actinin and cTNT. Nuclei were stained with DAPI (blue). Scale bars, 50 μm and 10 μm (inset).
(C) Quantification of α-actinin, CNN, and vWF in CD90− (16 clones) and CD90+ (11 clones) CS-derived clones.
(D) Differentiation potential of CD90− and CD90+ CS-derived clones: tripotent (CM/SMC/EC lineages), bipotent (CM/SM; SMC/EC lineages), or unipotent (SMC or EC lineages).
(E–I) Quantitative real-time PCR analysis for ISL-1, cTNT, INTA7, Periostin, and CD90 in CD90− and CD90+ CS-derived clones (n = 26). ∗p < 0.05. undiff., undifferentiated.
We determined the expression of candidate genes in CD90− versus CD90+ clones. ISL-1 mRNA was elevated 4-fold in undifferentiated CD90− clones, whereas Periostin was 3-fold elevated in CD90+ clones. Late CM gene cTNT was elevated 2-fold in CD90− clones after differentiation, but no increase was observed in CD90+ clones (Figure 6G). We also analyzed expression of integrins associated with maturation of cardiomyocytes (Maitra et al., 2000). Integrin α7 (INTA7) was elevated 6-fold in differentiated CD90− clones versus CD90+ clones (Figure 6H). Integrin α5 (INTA5) was expressed at similar levels in both clones (Figure S6E). Skeletal muscle differentiation could not explain our results because myogenin expression was not observed before or after differentiation in CD90− and CD90+ clones (Figure S6F). Expression of CD90 did not change with differentiation (Figure 6I).
To confirm that the CD90− population contains an aCPC, we made CS directly from sorted CD90− and CD90+ fractions obtained from fetal HDCs (Figure 7A). CS-derived cells from the CD90− fraction were more cardiogenic and more likely to undergo complete CM differentiation (Figures 7B–7D): cTNI, 10% ± 8% versus 2.6% ± 1.5% and 2.3% ± 1.5% for CD90− versus CD90+ and unsorted cells, respectively (p < 0.05); cTNT, 25% ± 11% versus 6.9% ± 3.7% and 11% ± 4% for CD90− versus CD90+ and unsorted cells, respectively (p < 0.05).
Figure 7.
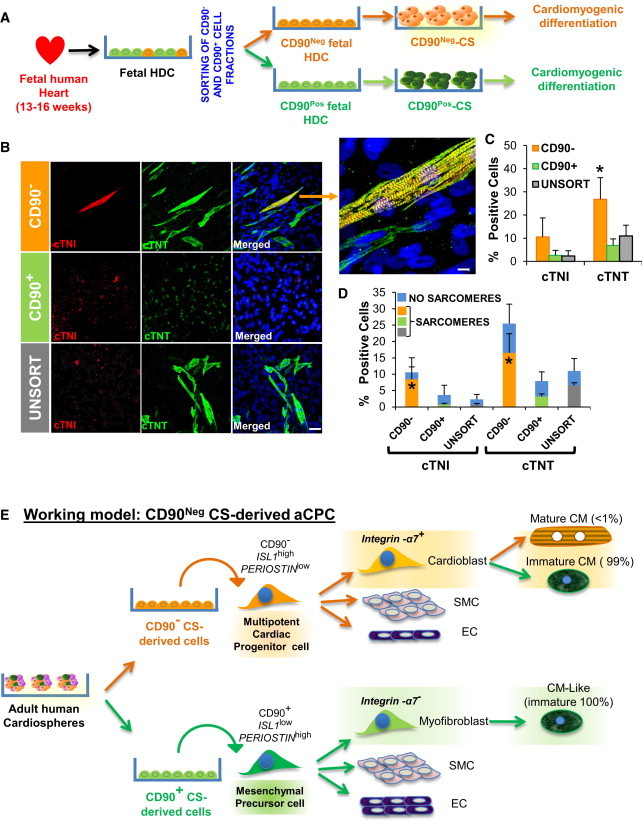
The CD90− Cell Fraction from Fetal Human Hearts Is Enriched in CPCs with Capacity to Differentiate into Mature Cardiomyocytes
(A) CD90+ versus CD90− subpopulations were sorted from fetal HDCs.
(B) After CS formation and differentiation of CD90+, CD90−, and unsorted (UNSORT) cell fractions, the cells were stained for cardiomyocyte proteins cTNI (red) and cTNT (green). Nuclei were counterstained with DAPI (blue). Scale bars, 40 μm.
(C) Quantification of the percentage of cells positive for cTNI and cTNT after differentiation. ∗p < 0.05.
(D) Quantification of the percentage of cTNI and cTNT+ cells with (orange bars) or without (black bars) sarcomere formation. All experiments were performed on three independent human heart samples. ∗p < 0.05.
(E) Working model of CS-derived cell lineages from adult human heart. CD90 differentiates adult human CS-derived cells with differing cardiomyogenic differentiation; CD90−/ISL-1hi/Periostinlo with limited potential to differentiate into CMs with sarcomere formation along with ECs and SMCs and a cardiac mesenchymal/fibroblast cell that was CD90+/ISL-1low/Periostinhi capable of partial differentiation into CM-like cells without sarcomere formation (CM-like immature), ECs, and SMCs.
Discussion
Tissue-specific stem or progenitor cells are notoriously difficult to expand and propagate in vitro in an undifferentiated state. Various culture systems have been developed in an attempt to maintain stemness in these cells, including a 3D culture system based on sphere formation. We utilized CS generated from fetal and adult hearts to attempt to identify an aCPC that has the capacity to differentiate into CMs, SMCs, and ECs. Fetal and adult CS-derived cells differentiated into vascular lineages like SMCs and ECs with similar efficiencies. However, whereas functional CMs were easily derived from fetal CS, the majority of adult CS-derived CMs did not form sarcomeres or exhibit calcium cycling despite expression of relevant cardiac-specific genes. We cloned cells from adult CS and demonstrated two distinct populations of cells with differing cardiovascular differentiation potential. The first cell type has properties consistent with a CPC (CD90−/ISL1hi/Periostinlo), including potential to differentiate into more mature CMs, although the percentage of mature CMs obtained was very low (<1%). This was not simply a technical issue related to our differentiation conditions because multiple published protocols were tested (Figures S7A–S7C). Likewise, the fact that cells isolated from CS, which were derived directly from CD90− cells, demonstrated cardiogenic differentiation suggests that CM dedifferentiation cannot explain these results. Although these results support the conclusion that CSs contain an aCPC with cardiogenic potential, it is unlikely that this differentiation could explain any therapeutic effect given its rarity. Furthermore, the explanation for why formation of mature CMs occurs at such a low efficiency even from clones with cardiogenic potential is unclear. Whether it is related to limitations of the artificial in vitro differentiation conditions or represents a block to differentiation in these cells will require further studies. Regardless, generation of sufficient aCPCs in vitro to regenerate injured hearts will require the discovery of novel techniques to expand and differentiate these cells.
The second cell type appeared to be a cardiac mesenchymal/fibroblast-like cell that was CD90+/ISL1low/Periostinhi and capable of forming SMCs and ECs and partial differentiation into cardiac protein-expressing cells without sarcomere formation. CD90+ clones demonstrated higher expression of fibroblast-associated genes including Periostin compared to CD90− clones. Periostin is the marker most commonly used to identify cardiac fibroblasts (Norris et al., 2008; Snider et al., 2009). Multipotent human fibroblasts have been described in other tissues such as dermis (Chen et al., 2007), but in this study we describe a multipotent cardiac fibroblast (CD90+/Periostin+) in the human heart. These data are consistent with multiple studies that have described “cardiac progenitor/precursor cells” from multiple tissues such as heart (Simpson et al., 2012), bone marrow (Deb et al., 2003; Toma et al., 2002), adipose tissue (Fraser et al., 2006; Armiñán et al., 2009), or dental pulp (Armiñán et al., 2009) that undergo incomplete cardiac differentiation. This has also been observed in other systems because human embryonic stem cell-derived mesenchymal progenitors express cardiac markers but do not form contractile cardiomyocytes (Raynaud et al., 2013; Ramkisoensing et al., 2011). Given the presence of cells in the adult heart with incomplete cardiogenic potential, which are not true CPCs, the ability to form sarcomeres and display calcium cycling should be an essential component for defining an “authentic” cardiac progenitor cell.
Human CS-derived CD90− clones are multipotent and could form mature CMs. These cells displayed a distinct cell surface marker phenotype and a unique stem cell-like transcriptome. Clones negative for CD90 also expressed cardiac progenitor transcription factors NKX2.5 and ISL-1 (Bu et al., 2009; Moretti et al., 2006; Wu et al., 2006). These results are in concordance with earlier studies suggesting that a SCA1+ cardiogenic subpopulation in CS-derived cells is enriched for Isl1 expression (Ye et al., 2012). In fact, genome-wide expression analysis using next-generation sequencing revealed differential gene expression in CD90− clones involved in pluripotency and stemness (Pardo et al., 2010). Interestingly, we observed a dramatic increase in INTA7 expression in CD90− clones with differentiation. α7-integrin forms a heterodimer with β1-integrin in terminal-differentiated cardiomyocytes in heart development and is progressively increased in adult stage (Maitra et al., 2000). The importance of the integrin receptor, α7β1, in the correct assembly of cytoskeleton proteins to form mature and functional CMs is an important unresolved issue.
CD90− and CD90+ clones share expression of some mesenchymal genes such as NG2, CNN, and CD105. Whether this indicates two cell types representing different developmental stages of a similar mesenchymal precursor will need to be determined. The exact identity of the CS-derived CD90− aCPC remains to be determined. Although C-KIT has been reported to identify resident cardiac stem cells (Bearzi et al., 2007; Beltrami et al., 2003), it was not expressed on most NKX2.5+ cells in CSs nor was it expressed on the CD90− cardiogenic clones we analyzed. Conversely, the majority of C-KIT+ cells in fetal and adult hearts did not express NKX2.5. Our results suggest that the majority of the C-KIT population in human heart expresses markers consistent with a hematopoietic/endothelial lineage (C-KIT+/CD90+/CD34+; C-KIT+/CD90+/CD144+). C-KIT+/CD90− CS-derived cells were cardiogenic, but they did not demonstrate sarcomere formation (Figures S7D and S7E). Consistent with our results, it has been reported that unselected human CS-derived cells are functionally superior to C-KIT+/CD90− or CD90+ purified CS-derived cells (K. Cheng et al., 2012, Circulation, abstract; R.R. Smith et al., 2008, Circulation, abstract). Whether C-KIT+/NKX2.5+ cells represent the endogenous aCPC and the CD90−/C-KIT−/NKX2.5low we cloned are related or this CD90−/C-KIT−/NKX2.5low clone is a unique CPC is impossible for us to determine because identification of NKX2.5+ cells required fixing and permeabilizing the cells. The neural crest marker CD271 was able to identify the largest proportion of the NKX2.5 population in the heart but was not expressed on our CD90 clones. Resolution of this issue will require identifying additional cell surface markers so that endogenous, live cells can be isolated directly. Interestingly, dramatic changes in the expression of surface markers including C-KIT were observed in cell culture. The surface marker changing most dramatically was CD90, decreasing from 80% to 10% in fetal and 40% to 10% in adult freshly dissociated cells to CSs, respectively (Figure S2). An epithelial-to-mesenchymal transition mechanism has been reported to occur within CS that is associated with downregulation of CD90 expression (Forte et al., 2012). Thus, whether the CS-derived CD90− aCPC clones we isolated exist endogenously in human hearts or are a byproduct of the CS culture system remains to be determined.
Taken together, these findings suggest a working model to define CS-derived cell lineages with cardiovascular differentiation potential from the adult human heart (Figure 7E). In this model, CD90 expression segregates adult human CS-derived cells with differing cardiomyogenic differentiation. Our study suggests the presence of a “true” adult CPC versus a mesenchymal/myofibroblast cell that can express cardiac proteins but not differentiate into a true CM.
Experimental Procedures
Derivation of Primary Cardiac Cell Culture from Human Heart Samples
A total of 32 human biopsies were obtained from 32 donors (16 fetal and 16 adult). Fetal heart tissue was harvested under the supervision of the UCLA institutional review board, and adult human heart tissue was harvested from discarded pathological specimens at the time of heart transplantation under the supervision of the UCLA institutional review board or at the time of left ventricular assist device (LVAD) placement under the supervision of the UW institutional review board (Table S4). Biopsies were stored in Ca2+/Mg++-free Dulbecco’s PBS (DPBS; Cellgro) supplemented with 2% penicillin/streptomycin (P/S) and processed at a maximum of 4 hr postsurgery. Further details of heart dissociation and primary culture of HDCs are described in the Supplemental Experimental Procedures.
Generation of CS
Confluent HDCs were washed with DPBS, trypsinized with a 0.1% trypsin solution (Sigma-Aldrich) for 4 min at 37°C, neutralized with serum, centrifuged at 1,200 rpm for 5 min, and finally resuspended in CS growth media (CGM): 70% DMEM:F12 1:1 (Invitrogen)/30% IMDM (Lonza), 0.001% 2-mercaptoethanol (Sigma-Aldrich), 40 nM thrombin (Sigma-Aldrich), 40 nM cardiotrophin (PeproTech), 2% B27 (Gibco), 10 ng/ml epidermal growth factor (EGF; R&D Systems), 20 ng/ml basic fibroblast growth factor (bFGF; PeproTech), and antibiotics and L-glutamine into 0.8% methylcellulose (R&D Systems). Cells were seeded at a density of 25,000 cells/cm2 in an untreated 60 mm culture dish (BD Falcon). Every 48 hr, EGF and bFGF were added to the culture medium. For ADH cultures, the pellet was resuspended and cultured at a density of 5,000 cells/cm2 in T-75 culture flasks (BD Falcon) precoated with fibronectin (20 μg/ml) in growth media (Figure 1A).
Cardiovascular Differentiation In Vitro
Primary CS (5–10 U) or ADHs (10,000 cells/cm2) were seeded on 12-mm-diameter laminin-coated (Sigma-Aldrich) coverslips (Menzel-Gläser, http://www.menzel.de). Four distinct differentiation media were used for cardiovascular differentiation: basal medium, cardiomyogenic medium, smooth-muscle medium, and endothelial medium. For EC tube formation assay, we followed the BD protocol (BD Matrigel matrix catalog number 354234). All differentiation experiments were performed for 2–3 weeks, replacing differentiation media every 3 days. Further details are described in the Supplemental Experimental Procedures.
Immunofluorescence in Cover Slides and Cytospin
We performed immunofluorescence as described in the Supplemental Experimental Procedures. All slides were analyzed by confocal microscopy (LEIKA SP2 1P-FCS Confocal and NIKON A1R Confocal).
Calcium Fluorescence Imaging
Cell cultures were incubated with the fluorescent calcium indicator dye Fluo-4 AM (10 μM; Invitrogen) and nonionic surfactant Pluronic F-127 (0.02%; Invitrogen) in cardiomyogenic medium. Spontaneous calcium fluorescence activity was imaged and recorded. Further details are described in the Supplemental Experimental Procedures.
Flow Cytometry
We performed FACS analysis as described in the Supplemental Experimental Procedures. Data were acquired in a FACS LSRII flow cytometer (BD Biosciences). Gating strategy is described in Figure S1. FACSDIVA software (BD Biosciences) was used for acquisition of data. For later analyses, we used FlowJo software (version 7.6.5).
Cell Sorting
We performed FACS as described in the Supplemental Experimental Procedures. Sorting experiments were done on a FACSAria III cell sorter (BD Biosciences), at low sheath pressure and by using a nozzle of 80 μm in diameter.
Clonal Assay
CS-derived cells were trypsinized and centrifuged. Pellet was resuspended in growth medium, and cell number was determined using a hemocytometer. We performed clonal assay as described in the Supplemental Experimental Procedures. All the cells from individual clones were analyzed by FACS flow cytometer, RNA isolation for qPCR and RNA-seq, and for differentiation potential.
RNA Isolation and Quantitative Real-Time-PCR
We performed RNA isolation and quantitative real-time PCR as described in the Supplemental Experimental Procedures.
RNA-Seq
We performed whole-genome transcriptomic analysis as described in the Supplemental Experimental Procedures.
Statistical Analyses
Results are presented as the mean ± SEM of a number (n) of independent experiments. p values were calculated using GraphPad software. Differences in multiple groups were evaluated using ANOVA. Correlation was deemed significant at the p < 0.05 level (two tailed).
Author Contributions
N.G.-L. carried out, designed, analyzed experiments, and wrote the manuscript. O.A. carried out heart-processing and immunofluorescence experiments. C.K. carried out calcium-imaging assays. Y.Z. analyzed RNA-seq experiments. A.N., D.L., and A.S.-O. carried out clinical work. W.R.M. designed and supervised this work and edited the manuscript.
Acknowledgments
We thank Iris Williams (Flow Cytometry Core Laboratory at UCLA), Matt Schibler (CNSI Advanced Light Microscopy/Spectroscopy Facility, UCLA, CA), Ron Seifert (ISCRM Lynn and Mike Garvey Cell Imaging Laboratory, UW, WA), and Jeffrey Boyd (ISCRM Flow Cytometry Core, UW, WA) for their technical assistance. As well, we thank the Genome Technology Access Center at Washington University for assistance with next-generation sequencing experiments. Kyohei Oyama, Daniel El-Nachef, and Ander Izeta provided helpful experimental suggestions and comments on the manuscript. This work was supported by grants from the NIH (R01 HL70748 to W.R.M. and R01HL094384 to A.S.-O.).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
Cells were incubated with the fluorescent calcium indicator dye Fluo-4 AM and nonionic surfactant Pluronic F-127 in cardiomyogenic medium for 30 min. Spontaneous calcium fluorescence activity in cell cultures was imaged and recorded. The acquired video image data were processed using ImageJ software.
Cell culture was incubated with the fluorescent calcium indicator dye Fluo-4 AM and nonionic surfactant Pluronic F-127 in cardiomyogenic medium for 30 min. Spontaneous calcium fluorescence activity in cell cultures was imaged and recorded, but no calcium transients were observed. The acquired video image data were processed using ImageJ software.
References
- Abo A., Qu J., Cammarano M.S., Dan C., Fritsch A., Baud V., Belisle B., Minden A. PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 1998;17:6527–6540. doi: 10.1093/emboj/17.22.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armiñán A., Gandía C., Bartual M., García-Verdugo J.M., Lledó E., Mirabet V., Llop M., Barea J., Montero J.A., Sepúlveda P. Cardiac differentiation is driven by NKX2.5 and GATA4 nuclear translocation in tissue-specific mesenchymal stem cells. Stem Cells Dev. 2009;18:907–918. doi: 10.1089/scd.2008.0292. [DOI] [PubMed] [Google Scholar]
- Arsalan M., Woitek F., Adams V., Linke A., Barten M.J., Dhein S., Walther T., Mohr F.W., Garbade J. Distribution of cardiac stem cells in the human heart. ISRN Cardiol. 2012;2012:483407. doi: 10.5402/2012/483407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barile L., Messina E., Giacomello A., Marbán E. Endogenous cardiac stem cells. Prog. Cardiovasc. Dis. 2007;50:31–48. doi: 10.1016/j.pcad.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Baum C.M., Weissman I.L., Tsukamoto A.S., Buckle A.M., Peault B. Isolation of a candidate human hematopoietic stem-cell population. Proc. Natl. Acad. Sci. USA. 1992;89:2804–2808. doi: 10.1073/pnas.89.7.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearzi C., Rota M., Hosoda T., Tillmanns J., Nascimbene A., De Angelis A., Yasuzawa-Amano S., Trofimova I., Siggins R.W., Lecapitaine N. Human cardiac stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearzi C., Leri A., Lo Monaco F., Rota M., Gonzalez A., Hosoda T., Pepe M., Qanud K., Ojaimi C., Bardelli S. Identification of a coronary vascular progenitor cell in the human heart. Proc. Natl. Acad. Sci. USA. 2009;106:15885–15890. doi: 10.1073/pnas.0907622106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Beltrami A.P., Barlucchi L., Torella D., Baker M., Limana F., Chimenti S., Kasahara H., Rota M., Musso E., Urbanek K. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Campbell P.A., Perez-Iratxeta C., Andrade-Navarro M.A., Rudnicki M.A. Oct4 targets regulatory nodes to modulate stem cell function. PLoS One. 2007;2:e553. doi: 10.1371/journal.pone.0000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.G., Zhang W.J., Bi D., Liu W., Wei X., Chen F.F., Zhu L., Cui L., Cao Y. Clonal analysis of nestin(-) vimentin(+) multipotent fibroblasts isolated from human dermis. J. Cell Sci. 2007;120:2875–2883. doi: 10.1242/jcs.03478. [DOI] [PubMed] [Google Scholar]
- Coultas L., Chawengsaksophak K., Rossant J. Endothelial cells and VEGF in vascular development. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- Davis D.R., Zhang Y., Smith R.R., Cheng K., Terrovitis J., Malliaras K., Li T.S., White A., Makkar R., Marbán E. Validation of the cardiosphere method to culture cardiac progenitor cells from myocardial tissue. PLoS One. 2009;4:e7195. doi: 10.1371/journal.pone.0007195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D.R., Ruckdeschel Smith R., Marbán E. Human cardiospheres are a source of stem cells with cardiomyogenic potential. Stem Cells. 2010;28:903–904. doi: 10.1002/stem.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb A., Wang S., Skelding K.A., Miller D., Simper D., Caplice N.M. Bone marrow-derived cardiomyocytes are present in adult human heart: a study of gender-mismatched bone marrow transplantation patients. Circulation. 2003;107:1247–1249. doi: 10.1161/01.cir.0000061910.39145.f0. [DOI] [PubMed] [Google Scholar]
- Fernandes K.J., McKenzie I.A., Mill P., Smith K.M., Akhavan M., Barnabé-Heider F., Biernaskie J., Junek A., Kobayashi N.R., Toma J.G. A dermal niche for multipotent adult skin-derived precursor cells. Nat. Cell Biol. 2004;6:1082–1093. doi: 10.1038/ncb1181. [DOI] [PubMed] [Google Scholar]
- Forte E., Miraldi F., Chimenti I., Angelini F., Zeuner A., Giacomello A., Mercola M., Messina E. TGFβ-dependent epithelial-to-mesenchymal transition is required to generate cardiospheres from human adult heart biopsies. Stem Cells Dev. 2012;21:3081–3090. doi: 10.1089/scd.2012.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J.K., Schreiber R., Strem B., Zhu M., Alfonso Z., Wulur I., Hedrick M.H. Plasticity of human adipose stem cells toward endothelial cells and cardiomyocytes. Nat. Clin. Pract. Cardiovasc. Med. 2006;3(Suppl 1):S33–S37. doi: 10.1038/ncpcardio0444. [DOI] [PubMed] [Google Scholar]
- Gago N., Pérez-López V., Sanz-Jaka J.P., Cormenzana P., Eizaguirre I., Bernad A., Izeta A. Age-dependent depletion of human skin-derived progenitor cells. Stem Cells. 2009;27:1164–1172. doi: 10.1002/stem.27. [DOI] [PubMed] [Google Scholar]
- Goumans M.J., de Boer T.P., Smits A.M., van Laake L.W., van Vliet P., Metz C.H., Korfage T.H., Kats K.P., Hochstenbach R., Pasterkamp G. TGF-beta1 induces efficient differentiation of human cardiomyocyte progenitor cells into functional cardiomyocytes in vitro. Stem Cell Res. 2007;1:138–149. doi: 10.1016/j.scr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- He J.Q., Vu D.M., Hunt G., Chugh A., Bhatnagar A., Bolli R. Human cardiac stem cells isolated from atrial appendages stably express c-kit. PLoS One. 2011;6:e27719. doi: 10.1371/journal.pone.0027719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E.W., Crider B.J., Updike D.L., Bullen E.C., Parks E.E., Haaksma C.J., Sherry D.M., Tomasek J.J. MMP-2 expression by fibroblasts is suppressed by the myofibroblast phenotype. Exp. Cell Res. 2012;318:1542–1553. doi: 10.1016/j.yexcr.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Jaleel N., Kumarapeli A., Berretta R.M., Bratinov G., Shan X., Wang H., Houser S.R., Margulies K.B. Increased cardiac myocyte progenitors in failing human hearts. Circulation. 2008;118:649–657. doi: 10.1161/CIRCULATIONAHA.107.761031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme M.A., Myerson D., Saffitz J.E., Murry C.E. Evidence for cardiomyocyte repopulation by extracardiac progenitors in transplanted human hearts. Circ. Res. 2002;90:634–640. doi: 10.1161/01.res.0000014822.62629.eb. [DOI] [PubMed] [Google Scholar]
- Li T.S., Cheng K., Lee S.T., Matsushita S., Davis D., Malliaras K., Zhang Y., Matsushita N., Smith R.R., Marbán E. Cardiospheres recapitulate a niche-like microenvironment rich in stemness and cell-matrix interactions, rationalizing their enhanced functional potency for myocardial repair. Stem Cells. 2010;28:2088–2098. doi: 10.1002/stem.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo F.S., Steinhauser M.L., Gannon J., Lee R.T. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell. 2011;8:389–398. doi: 10.1016/j.stem.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra N., Flink I.L., Bahl J.J., Morkin E. Expression of alpha and beta integrins during terminal differentiation of cardiomyocytes. Cardiovasc. Res. 2000;47:715–725. doi: 10.1016/s0008-6363(00)00140-1. [DOI] [PubMed] [Google Scholar]
- Makkar R.R., Smith R.R., Cheng K., Malliaras K., Thomson L.E., Berman D., Czer L.S., Marbán L., Mendizabal A., Johnston P.V. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Montserrat N., Okamura D., Suzuki K., Izpisua Belmonte J.C. Cardiosphere-derived cells for heart regeneration. Lancet. 2012;379:2425–2426. doi: 10.1016/S0140-6736(12)61062-9. [DOI] [PubMed] [Google Scholar]
- Messina E., De Angelis L., Frati G., Morrone S., Chimenti S., Fiordaliso F., Salio M., Battaglia M., Latronico M.V., Coletta M. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ. Res. 2004;95:911–921. doi: 10.1161/01.RES.0000147315.71699.51. [DOI] [PubMed] [Google Scholar]
- Mishra R., Vijayan K., Colletti E.J., Harrington D.A., Matthiesen T.S., Simpson D., Goh S.K., Walker B.L., Almeida-Porada G., Wang D. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011;123:364–373. doi: 10.1161/CIRCULATIONAHA.110.971622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A., Caron L., Nakano A., Lam J.T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Murry C.E., Soonpaa M.H., Reinecke H., Nakajima H., Nakajima H.O., Rubart M., Pasumarthi K.B., Virag J.I., Bartelmez S.H., Poppa V. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004;428:664–668. doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- Nelson T.J., Faustino R.S., Chiriac A., Crespo-Diaz R., Behfar A., Terzic A. CXCR4+/FLK-1+ biomarkers select a cardiopoietic lineage from embryonic stem cells. Stem Cells. 2008;26:1464–1473. doi: 10.1634/stemcells.2007-0808. [DOI] [PubMed] [Google Scholar]
- Norris R.A., Borg T.K., Butcher J.T., Baudino T.A., Banerjee I., Markwald R.R. Neonatal and adult cardiovascular pathophysiological remodeling and repair: developmental role of periostin. Ann. N Y Acad. Sci. 2008;1123:30–40. doi: 10.1196/annals.1420.005. [DOI] [PubMed] [Google Scholar]
- Oh H., Chi X., Bradfute S.B., Mishina Y., Pocius J., Michael L.H., Behringer R.R., Schwartz R.J., Entman M.L., Schneider M.D. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann. N Y Acad. Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M.M., Choudhary J. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramkisoensing A.A., Pijnappels D.A., Askar S.F., Passier R., Swildens J., Goumans M.J., Schutte C.I., de Vries A.A., Scherjon S., Mummery C.L. Human embryonic and fetal mesenchymal stem cells differentiate toward three different cardiac lineages in contrast to their adult counterparts. PLoS One. 2011;6:e24164. doi: 10.1371/journal.pone.0024164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C.M., Halabi N., Elliott D.A., Pasquier J., Elefanty A.G., Stanley E.G., Rafii A. Human embryonic stem cell derived mesenchymal progenitors express cardiac markers but do not form contractile cardiomyocytes. PLoS One. 2013;8:e54524. doi: 10.1371/journal.pone.0054524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Sandstedt J., Jonsson M., Lindahl A., Jeppsson A., Asp J. C-kit+ CD45- cells found in the adult human heart represent a population of endothelial progenitor cells. Basic Res. Cardiol. 2010;105:545–556. doi: 10.1007/s00395-010-0088-1. [DOI] [PubMed] [Google Scholar]
- Shakhova O., Sommer L. Neural crest-derived stem cells. In: Gage F., Watt F., editors. StemBook. The Stem Cell Research Community; Boston: 2010. [Google Scholar]
- Shimoji K., Yuasa S., Onizuka T., Hattori F., Tanaka T., Hara M., Ohno Y., Chen H., Egasgira T., Seki T. G-CSF promotes the proliferation of developing cardiomyocytes in vivo and in derivation from ESCs and iPSCs. Cell Stem Cell. 2010;6:227–237. doi: 10.1016/j.stem.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Simpson D.L., Mishra R., Sharma S., Goh S.K., Deshmukh S., Kaushal S. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 2012;126(11, Suppl 1):S46–S53. doi: 10.1161/CIRCULATIONAHA.111.084699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.R., Barile L., Cho H.C., Leppo M.K., Hare J.M., Messina E., Giacomello A., Abraham M.R., Marbán E. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- Smits A.M., van Laake L.W., den Ouden K., Schreurs C., Szuhai K., van Echteld C.J., Mummery C.L., Doevendans P.A., Goumans M.J. Human cardiomyocyte progenitor cell transplantation preserves long-term function of the infarcted mouse myocardium. Cardiovasc. Res. 2009;83:527–535. doi: 10.1093/cvr/cvp146. [DOI] [PubMed] [Google Scholar]
- Snider P., Standley K.N., Wang J., Azhar M., Doetschman T., Conway S.J. Origin of cardiac fibroblasts and the role of periostin. Circ. Res. 2009;105:934–947. doi: 10.1161/CIRCRESAHA.109.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawowy P., Margeta C., Kallisch H., Seidah N.G., Chrétien M., Fleck E., Graf K. Regulation of matrix metalloproteinase MT1-MMP/MMP-2 in cardiac fibroblasts by TGF-beta1 involves furin-convertase. Cardiovasc. Res. 2004;63:87–97. doi: 10.1016/j.cardiores.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Toma C., Pittenger M.F., Cahill K.S., Byrne B.J., Kessler P.D. Human mesenchymal stem cells differentiate to a cardiomyocyte phenotype in the adult murine heart. Circulation. 2002;105:93–98. doi: 10.1161/hc0102.101442. [DOI] [PubMed] [Google Scholar]
- Tomita Y., Matsumura K., Wakamatsu Y., Matsuzaki Y., Shibuya I., Kawaguchi H., Ieda M., Kanakubo S., Shimazaki T., Ogawa S. Cardiac neural crest cells contribute to the dormant multipotent stem cell in the mammalian heart. J. Cell Biol. 2005;170:1135–1146. doi: 10.1083/jcb.200504061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.M., Fujiwara Y., Cibulsky S.M., Clapham D.E., Lien C.L., Schultheiss T.M., Orkin S.H. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- Wu S.M., Chien K.R., Mummery C. Origins and fates of cardiovascular progenitor cells. Cell. 2008;132:537–543. doi: 10.1016/j.cell.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Boyle A., Shih H., Sievers R.E., Zhang Y., Prasad M., Su H., Zhou Y., Grossman W., Bernstein H.S., Yeghiazarians Y. Sca-1+ cardiosphere-derived cells are enriched for Isl1-expressing cardiac precursors and improve cardiac function after myocardial injury. PLoS One. 2012;7:e30329. doi: 10.1371/journal.pone.0030329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells were incubated with the fluorescent calcium indicator dye Fluo-4 AM and nonionic surfactant Pluronic F-127 in cardiomyogenic medium for 30 min. Spontaneous calcium fluorescence activity in cell cultures was imaged and recorded. The acquired video image data were processed using ImageJ software.
Cell culture was incubated with the fluorescent calcium indicator dye Fluo-4 AM and nonionic surfactant Pluronic F-127 in cardiomyogenic medium for 30 min. Spontaneous calcium fluorescence activity in cell cultures was imaged and recorded, but no calcium transients were observed. The acquired video image data were processed using ImageJ software.


