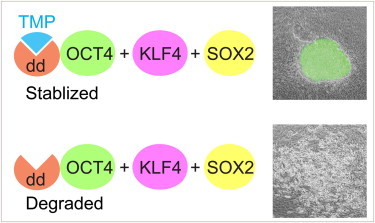
Summary
Induced pluripotent stem cells (iPSCs) generated from somatic cells by ectopic expression of reprogramming factors, e.g., POU5F1 (OCT4), KLF4, and SOX2, have great potential for regenerative medicine. However, before they can be used in a clinical setting, the mechanism of reprogramming needs to be better understood. Here, by engineering reprogramming factors to a destabilizing protein domain, we achieved inducible generation of mouse and pig iPSCs. Stability of the fusion protein was precisely regulated by the addition of the cell-permeable small molecule trimethoprim (TMP) in a dose-dependent manner. With these tools, we found that during the early and middle stages of reprogramming, exogenous OCT4 or KLF4 could be omitted, whereas exogenous SOX2 expression at early and middle stages was required for successful reprogramming. Our TMP reprogramming system is useful for defining the stoichiometry and temporal requirements of transcription factors for reprogramming.
Graphical Abstract
Highlights
-
•
TMP reprogramming system can be used for inducible generation of iPSCs
-
•
Stability of reprogramming factors is precisely regulated by TMP
-
•
During early/middle stages of reprogramming, exogenous OCT4 or KLF4 can be omitted
-
•
During early/middle stages of reprogramming, exogenous SOX2 expression is required
By engineering reprogramming factors to a destabilizing protein domain, Li, Wu, and colleagues achieved inducible generation of iPSCs. Stability of the fusion protein was precisely regulated by small molecule TMP. With these tools, we found that during the early and middle stages of reprogramming, exogenous OCT4 or KLF4 could be omitted, whereas exogenous SOX2 expression at early/middle stages was required for successful reprogramming.
Introduction
Mouse and human somatic cells can be reprogrammed to embryonic stem cell (ESC)-like cells, known as induced pluripotent stem cells (iPSCs), classically by ectopic expression of transcription factors, Oct4, Klf4, Sox2, and c-Myc (Okita et al., 2007; Takahashi and Yamanaka, 2006), or by other combinations (Nakagawa et al., 2008; Wernig et al., 2008b). The Cre/loxP and the Doxycycline (Dox)-inducible systems have proven useful for studying reprogramming mechanisms (Hanna et al., 2009; Soldner et al., 2009; Yamanaka, 2009). For instance, the Dox system has been used to demonstrate that reprogramming somatic cells to iPSCs occurs in a sequential and stochastic manner (Hanna et al., 2009; Yamanaka, 2009). However, limitations of the Dox system, such as the need for a transactivating protein as well as limited control of gene expression levels (Gossen et al., 1995), clearly demonstrate the need for a more flexible system with fine-tuning capabilities to further dissect the molecular mechanisms of reprogramming. By determining the optimal levels of reprogramming factors necessary, we may be able to improve the efficiency and quality of reprogramming in multiple species in which reprogramming remains elusive.
Recently, an inducible system to regulate protein stability by a commercially available small molecule, trimethoprim (TMP), was reported (Iwamoto et al., 2010; Sando et al., 2013). In this system, protein instability is conferred by engineering a fusion protein with the destabilizing domain (dd) derived from Escherichia coli dihydrofolate reductase, which targets the fusion protein to the proteasome for degradation (Iwamoto et al., 2010). The addition of TMP stabilizes the fusion protein in a rapid, reversible, and dose-dependent manner, thereby altering the protein-turnover rate to transform a short-lived or nondetectable protein into a protein that functions for a precisely controlled period of time (Iwamoto et al., 2010).
We have incorporated the TMP-regulated dd into piggyBac transposon-based reprogramming vectors to allow inducible generation of mouse and pig iPSCs. We are able to fine-tune the level and duration of reprogramming protein stability and analyze the stoichiometry and temporal requirements in detail. A recent report that used single-cell expression analyses revealed the essential role of SOX2 during the late phase of reprogramming (Buganim et al., 2012). By using the TMP system, we further found that Sox2 is also essential during early and middle phases of reprogramming.
Results
Reprogramming of Mouse Fibroblasts with the TMP-Inducible System
To create the TMP-inducible reprogramming system, we constructed two piggyBac transposon overexpression vectors, OddKS and OKS (Figure 1A; Table S1 available online), both of which encode the human OCT4, KLF4, and SOX2 (hOCT4, hKLF4, and hSOX2) cDNA sequences as a single open reading frame linked by the F2A and T2A sequences, respectively (Okita et al., 2008; Szymczak et al., 2004). The destabilizing domain (dd) was encoded in frame at the 5′ end of the OCT4 cDNA in the OddKS construct and represented the only difference between the two constructs. These constructs were individually introduced by electroporation into Oct4-GFP mouse embryonic fibroblasts (MEFs) that carried a transgene composed of the 18 kb Oct4 promoter sequence upstream of the GFP coding sequence. OddKS -transfected fibroblasts were grown in the absence of TMP or in 1 μM TMP. By day 6 posttransfection, colonies with an ESC-like morphology started to appear among the MEFs transfected with OKS and OddKS + 1 μM TMP, but not among the OddKS MEFs without TMP. By day 8, the ESC-like colonies showed distinct edges, a hallmark of ESC colonies, and were clearly GFP positive under fluorescence microscope. At day 14, all of the ESC-like colonies uniformly expressed GFP (Figure 1B). About 287 ± 23 GFP-positive colonies arose from 7.2 × 104 cells that were transfected with the OddKS vector and treated with 1 μM TMP, representing a reprogramming efficiency of ∼0.40%, similar to the efficiency of cells transfected with the OKS control vector, ∼0.38% (279 ± 26 GFP-positive colonies per 7.2 × 104 cells transfected) (Figure 1C; Table S2). No GFP-positive colonies were detected in the absence of TMP at day 14 or day 20, when cells became too confluent to be cultured further. Taken together, these data demonstrated that our TMP-inducible reprogramming system is fully TMP dependent, with a clear all-or-none effect. GFP-positive clones were picked at day 15 and expanded, which showed similar behavior during passaging as other iPSC lines obtained from more conventional protocols. These iPSCs have been continuously maintained for more than 35 passages by standard trypsinization. They homogenously expressed the stem cell markers OCT4, NANOG, and SSEA1 (Figure S1). Transplantation of these cells into nude mice resulted in teratomas that consisted of tissues derived from all three germ layers, indicating that these cells are pluripotent (Figure S2).
Figure 1.
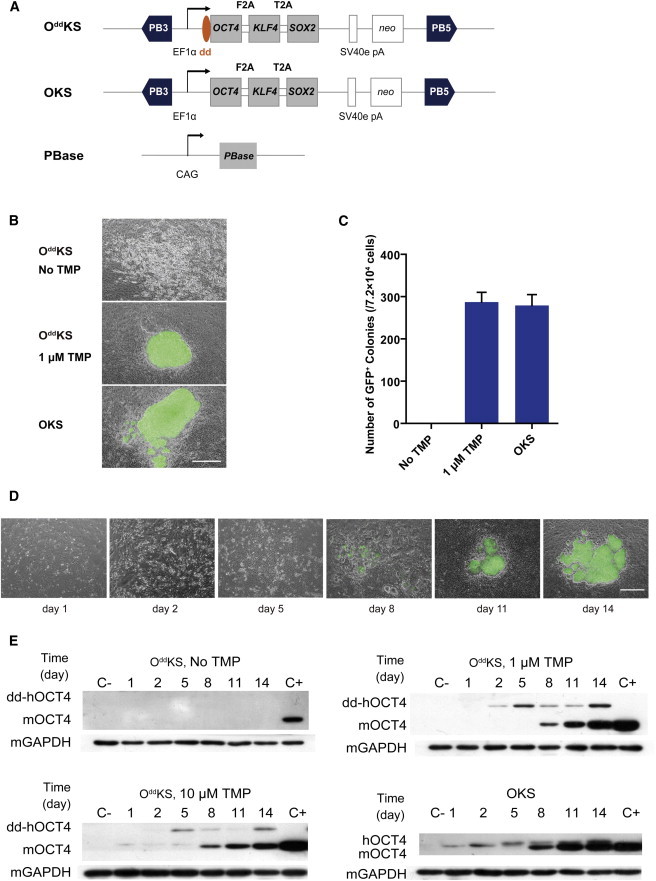
Generation of Mouse iPSCs by a TMP-Inducible piggyBac Vector
(A) Schematic representation of the piggyBac transposon reprogramming vectors. Three factors, hOCT4, hKLF4, and hSOX2, linked by 2A peptides, were coexpressed and driven by the constitutively active EF1α promoter. The destabilizing domain (dd) was fused to the 5′ end of the hOCT4 gene in the OddKS vector. The OKS vector with unfused hOCT4 was used as the control. SV40E pA, the SV40 early polyadenylation signal; PB3′ and PB5′, terminals of the piggyBac transposon.
(B) Micrographs of iPSCs in the absence or presence of 1 μM TMP 14 days after electroporation with the OddKS or OKS constructs. TMP-regulated stability of OCT4 led to reprogrammed GFP-positive iPS colonies. Top: OddKS, no TMP treatment; middle: OddKS + 1 μM TMP treatment; bottom: OKS vector, no TMP treatment. Scale bar, 500 μm.
(C) The number of GFP-positive colonies obtained by transposon-based reprogramming per 7.2 × 104 cells were scored at day 14 postelectrotransfection. Experiments were performed at least three times per condition. Error bars, SD.
(D) Micrographs of iPSCs in the presence of 1 μM TMP with the OddKS or OKS constructs at different days after electroporation. Scale bar, 500 μm.
(E) Western blots of OddKS- and OKS-transfected MEFs monitoring the presence of exogenous OCT4 (hOCT4) and endogenous OCT4 (mOCT4) expression at multiple time points in the presence or absence of TMP. Oct4-GFP MEFs were used as a negative control (C−) for OCT4 expression, and G4-56 ES cells were used as the positive control (C+).
To follow the dynamics of TMP-regulated OCT4 protein expression, we monitored exogenous dd-hOCT4 and endogenous mOCT4 protein levels in transfected MEFs at six time points covering the three phases of reprogramming: early stage (day 1, 2), middle stage (day 5, 8), and late stage (day 11, 14). The anti-OCT4 antibody used can detect OCT4 of both mouse and human origin. Mouse OCT4 has a predicted molecular weight of approximately 43 kDa, whereas hOCT4 has a higher molecular weight (∼63.6 kDa) because of the dd addition (∼20 kDa) to the N terminus of OCT4 and residual 2A peptide (∼0.6 kDa) linked to the C terminus of OCT4. Multiple replicate experiments showed similar protein expression dynamics for hOCT4 and mOCT4, which was consistent with the timing of colony formation and morphological changes (Figures 1D and 1E). In the TMP induction group or OKS group, no GFP-positive colonies were observed until day 8 (Figure 1D), which coincided with initial endogenous mOCT4 detection (Figure 1E). The group without TMP treatment did not form iPS colonies and showed no detectable mOCT4 expression at any time point (Figure 1E). The RNA levels of hOCT4 and mOct4 at these time points are shown in Figure S3. Intriguingly, we found that during reprogramming mRNA levels of hOCT4 did not correlate with the protein levels. Similarly, in the group of OKS induction, peak hOCT4 RNA levels were seen at day 8 (Figure S3), whereas the time of peak of protein levels appeared at day 5 (Figure 1E). These results suggest that OCT4 expression is also regulated at the protein level during reprogramming, independent of TMP induction.
In order to compare reprogramming efficiency by limiting the availability of stable KLF4 or SOX2, we constructed three additional vectors, OKddS and OKSdd, which have the dd fused to the 5′ end of hKLF4 or hSOX2, respectively (Figure 2A), and dd-3, where all three genes have a fused dd domain (Figure 2A). After electroporation into Oct4-GFP MEFs and treatment with TMP, all three vectors were competent to give rise to iPSCs, which exhibited strong alkaline phosphatase activity (Figure 2B). Whereas OddKS or OKSdd had a reprogramming efficiency comparable to OKS (∼0.38%), OKddS had a 30% increased efficiency (∼0.56%, 402 ± 21 GFP-positive colonies per 7.2 × 104 transfected cells). In contrast, we only obtained 62 ± 3 GFP-positive colonies from 7.2 × 104 cells transfected with the dd-3 vector, a much lower reprogramming efficiency of ∼0.08% (Figure 2C). These data demonstrated that all vectors with a single factor fused to the dd domain have a similar reprogramming efficiency.
Figure 2.
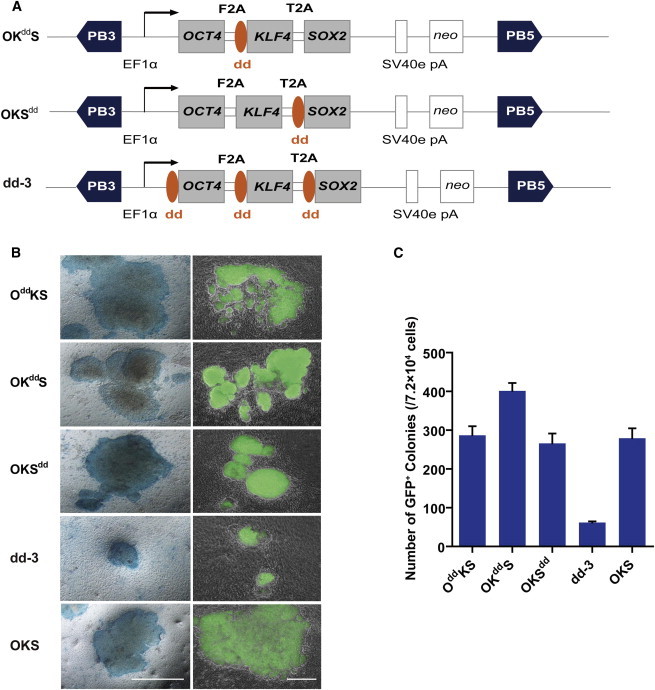
Comparison of Mouse iPSC Generation by Different TMP-Inducible Vectors
(A) Schematic representation of alternate piggyBac transposon reprogramming vectors. The destabilizing domain (dd) was fused to the 5′ end of the KLF4, SOX2, or all three factors (OKddS, OKSdd, and dd-3, respectively).
(B) Micrographs of Oct4-GFP MEFs 14 days after stable transfection, with OddKS, OKddS, OKSdd, and dd-3, treated with 1 μM TMP, or OKS untreated cells. Scale bar, 500 μm.
(C) The number of GFP-positive colonies obtained following transfection of the various vectors in 7.2 × 104 cells were scored at day 14 after transfection and 1 μM TMP treatment. Experiments were performed at least three times per condition. Error bars, SD.
Effect of TMP Dose and Duration on OCT4 Expression
To characterize in detail the dose dependence of OCT4 protein stability on the concentration of TMP, we transfected Oct4-GFP MEFs with the OddKS or control OKS vector and treated with various concentrations of TMP for 5 days. As determined by qRT-PCR, RNA levels of dd-hOCT4 from each group were very similar (Figure 3A), whereas protein levels increased in a dose-dependent manner as measured by western blot (Figures 3B and 3C). Treatment with as little as 10 nM TMP was able to confer significant protein stability, and the entire pool of dd-hOCT4 could be stabilized by 1 μM TMP (Figures 3B and 3C).
Figure 3.
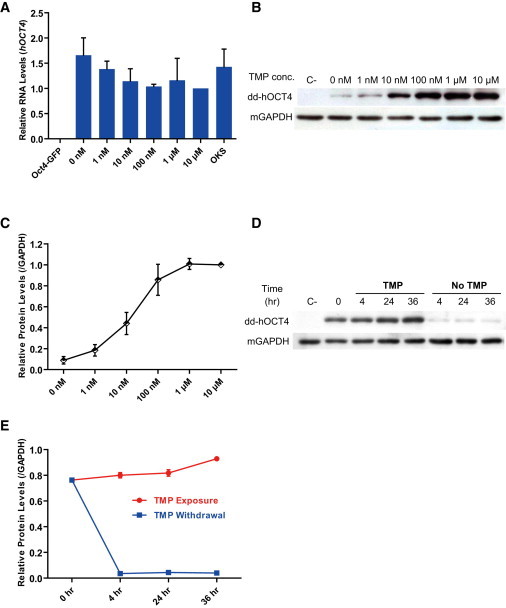
Characterization of TMP-Dependent Protein Stability
(A) OddKS-transfected Oct4-GFP MEFs were treated with varying concentrations of TMP for 5 days and analyzed by quantitative PCR (qPCR) for hOCT4 RNA levels. For comparison, the RNA level of hOCT4 with 10 μM TMP treatment was arbitrarily set to one. Oct4-GFP MEFs were used as a negative control, and OKS was used as the positive control.
(B) Western blot analysis of dd-hOCT4 protein 5 days after TMP exposure. Mouse GAPDH acted as a loading control. Oct4-GFP MEFs were used as the negative control (C−).
(C) The protein levels of dd-hOCT4 at day 5 were quantified using Gelpro32 software, and the indicated ratios normalized against the dd-hOCT4 protein levels seen when cells were treated with 10 μM TMP treatment cells are graphed.
(D) OddKS-transfected Oct4-GFP MEFs were exposed to 1 μM TMP for 5 days and then TMP was withdrawn from a subset of plates. Western blot analysis for dd-hOCT4 protein was performed at the indicated time points under continued TMP exposure or following withdrawal. Oct4-GFP MEFs were used as the negative control (C−).
(E) The levels of dd-hOCT4 protein from (D) were quantified using Gelpro32 software, and the indicated ratios were graphed.
Data in (A), (C), and (E) indicate the means ± SD of three independent experiments.
To determine if the dd-hOCT4 stability is dependent on the continued presence of TMP, we analyzed stabilized protein levels after TMP removal. Because OddKS-transfected Oct4-GFP MEFs showed that dd-hOCT4 stability was maximized at day 5 after 1 μM TMP treatment (Figure 1E), we repeated this experiment and analyzed protein levels following TMP removal. As little as 4 hr after TMP was removed from the medium, dd-hOCT4 protein level decreased by more than 95% (Figures 3D and 3E). In summary, TMP-mediated dd-hOCT4 stability strongly correlated with TMP dosage and dd-hOCT4 was rapidly degraded following TMP withdrawal.
Stoichiometry and Temporal Requirement of Reprogramming Factors
Because dd-hOCT4 stability was dependent on TMP dosage, we set out to determine if its reprogramming function is also under the same dosage constraints. Therefore, we treated OddKS-transfected Oct4-GFP MEFs with different concentrations of TMP (1 nM, 10 nM, 100 nM, 1 μM, and 10 μM) and scored GFP-positive colonies at day 14. TMP concentration of 1 or 10 nM was not sufficient to reach the same reprogramming efficiency as the OKS construct lacking the destabilizing domain, and only a few iPS clones were induced (Figure 4A). As TMP concentration increased, both protein levels and the number of GFP-expressing colonies increased (Figures 4A–4C). The lowest concentration of TMP capable of reaching the maximum reprogramming efficiency of ∼0.40%, was 100 nM (∼0.38%; 45 ± 6 GFP-positive colonies per 1.2 × 104 transfected cells). Furthermore, total OCT4 protein expression level in these reprogrammed colonies was not significantly different from blastocyst-derived mouse G4 ES cells (Figures 4A–4C). Although TMP does not regulate dd-hOCT4 at the mRNA level, the dd-hOCT4 mRNA expression appeared to increase along with the increase of TMP concentration (Figure 4D). Because higher TMP produces more iPS colonies, dd-hOCT4 mRNA would make up a greater proportion of the total RNA.
Figure 4.
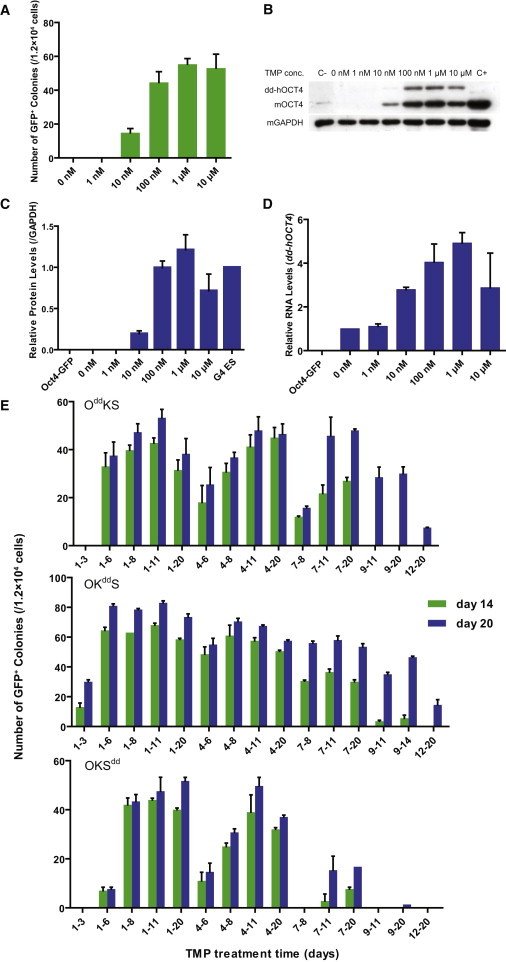
Stoichiometry and Temporal Requirement of Protein Expression of Classic Reprogramming Factors
(A) OddKS-transfected Oct4-GFP MEFs were treated with varying concentrations of TMP for 14 days. The number of GFP-positive colonies obtained by transposon-based reprogramming per 1.2 × 104 cells were scored at day 14 posttransfection. Experiments were performed three times for each condition. Error bars, SD.
(B) Western blot analysis of dd-hOCT4 and mOCT4 protein at day 14. Mouse GAPDH was used as a loading control. Oct4-GFP MEFs were used as a negative control (C−). G4 mouse ES cells were used as a positive control (C+).
(C) The levels of total OCT4 protein at day 14 were quantified using Gelpro32 software, and the indicated ratios normalized against the OCT4 protein expression levels of G4-56 mouse ES cells are plotted. Data indicate the means ± SD of three independent experiments.
(D) qPCR analysis of dd-hOCT4 RNA at day 14. The RNA level of dd-hOCT4 in the transfected cells with 0 nM TMP treatment was set to one. Oct4-GFP MEFs were used as a negative control. Data indicate the means ± SD of three independent experiments.
(E) OddKS-, OKddS-, or OKSdd-transfected Oct4-GFP MEFs were treated with 1 μM TMP during the varying time periods indicated. The number of GFP-positive colonies obtained by transposon-based reprogramming per 1.2 × 104 cells were scored at day 14 and day 20 postelectrotransfection. Experiments were performed at least three times per condition. Error bars, SD.
Previously it was reported that continuous ectopic expression of OCT4 and KLF4 is required for at least 12 days with the Dox system. Ectopic SOX2 expression for only 5 days was enough to give rise to AP-positive colonies, and prolonged ectopic SOX2 expression greatly increased the number of AP-positive colonies (Sridharan et al., 2009). However, the Dox system is not versatile enough to allow for more detailed dissection of temporal requirements of the four factors. Taking advantage of our TMP system, we sought to investigate the temporal requirement for each of these ectopic factors during reprogramming. Fibroblasts were transfected with OddKS, OKddS, or OKSdd, and 1 μM TMP treatment was restricted to a specific phase, or phases, of reprogramming (Figure 4E). Fibroblasts induced to express dd-hOCT4 for 6 days gave rise to GFP-positive colonies, and the efficiency of iPSC generation increased to ∼0.36% (43 ± 2 colonies per 1.2 × 104 cells transfected) after treating with TMP for an additional 5 days. No further increase in the number of iPS colonies was observed, even when dd-hOCT4 was maintained for longer periods. Interestingly, TMP induction of dd-hOCT4 for only 3 days during the early stage (days 1–4) did not generate any iPS colonies, suggesting that at least 6 days of ectopic OCT4 expression was required for iPSC generation. In contrast, only 2 days of TMP addition during the later phases (days 7–8) was able to give rise to iPSCs, suggesting that these cells had been in a partially reprogrammed state. When culture was maintained for 20 days, we found that TMP induction of dd-hOCT4 at the late stage could generate iPSCs at frequencies comparable to TMP induction at the early stage (Figure 4E).
Similar to dd-hOCT4, dd-hKLF4 induction, for any period tested, was able to effectively reprogram MEFs, and comparable efficiency could be reached with similar TMP treatment regimens. In contrast to the result obtained with dd-hOCT4, ectopic KLF4 expression for only 3 days during the early stage (days 1–4) was able to generate GFP-positive iPSCs (Figure 4E). When OKSdd was examined, surprisingly, we found that exogenous SOX2 expression from day 4 to day 8 was most important for iPSC induction. dd-hSOX2 induction from day 1 to day 3 failed to generate GFP positive colonies, as did induction on or after day 9. These results indicated that hSOX2 mainly acted at the early stage (day 1–4) and middle stage (days 5–8) of reprogramming, whereas exogenous SOX2 expression during the late stage only (days 9–14) was ineffective in iPSC generation. Taken together, these data demonstrated a different temporal requirement for each ectopic factor. Treatment with exogenous OCT4 and exogenous KLF4 during any reprogramming phase had the potential to generate iPSCs (Figure 4E), which is consistent with experiments reported by others (Hanna et al., 2009). However, exogenous SOX2 expression was required at the early stage (days 1–4) and middle stage (days 5–8) of reprogramming.
Regulation of Other Factors during Reprogramming
We next asked whether our TMP system could be extended to test other factors during reprogramming. Previous research suggested that reprogramming of somatic cells is accompanied by global epigenetic remodeling, including DNA demethylation (Cowan et al., 2005; Maherali et al., 2007; Tada et al., 2001). However, the effect of ectopic expression of DNA demethylation factors during iPSC generation was not well documented. We constructed four vectors in which dd was fused to the 5′ end of the genes involved in DNA demethylation, including human growth arrest and DNA-damage-inducible protein 45 alpha (hGADD45a) (Barreto et al., 2007), human methyl-CpG-binding domain (hMBD4) (Bhattacharya et al., 1999), human thymine DNA glycosylase (hTDG) (Maiti and Drohat, 2011), and human activation induced cytidine deaminase (hAID) (Morgan et al., 2004; Rai et al., 2008), to investigate the temporal requirements for the four demethylation factors on reprogramming efficiency (Figure 5A). Oct4-GFP MEFs were cotransfected with OKS and dd-Factor mixed with PBase. TMP treatment was applied at different time points and for various time periods. Interestingly, our results indicated that addition of these four factors did not improve reprogramming efficiency, no matter when these factors were added by TMP induction (Figure 5B). This result is opposite to the expectation that demethylation factors might enhance reprogramming. However, it complements a recent report that knockdown of another demethylation factor MBD3 increases reprogramming efficiency (Brumbaugh and Hochedlinger, 2013).
Figure 5.
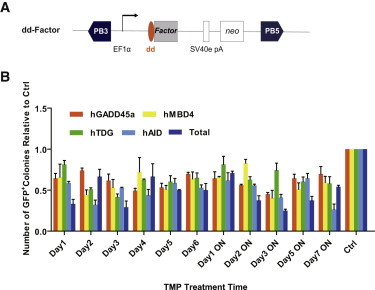
Regulation of DNA Demethylation-Related Factors during Reprogramming
(A) Schematic representation of piggyBac transposon reprogramming vectors. Each factor was fused with dd, and expression was driven by the constitutively active EF1α promoter.
(B) The Oct4-GFP MEFs were transfected with the varying TMP-inducible vectors indicated to generate mouse iPSCs. The number of GFP-positive colonies obtained per 7.2 × 104 cells were scored at day 14 posttransfection. Experiments were performed at least three times per condition. Error bars, SD.
Application of TMP Reprogramming System in Other Species
To test whether the TMP reprogramming system could be used in species other than the mouse, we reprogrammed pig embryonic fibroblasts with TMP-inducible vector OddKSM, where hOCT4 could be regulated by TMP. The four reprogramming genes on the vector (dd-hOCT4, hKLF4, hSOX2, and hc-MYC) were connected by F2A, T2A, and F2A (Figure 6A). TMP was added to the culture medium throughout the entire reprogramming process. Six days after transfection, primary iPS colonies appeared, with morphology similar to mouse ESC colonies (Figure 6B). At day 14, we picked individual colonies and transferred them to dual inhibition (PD0325901 plus CHIR99021) (2i) medium (Ying et al., 2008) supplemented with TMP (Figure 6C), and they could be passaged by standard trypsinization >30 times without any sign of deterioration. To test the dependence of established iPSC lines on exogenous dd-hOCT4 stabilization, TMP was removed from the culture media. Upon TMP removal, pig iPSCs lost their domed colony morphology and differentiated (Figure 6D), in contrast to the same iPSC lines cultured in 2i medium with TMP (Figure 6E). Although RNA expression levels of pig iPSCs cultured with and without TMP were similar, protein levels were markedly altered. Furthermore, pig iPSC lines cultured with TMP were AP+ and OCT4+, but in the absence of TMP they lost most of the AP staining and were OCT4− (Figures 7A–7D). This was consistent with previous observations that maintenance of pig iPSCs is dependent on exogenous OCT4 (Ezashi et al., 2009; Fan et al., 2013; Esteban et al., 2009; Wu et al., 2009).
Figure 6.
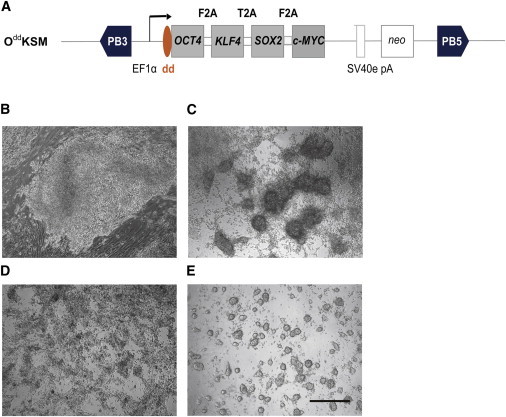
Generation of Pig iPSCs by TMP-Inducible Vector OddKSM
(A) Schematic representation of the piggyBac transposon reprogramming vectors. Four coexpressed factors, hOCT4, hKLF4, hSOX2, and hc-MYC, were linked by 2A peptides and driven by the constitutively active EF1α promoter. The destabilizing domain (dd) was fused to the 5′ end of the OCT4 gene, and this plasmid is designated OddKSM.
(B) A representative primary colony formed at day 9 of pig iPSC induction using the TMP-inducible vector.
(C) Pig iPSCs in 2i medium showed stably compacted colony with continuously added TMP.
(D) Pig iPSCs in 2i medium without TMP for 3 days lost compacted colony morphology and differentiated.
(E) Pig iPSCs in 2i medium with TMP maintained compacted colony morphology in further culture. Scale bar, 500 μm.
Figure 7.
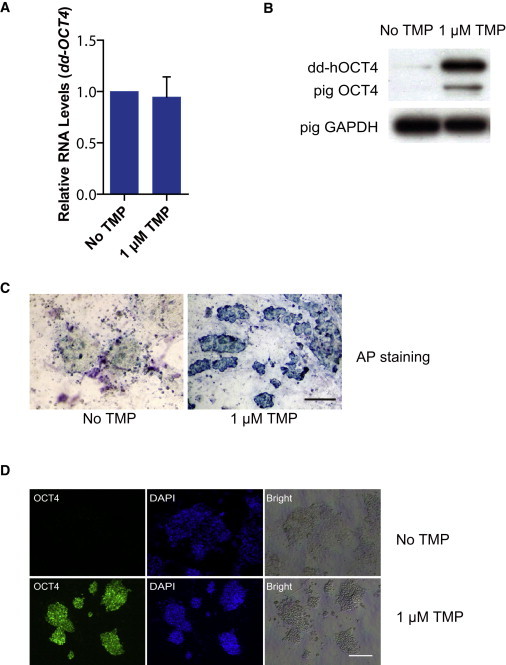
Characterization of the Effect of TMP Treatment on Pig iPSCs
(A) Quantitative PCR analysis of dd-hOCT4 RNA. For comparison, the RNA level of hOCT4 in pig iPSCs with no TMP treatment was arbitrarily set to one. Data indicate the means ± SD of three independent experiments. Pig embryonic fibroblasts were used as a negative control.
(B) Western blot analysis of dd-hOCT4 and pOCT4 protein. Pig GAPDH was used as a loading control.
(C) AP staining of pig iPSC lines cultured with 1 μM TMP or no TMP. Scale bar, 500 μm.
(D) Immunofluorescent analysis of pig iPSC lines cultured with 1 μM TMP or no TMP. Scale bar, 500 μm.
Discussion
In this study, we used a TMP-inducible reprogramming system to regulate gene expression at the protein level. This was a convenient and effective system for controlling the temporal expression and stoichiometry of reprogramming factors.
Among different methods, the Dox-inducible system has been widely used to dissect reprogramming mechanisms (Hanna et al., 2009; Maherali et al., 2008; Wernig et al., 2008a). However, Dox-inducible systems have limitations, such as leaky expression and requirement of additional components (tTA or rtTA). The TMP-inducible system is designed to directly regulate protein stability, instead of regulating expression at the RNA level (Iwamoto et al., 2010). By adding a small molecule drug TMP, proteins fused to the dd domain are shielded from degradation. As a bacteriostatic antibiotic, TMP is mainly used in the prophylaxis and treatment of urinary tract infections and can pass the blood-brain barrier (Barling and Selkon, 1978) and the placental barrier (Schulz, 1972). No adverse effects have been observed, even when doses twice the concentration used to regulate transgenes (0.4 mg/ml; 3 mM) are given for 6 weeks in vivo (Iwamoto et al., 2010). Furthermore, compared with a Dox-inducible system, the TMP-inducible system has several additional advantages. First, the TMP system is more convenient and does not require additional molecular control components, allowing for direct application in various species and cell lines. Second, the levels of dd fusion proteins increase in a strict dose-dependent manner (Figures 3B and 3C). Third, the TMP system is highly sensitive; a dose as low as 10 nM TMP is able to induce protein expression, whereas 1 μM of TMP can fully stabilize dd fused proteins (Figures 3B and 3C). Four hours after TMP removal, protein levels quickly fall to undetectable levels (Figures 3D and 3E).
To date, the derivation and maintenance of high-quality pig iPSCs that have reproducible chimera formation ability or germline competency have not been achieved. Pig iPSCs generated with retroviruses/lentiviruses maintain expression of exogenous genes and depend on the expression of exogenous factors to maintain pluripotency, suggesting that reprogramming is incomplete. Therefore, these pig iPSCs are more like epiblast-derived stem cells. This has been confirmed by several groups including ourselves (Ezashi et al., 2009; Fan et al., 2013; Esteban et al., 2009; Wu et al., 2009). By use of TMP to temporally control exogenous OCT4 stability, we found that, consistent with previous reports, pig iPSCs derived by our piggyBac vectors containing OSKM factors were also incompletely reprogrammed and needed exo-OCT4 to sustain an undifferentiated morphology in 2i medium. When expression of exo-OCT4 was terminated by removing TMP, these pig iPSCs could no longer maintain pluripotency. By regulation of exogenous protein stability, the TMP-inducible system may provide a useful method for the design of screening strategies to identify factors, chemicals, or small hairpin RNA inhibitors that could rescue the iPSCs when TMP is not present. We hope that this may help solve the problem of incomplete reprogramming of pig iPSCs in the future.
Recent studies show that changes in mRNA levels do not always correlate with the amount of protein produced in ESCs (Lu et al., 2009; Thomson et al., 2011). Similarly, we observed that OCT4 protein levels during reprogramming do not always correlate with mRNA levels (Figure 1E and S3). These results emphasize the importance of understanding the regulation of genes at the protein level during reprogramming. At the same time, there is increasing evidence that core transcription factors in ESCs, OCT4 (Liao and Jin, 2010), SOX2 (Van Hoof et al., 2009), NANOG (Ramakrishna et al., 2011), KLF4 (Kim et al., 2012), and c-MYC (Gregory and Hann, 2000) undergo posttranslational modifications, including phosphorylation (Kim et al., 2012; Van Hoof et al., 2009), sumoylation (Wei et al., 2007), and ubiquitination (Gregory and Hann, 2000; Liao and Jin, 2010). Recent studies have identified that ligases and deubiquitinases of UPS members are essential for both ESC self-renewal and differentiation (Buckley et al., 2012). We speculate that, similar to the situation in ESCs, posttranslational regulation of core factors may also play a role in iPSC induction. The TMP system has offered a unique tool to study this phenomenon in detail.
Previous reports have shown that the stoichiometry and temporal requirements of reprogramming factors play a crucial role in determining the epigenetic and pluripotent state of iPSCs, as well as in determining reprogramming efficiency (Carey et al., 2011; Nagamatsu et al., 2012; Tiemann et al., 2011; Xu et al., 2008). However, the results from different groups often vary. The cause for this discrepancy may be due to two limitations of viral systems: heterogeneous expression (Wernig et al., 2008a) and autoimmune regulation (Lee et al., 2012). Perhaps these systems used do not reflect the true status of factor gene expression (Lu et al., 2009). Here, we report the direct correlation of OCT4 protein levels and efficiency of iPSC generation, and provide an inducible system to investigate the dynamic molecular mechanism of reprogramming. We found that exogenous SOX2 expression in the early stage (day 1–4) and middle stage (day 5–8) was essential for reprogramming. The absence of exogenous SOX2 expression during the early and middle stages cannot be rescued by exogenous SOX2 expression at the late stage (day 9–14). Whereas a recent report emphases the upstream role of endogenous SOX2 in the gene expression hierarchy in the late phase (Buganim et al., 2012), our functional data further reveal the essential role of SOX2 during the initial stages of reprogramming. We propose that reprogramming is a deterministic process, although in the early stages it appears to be stochastic. The TMP system is a useful platform for interrogating reprogramming mechanisms and determining how individual factors contribute to the reprogramming process. The versatility of the TMP system allows testing multiple factors simultaneously. This should open up more uses of the TMP system to study detailed mechanism of factor interactions. In the current study, we focused on characterizing the temporal requirement of individual OSK factors. For future studies, detailed interactions should be further characterized.
In summary, we have shown that the TMP-inducible system could be used for efficient iPSC generation. This system regulates ectopic protein stabilization in a convenient, highly sensitive, and dose-dependent manner. It provides a simple and practical tool for research into reprogramming mechanisms and a potential way to generate safer iPSCs. This system may be combined with the Dox-inducible system to allow for tighter and more precise control at both mRNA and protein levels.
Experimental Procedures
Plasmid Construction
We first synthesized a human codon-optimized gene that codes for the destabilizing domain (dd) derived from Escherichia coli dihydrofolate reductase, with the R12Y and Y100I mutations (Iwamoto et al., 2010). Fusion PCR was used to construct all of the dd-containing plasmids onto a piggyBac transposon vector backbone that contains the PB 5′ and 3′ terminals from the ZGs vector (Wu et al., 2007). The destabilizing domain (dd) was fused to the N-terminal of the genes used in the study. The EF1α promoter was placed upstream of the single open reading frame. The OKS genes with or without related dd sequences were linked with the 2A sequences by PCR. The CAG promoter-driven PBase was from the CAG-PBase vector.
Cell Culture
MEFs were isolated from 13.5 days postcoitum. F1 embryos (50% C57BL/6 and 50% 129sv) from homozygous Oct4-GFP mice (JaxMice strain name: B6;CBA-Tg(Pou5f1-EGFP)2Mnn/J; stock number 004654) crossed to 129sv and cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen) containing 15% FBS (Invitrogen), 0.5 mM GlutaMAX, 1 × nonessential amino acid, and 1 mM Sodium Pyruvate. A germline-competent mouse ESC line, G4, was cultured in DMEM (Invitrogen) containing 20% FBS (Invitrogen, catalog no. 16141, lot. 821754), 0.5 mM GlutaMAX, 1 × nonessential amino acid, 1 mM Sodium Pyruvate, 0.1 mM 2-mercaptoethanol, and 500 U/ml LIF (Millipore). Mouse iPSC lines were cultured on β2 feeder layers that were 40 Gy 60Co γ-ray treated. Pig iPSC lines were cultured on β2 feeder layers in the 2i medium as described previously (Ying et al., 2008).
Reprogramming of MEFs Using piggyBac Vectors
On the first day (day 0), MEFs were transfected with various piggyBac vector combinations, using the Amaxa Basic Nucleofector Kit for primary mammalian fibroblasts (Lonza), in accordance with the manufacturer’s instructions. The piggyBac vectors (pEF1α-OddKS, pEF1α-OKddS, pEF1α-OKSdd, pEF1α-OddKddSdd, named OddKS, OKddS, OKSdd, and dd-3, respectively) were combined with CAG-PBase plasmid in a 3:1 ratio, 1.5 μg piggyBac vectors, 0.5 μg PBase, per 106 cells. Posttransfected cells were seeded onto feeder layers. After 24 hr, MEF medium was replaced by ES medium with 350 μg/ml G418 for 5 days. Different concentrations of TMP were added to test groups from the first day. The medium was refreshed every day. On day 6, medium was changed to ESC medium without G418. On day 14, colonies were either counted by GFP-positive numbers and stained by alkaline phosphatase or picked and further expanded.
AP Staining
iPSCs were fixed in 4% PFA for 30 min and then washed with PBS three times, followed by the addition of BM purple (Roche).
Assay for Teratoma Formation
For teratoma formation, three mouse iPSC lines (derived with OddKS, OKddS, and OKSdd separately, passage 10) were cultured with ES medium on β2 feeder layers for 2 days (∼60% to 70% confluent). Approximately 1 × 106 iPSCs were resuspended in PBS and injected subcutaneously into nude mice. All three iPSc lines injected formed tumors within 4 weeks after initial transduction, and paraffin sections were stained with hematoxylin and eosin for all histological determinations. Control mice injected with 1 × 106 β2 feeder cells failed to form teratomas.
Western Blot Analysis
Transfected cells were washed with PBS, and each well was harvested and suspended in cell lysis buffer for western blot analysis at the different indicated times. The concentrations of total cell protein were measured. Ten micrograms of each lysate sample was boiled at 95°C–100°C for 5 min in sample buffer, and proteins were separated by10% SDS-PAGE and transferred onto polyvinylidene fluoride membrane (GE Healthcare). Nonspecific reactivity was blocked in 5% nonfat dry milk in Tris-buffered saline and Tween 20 (1 mM Tris-HCl [pH = 7.5], 150 mM NaCl, and 0.05% Tween-20) for 1 hr at room temperature. The specific proteins were analyzed using anti-OCT4 (1:1,000, rabbit monoclonal, ab109187, abcam), anti-OCT4 (1:3,000, rabbit polyclonal, ab19857, abcam), or anti-mGAPDH (1:2,000, rabbit monoclonal, #2118, CST). The membranes were washed five times and incubated with goat anti-rabbit IgG HRP (1:20,000) for 1 hr at room temperature. The membranes were washed again as before and incubated with substrates that were prepared in accordance with the manufacturer’s instructions. Each membrane was exposed to X-ray film for 30 s to ∼5 min.
qRT-PCR Analysis
Total mRNA was extracted using Trizol reagent (Invitrogen). Total RNA (2 μg) was reverse transcribed to cDNA using a QuantiTect Reverse Transcription Kit (205213, QIAGEN), in accordance with the manufacturer’s protocol. qRT-PCR was performed using TaqMan Gene Expression Master Mix (Applied Biosystems) on the ABI7900HT sequence detector (Applied Biosystems). Primer and probe sequences used in qRT-PCR are as follows:
hOCT4-Forward primer: 5′-AGTCGGGGTGGAGAGCAAC-3′;
hOCT4-Reverse primer: 5′-GGCAAATTGCTCGAGTTCTTTC-3′
hOCT4-BHQ-probe: 5′-CCCTGCACCGTCACCCCTGG-3′
mOct4-Forward primer: 5′-AGGCAGGAGCACGAGTGG-3′
mOct4-Reverse primer: 5′-GGACTCCTCGGGAGTTGGTT-3′
mOct4-BHQ-probe: 5′-CTGTGCCGACCGCCCCAATG-3′
Total-Oct4-Forward primer: 5′-CCTGGGGSCAGAGGAAAG-3′ S = G/C
Total-Oct4-Reverse primer: 5′-AGCTTGGGCTMGAGAAGGAT-3′ M = A/C
Total-Oct4-BHQ-probe: 5′-TGCCCTTCTGGCGCCGGTTAC-3′
mGAPDH-Forward primer: 5′-GCACAGTCAAGGCCGAGAA-3′
mGAPDH-Reverse primer: 5′-CCTCACCCCATTTGATGTTAGTG-3′
mGAPDH-BHQ-probe: 5′-CATCACCATCTTCCAGGAGCGAGACC-3′
pGAPDH-Forward primer: 5′-GTCAAGCTCATTTCCTGGTACGA-3′
pGAPDH-Reverse primer: 5′-GGCCTCTCTCCTCCTCGC-3′
pGAPDH-BHQ-probe: 5′-CCTCATGGTCCACATGGCCTCCA-3′.
Acknowledgments
We thank Drs. Anne Boulet and Simon Titen for critically reading the manuscript. We thank Shuangyu Ma, Linyuan Ma, and Hanshi Xu for their kind help with cell cultures and western blots. We thank Dr. Klaus Wilbrandt Kjær for his helpful discussion during the early stages of this study. We thank Qian Zhao for her help with illustrations. This work was supported by the National Key Basic Research Program (2011CBA01002) and the National High Technology Research and Development Program (2013AA102502).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Sen Wu, Email: swu@cau.edu.cn.
Ning Li, Email: ninglcau@cau.edu.cn.
Supplemental Information
References
- Barling R.W., Selkon J.B. The penetration of antibiotics into cerebrospinal fluid and brain tissue. J. Antimicrob. Chemother. 1978;4:203–227. doi: 10.1093/jac/4.3.203. [DOI] [PubMed] [Google Scholar]
- Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S.K., Handa V., Döderlein G., Maltry N., Wu W., Lyko F., Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S.K., Ramchandani S., Cervoni N., Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Brumbaugh J., Hochedlinger K. Removing reprogramming roadblocks: Mbd3 depletion allows deterministic iPSC generation. Cell Stem Cell. 2013;13:379–381. doi: 10.1016/j.stem.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Buckley S.M., Aranda-Orgilles B., Strikoudis A., Apostolou E., Loizou E., Moran-Crusio K., Farnsworth C.L., Koller A.A., Dasgupta R., Silva J.C. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., Creyghton M.P. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Cowan C.A., Atienza J., Melton D.A., Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J. Biol. Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T., Telugu B.P., Alexenko A.P., Sachdev S., Sinha S., Roberts R.M. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan N., Chen J., Shang Z., Dou H., Ji G., Zou Q., Wu L., He L., Wang F., Liu K. Piglets cloned from induced pluripotent stem cells. Cell Res. 2013;23:162–166. doi: 10.1038/cr.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M., Freundlieb S., Bender G., Müller G., Hillen W., Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Gregory M.A., Hann S.R. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt’s lymphoma cells. Mol. Cell. Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P., van Oudenaarden A., Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M., Björklund T., Lundberg C., Kirik D., Wandless T.J. A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 2010;17:981–988. doi: 10.1016/j.chembiol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.O., Kim S.H., Cho Y.Y., Nadas J., Jeong C.H., Yao K., Kim D.J., Yu D.H., Keum Y.S., Lee K.Y. ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4. Nat. Struct. Mol. Biol. 2012;19:283–290. doi: 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- Lee J., Sayed N., Hunter A., Au K.F., Wong W.H., Mocarski E.S., Pera R.R., Yakubov E., Cooke J.P. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B., Jin Y. Wwp2 mediates Oct4 ubiquitination and its own auto-ubiquitination in a dosage-dependent manner. Cell Res. 2010;20:332–344. doi: 10.1038/cr.2009.136. [DOI] [PubMed] [Google Scholar]
- Lu R., Markowetz F., Unwin R.D., Leek J.T., Airoldi E.M., MacArthur B.D., Lachmann A., Rozov R., Ma’ayan A., Boyer L.A. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K., Stadtfeld M., Yachechko R., Tchieu J., Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Nagamatsu G., Saito S., Kosaka T., Takubo K., Kinoshita T., Oya M., Horimoto K., Suda T. Optimal ratio of transcription factors for somatic cell reprogramming. J. Biol. Chem. 2012;287:36273–36282. doi: 10.1074/jbc.M112.380683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K., Nakagawa M., Hyenjong H., Ichisaka T., Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Suresh B., Lim K.H., Cha B.H., Lee S.H., Kim K.S., Baek K.H. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20:1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- Sando R., 3rd, Baumgaertel K., Pieraut S., Torabi-Rander N., Wandless T.J., Mayford M., Maximov A. Inducible control of gene expression with destabilized Cre. Nat. Methods. 2013;10:1085–1088. doi: 10.1038/nmeth.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R. Distribution and elimination of trimethoprim in pregnant and newborn rats. Naunyn Schmiedebergs Arch. Pharmacol. 1972;272:369–377. doi: 10.1007/BF00501243. [DOI] [PubMed] [Google Scholar]
- Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G.W., Cook E.G., Hargus G., Blak A., Cooper O., Mitalipova M. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., Vignali D.A. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Tada M., Takahama Y., Abe K., Nakatsuji N., Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thomson M., Liu S.J., Zou L.N., Smith Z., Meissner A., Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145:875–889. doi: 10.1016/j.cell.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemann U., Sgodda M., Warlich E., Ballmaier M., Schöler H.R., Schambach A., Cantz T. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–435. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Muñoz J., Braam S.R., Pinkse M.W., Linding R., Heck A.J., Mummery C.L., Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Wei F., Schöler H.R., Atchison M.L. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J. Biol. Chem. 2007;282:21551–21560. doi: 10.1074/jbc.M611041200. [DOI] [PubMed] [Google Scholar]
- Wernig M., Lengner C.J., Hanna J., Lodato M.A., Steine E., Foreman R., Staerk J., Markoulaki S., Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wu S., Ying G., Wu Q., Capecchi M.R. Toward simpler and faster genome-wide mutagenesis in mice. Nat. Genet. 2007;39:922–930. doi: 10.1038/ng2060. [DOI] [PubMed] [Google Scholar]
- Wu Z., Chen J., Ren J., Bao L., Liao J., Cui C., Rao L., Li H., Gu Y., Dai H. Generation of pig induced pluripotent stem cells with a drug-inducible system. J. Mol. Cell Biol. 2009;1:46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]
- Xu J., Ho A.Y., Yin K., Yuan X., Anderson D.M., Lee J.H., Harrison P.J. Temporal and spatial variations in nutrient stoichiometry and regulation of phytoplankton biomass in Hong Kong waters: influence of the Pearl River outflow and sewage inputs. Mar. Pollut. Bull. 2008;57:335–348. doi: 10.1016/j.marpolbul.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


