Abstract
Regulated intramembrane proteolysis (RIP) is a highly conserved signaling paradigm whereby membrane-bound signaling proteins are cleaved in their transmembrane region and then released into the cytoplasm to act as signaling molecules. In most if not all cases intramembrane cleavage is preceded and regulated by a membrane proximal cleavage step called “ectodomain shedding”. Here we will review the role of ectodomain shedding in RIP of the NOTCH signaling pathway, a highly conserved cell-cell communication pathway that mediates cell fate decisions during development and in adult tissues.
INTRODUCTION
RIP is a widespread signaling paradigm where transmembrane proteins are cleaved within their transmembrane domain to release a cytosolic signaling fragment that can enter the nucleus to control gene transcription.1 The RIP is conserved from bacteria to humans2 and controls many diverse processes from lipid metabolism,3 the unfolded protein response,4 Epidermal Growth Factor (EGF) signaling,5 antigen presentation,6 production of the beta Amyloid Precursor Protein (APPβ)7 and cell fate determination by the Notch signaling pathway.8 There are 4 classes of Intramembrane Cleaving Proteases (I-CLIPs), Aspartyl Proteases, Presenilins (PS) and Signal Peptide Peptidases (SPP), Serine Proteases (Rhomboid) and Metalloproteases (Site-2 Protease (S2P)). No cysteine or threonine proteases have been identified that exhibit I-CLIP activity.9 I-CLIPs are polytopic membrane proteins with catalytic residues within their transmembrane domains (TMD) that hydrolyze peptide bonds within the lipid bilayer.10 PS and Rhomboids cleave Type I transmembrane proteins whereas SPP and S2P cleave Type II substrates. A common but not exclusive feature of RIP is that intramembrane proteolysis precedes ligand binding and ectodomain shedding.11 Cleavage by Rhomboids is an exception and does not require juxtamembrane cleavage prior to intramembranous cleavage. Here we will review our current understanding of how a membrane bound family of metalloproteinases regulate ectodomain shedding and RIP of the NOTCH signaling pathway under physiological and pathological conditions and how this knowledge may be applied for therapeutic intervention.
NOTCH SIGNALING
The Notch pathway is a highly conserved signaling cascade in multicellular eukaryotes and controls spatial patterning, morphogenesis and homeostasis in embryonic and adult tissues.12,13 Notch proteins orchestrate tissue homeostasis through receptor ligand interactions on adjacent cells.14,15 Disruption of this homeostatic control by a deregulated Notch cascade underlies cancer formation in several organs.15,17 Notch receptors (N1 to N4) and ligands (i.e., Delta, Jagged) are Type I transmembrane glycoproteins that transduce signals by binding to membrane bound ligands on adjacent cells. Most if not all Notch functions reported require RIP and constitute the cleavage-dependent or canonical Notch signaling pathway. Upon ligand binding, Notch receptors undergo two successive proteolytic cleavages: an ectodomain cleavage followed by intramembrane proteolysis by γ-secretase18. This process releases the Notch intracellular domain (NICD), which translocates to the nucleus and binds CBF1/Suppressor of Hairless/Lag-1 or CSL (RBP-Jk in mice) to activate its target genes.19,23 In the absence of ligand the Notch juxtamembrane localized heterodimerization domain (HD) inhibits extracellular proteolysis and activation.24,25 The Notch cascade is deregulated in many human cancers and oncogenic mutations in Notchi are frequently found in human T-cell leukemia that map to the HD and PEST domain,26 making Notch proteolysis an attractive therapeutic target (Fig. 1).
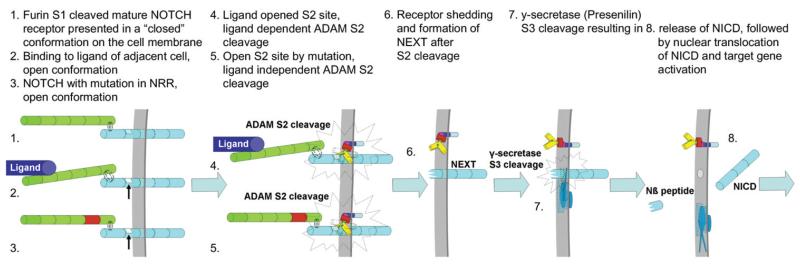
Figure 1. Regulated Intramembrane Proteolysis Activates The Notch Pathway.
1) After SI cleavage in the secretory pathway the mature receptor is presented as a heterodimer at the cell surface in a “closed” conformation. 2) and 3) The notch receptor is activated or “opened” (arrow head) by ligand binding on neighboring cells or by an activating mutation in the NNR. 4) and 5) ADAMs proteases (i.e. ADAMIO/Kuzbanian) access and cleave NOTCH at the juxtamembrane S2 site. 6) S2 cleavage results in the membrane tethered NEXT fragment and the notch ectodomain which is transendocytosed in signaling cells. 7) Within the plane of the membrane the NEXT fragment is cleaved at S3 by the aspartyle protease Presenilin which is part of the γ-secretase complex. 8) This releases the extracellular Nβ peptide an the intracellular NICD, which translocates to the nucleus to activate target genes..
In Drosophila melanogaster, a single prototypical Notch protein is found and two ligands Delta and Serrate (Jagged in mammals). In the nematode Caenorhabditis elegans, several Delta-Jagged type ligands (LAG-2/Apx2) are found and two Notch-like receptors, Linl2 and Glp-127. In mammals four Not chgenes (Notchl-4) and five ligands: Jagged1-2, Delta-like ((Dll) 1, 3 and 4) are known. These ligands are collectively called DSL ligands (Delta and Serrate/ in Drosophila and LAG-2 in C. elegans).8 Notch Ligand receptor interactions are regulated by O-linked glysolation/fucosylation of EGF-like repeats in Notch receptor and DSL ligands by Fringe proteins during ER-Golgi transport.28-30 Glycosylated Notch receptors have higher affinity for Delta than for Jagged type ligands.31 Physiological Notch activation is triggered by ligand binding and governed by RIP. Most if not all function of Notch require proteolysis.32,33
NOTCH PROTEOLYSIS SITE-1 (S1, FURIN, SERINE PROTEASE)
During maturation in the trans-Golgi network, Notch precursors are first cleaved at Site-1 (S1) by Furin-like convertase producing a heterodimeric Type I receptor with the Notch extracellular domain (NECD) non-covalently bound to a transmembrane/intracellular fragment (TMIC).34-36 Whereas in mammals most canonical (CSL dependent) Notch1 signaling requires furin there are exceptions.37 S1 cleavage of dNotch is infrequent and not required in flies.38 Whereas mammalian Notch1 receptors are dependent on furin for proper cell surface expression furin cleavage appears dispensable for Notch2 trafficking and signaling. No information is available on the requirement of furin for Notch3 and Notch4 signaling although they are likely processed in a similar manner.39 In the absence of ligand mature Notch receptors are held into an inactive “proteolysis resistant” or closed state because the Negative Regulatory Region (NRR) composed of the HD domain and the globular Lin12/Notch repeats (LNR) inhibits Notch activation.34,40,41 Ligand binding to Notch receptors unfolds the NRR permitting cleavage by a metalloprotease at a site close to the membrane (S2) (Fig. 1).24,42 Another model involves transendocytosis of NECD upon ligand binding and dissociation of NECD and TMIC heterodimer followed by S2.43 The latter model however is not consistent with the lack of furin cleavage of dNotch in flies38 and the activity of ligand–independent gain of function mutations in the NRR although some ligand-requirement still exists.24,44,45 Not unexpectedly cancer-causing mutations localize to the HD in almost 50% of sporadic human T-ALL causing increased NOTCH1 cleavage and activity.26, 44
SITE-2 (S2, ADAM10, METALLOPROTEASE)
The S2 cleavage occurs close to the proximal juxtamembrane stalk of the extracellular domain and leads to the shedding of most of the NECD. S2 cleavage is performed by a zinc-dependent metalloprotease and occurs in response to ligand binding that induces a conformational change.46-48 The membrane-bound cleaved form is termed Notch Extracellular Truncation (NEXT) and is a rate-limiting substrate for the intramembrane cleaving aspartyl protease Presenilin. S2 cleaved Notch fragments are extremely short-lived and can only be observed when blocking γ-secretase processing.25,48
SITE-3 (S3, PRESENILIN, ASPARTYL PROTEASE)
Following S2 cleavage, NEXT proteins are cleaved within their transmembrane domain by the GxGD aspartyl protease Presenilin (S3), part of a multi-protein complex termed γ-secretase, releasing the Notch Intracellular Domain (NICD) and the juxtamembranous Nβ peptide.18 NICD translocates to the nucleus where it binds to the DNA-bound protein CSL together with co-activator proteins Mastermind, leading to target gene activation.49 The non-canonical signaling pathway that does not rely on CSL (in mice RBP-Jκ), is less well understood and therefore not further discussed here.50 There is very little evidence for a RIP independent Notch pathway activation. Excellent and more comprehensive reviews on the Notch activation cascade have appeared elsewhere.8 Here we will summarize our current understanding on the role of ADAM metalloproteases in the activation of Notch signaling cascade and how this may be exploited for therapeutic intervention in diseases with aberrant Notch signaling.
THE ADAM FAMILY OF PROTEASES
Disintegrin Metalloproteases (ADAMs) are a distinct class of metalloproteases that have both protease and adhesion domains and play key roles in cell-cell and cell-matrix interactions in diverse cell biological processes such as cell adhesion, fertilization and cytokine and growth factor receptor signaling. ADAMs have a broad tissue expression and are mainly regulated at the posttranslational level. They function as signaling scissors regulating the activity of transmembrane signaling molecules and release soluble ectodomains with an altered location and function.51,52 In total 40 ADAM family members have been identified in the animal kingdom. Annotated ADAMS of different species can be found at the White Laboratory (web site 1). Currently in humans 22 ADAM genes are known while in mice 34 ADAM genes have been identified.53 ADAMs are membrane bound zinc-dependent metalloprotease that belong to the class of astacin/adamalysin family (MEROPS, web site 2).54 The adamalysins subfamily is comprised of snake venom metalloproteases (SVMP), the membrane bound ADAM and secreted ADAM-TS family. The latter can be structurally distinguished from ADAMs by their various numbers of thrombospondin-like (TS) motifs.55 ADAM proteins are multidomain proteins that consist of several conserved domains. The amino terminus is composed of a pro-domain followed by the catalytic domain, a disintegrin domain, a cysteine-rich domain containing epidermal-growth factor (EGF)-like repeats, transmembrane domain, and cytoplasmic tail (Fig. 2). The prodomain keeps the protease in a latent state and is clipped off by proprotein convertases (furin) during maturation in the secretory pathway. Some ADAMs lack the furin cleavage site and are activated by autocatalysis.56,57 The metalloprotease domain encodes the catalytic core sequence HEXXHXXGXX(H/D),58 that is conserved among ADAM members and comprised of three histidine (H) residues that act as a ligand for the catalytic zinc atom (Zn2+) complexed with the O2 atom from a H2O molecule. During catalysis, the Zn2+ promotes nucleophilic attack on the carbonyl carbon by the oxygen atom of a water molecule at the active site. The conserved glutamate (E) facilitates this reaction by extracting a proton from the attacking water molecule. Zn2+ chelating drugs such as hydroxamate-type inhibitors can inhibit most ADAMs although these are mostly broad-spectrum inhibitors.59 An intramolecular complex can be formed through a cysteine residue in the prodomain and the Zn2+ in the catalytic domain keeping the ADAMs catalytic domain inactive, this is called the cysteine switch mechanism.6061 For some ADAMs the disintegrin domain (web link 3, PF00200) acts as a receptor antagonist and blocks integrin binding and platelet aggregation.51 The cysteine-rich domain (web link 3, PF08516) is thought to complement the disintegrin domain in substrate binding. For example the disintegrin and cysteine-rich domains of ADAM13 bind to both integrin receptors and to fibronectin.62 Besides binding of other proteins on the cell surface there is no evidence that supports another function of the cysteine-rich domain. ADAM proteins differ mostly in their cytoplasmic tails, which are involved in inside-out regulation of metalloprotease activity, outside-in regulation of cell signaling, maturation and subcellular localization and are highly variable in both length and sequence. There are several common motifs including a binding site for SH3-domain containing proteins and putative phosphorylation sites (Fig. 2).53 In mammals, many ADAMs are expressed exclusively in the male gonads, including ADAM2, 7, 18, 20, 21, 29, and 30. The ADAM8, 9, 10, 11, 12, 15, 17, 19, 22, 23, 28, and 33 have known catalytic activity and a more widespread expression patterns. ADAM-deficient mice have been generated by many groups which are discussed elsewhere.63 Loss of ADAM2 and 3 both lead to male sterility64,65 while ADAM9 deficient mice do not show any obvious pathology despite its ubiquitous expression.66 Moreover, mice that are deficient for ADAM 9, 12 and 15 (Meltrin KO) are viable and fertile and resembled wild type mice with no defects in shedding of EGFR ligands.67 This suggests a high functional redundancy between members of the ADAMs family (web link 4).68 The Tissue Inhibitors of Metalloproteases or TIMPs are a family of highly homologous proteins that are the major natural inhibitors of ADAMs.69 There are 4 TIMPs and all have been found to have some activity against one or more ADAMs and act in a 1:1 stochiometry. For example ADAM10 and ADAM17 are both inhibited by TIMP370,71 and mice lacking Timp3 have an impaired inflammatory response due to deregulated TACE/ADAM17 activity.72 In most cases the physiological relevance of TIMPs on ADAMs remains less clear and requires more study.52,68 There is ample experimental evidence from flies, worms, and mammals that implicate the ADAM sheddases in the direct cleavage and activation of Notch signaling pathway.
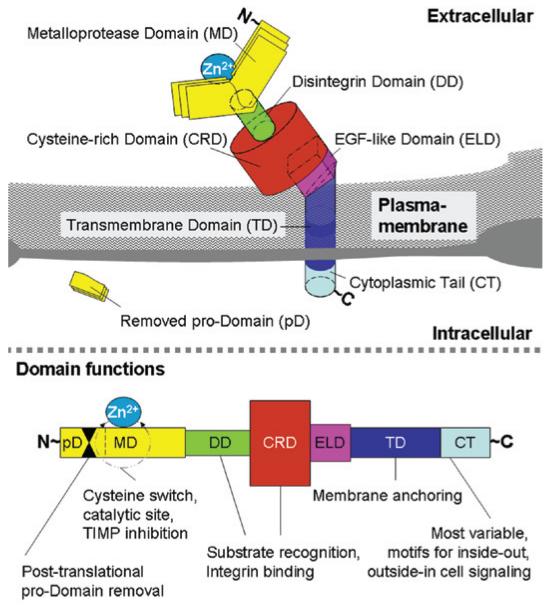
Figure 2. ADAMs structure and function.
Upper panel: schematic illustration of an ADAM with extra-and intracellular domains anchored in the plasma membrane. Typical domains from amino-(N~) to carboxy terminus (~C) are: the pro-Domain (pD) shown after removal during maturation of the ADAM, the Metalloprotease Domain (MD) with a zince atome (Zn2+) marking the catalytic site, the Disintegrin Domain (DD), the Cysteine-rich Domain (CRD), the EGF-like Domain (ELD), the Transmembrane Domain (TD) and the Cytoplasmic Tail (CT). Lower panel: ADAM domains and their function. For a detailed description see text. From N~ to ~C: the pD interacts with Zn2+ through a cysteine residue at the catalytic stie, called the “cysteine switch.” The catalytic site is activated by removal of the pD. ADAM activity can be inhibited by TIMPs. THe MP holds the Zn2+ for proteolytic processing of substrates. The DD and ELD recognize substrates and can bind to integrins. The ELD is indispensible but has no known function. The TD anchors the ADAM in the plasma-membrane. The cytoplasmic tail is most variable among ADAMs and has cell signaling motifs such as phosphorylation sites.
NOTCH RECEPTOR CLEAVAGE BY ADAMs
D. melanogaster express a single Notch protein, and a total of five ADAM metalloproteases.73 There are two homolog’s for ADAM10, Kuz and Kuzbanian-like (Kul) and for ADAM12, DMeltrin and mind-meld (Mmd) and one for tumor necrosis a (TNFa) converting enzyme (TACE)/ADAM17, dTACE.73-75 The nematode C. elegans has two Notch homologs LIN-12 and Glp-1.27 In worms two ADAMs have been reported to regulate LIN-12/Glp-1/Notch signaling, SUP-17/ADAM10 and ADM-4/ADAM17, respectively.76
Genetic studies in D. melanogaster and C. elegans have demonstrated that ADAM10 (Kuz)/Sup-17 plays a critical role in Notch/LIN-12 signaling upstream of S3 γ-secretase cleavage.77-79 Early fly studies had suggested involvement of Kuz in Notch signaling because of the similarity of the Kuz-mutant phenotype with Notch pathway loss of function phenotypes, caused by defects in lateral inhibition leading to central nervous system hyperplasia and a multiple bristle phenotype.80 Maternal and zygotic Kuz mutant flies have a more severe phenotype than Notch mutant flies suggesting that Kuz has other substrates as well. For instance, Kuzbanian plays an important role in neuronal guidance by cleavage of Eph/Ephrin receptors.81-83 Sotillos and Pan provided the first comprehensive analysis on the role of Kuz in the regulation of Notch receptor signaling in flies. Both studies revealed defects in Kuz-mutant flies in tissues whose fate is determined by canonical Notch activity.77, 78 Sotillos cloned the Kuz gene from a P-element insertion in mutant flies and demonstrated, by epistatic analysis were, it functions in a dose-dependent manner upstream of S3/NICD cleaved Notch. Notch ligand dependent hyperactive alleles (Abruptex) are less active in the absence of Kuz whilst NICD rescues partly Kuz phenotypes. Altogether these studies demonstrated that Kuz acts cell autonomously in the signal-receiving (Notch expressing) cell, downstream of ligand binding but upstream of S3/NICD cleavage. Since several of the Kuz phenotypes seemed less severe than the Notch phenotypes alternative ways for Notch activation must exist.77 Pan and Rubin demonstrated that Kuz phenotypes in flies and frogs could be rescued by wild type mammalian ADAM10 expression but not by catalytic-dead mutants demonstrating a requirement for proteolytic activity. Similar findings were reported by Wen and colleagues who showed that SUP-17/Kuz phenotypes mimicked Notch/LIN-12 phenotypes in worm.79 Several groups proposed that Kuz was the protease responsible for furin cleavage (S1) of Notch.35,78,79 It was only after studies in mammalian cells that demonstrated that the ectodomain cleavage of Notch1 receptor was regulated by ligand binding and is distinct from furin/S 1 cleavage during receptor maturation.48 This cleavage was coined S2 and sensitive to inhibitors of metalloproteases and not to furin inhibitors and occurred between the non-conserved residues Ala1710 and Val1711 (Fig. 3).47, 48 This finding was later confirmed on endogenous Notchl signaling as well.25 By biochemical purification ADAM17/TACE was shown to be required for Notch1 cleavage at Val1711 in vitro excluding Kuz/ADAM10.47 Also in ligand independent Notch signaling (using constitutively active receptors lacking the EGF-repeat and NRR domains) these proteins still underwent furin/S1 cleavage,34 which occurred in the absence of Kuz/ADAM10.48 Lieber et al., presented the first evidence for a direct role of Kuz in the S2 extracellular cleavage of Notch.74 Kuz was found to bind Notch directly and induce extracellular cleavage at S2 functioning downstream of ligand binding and upstream of Presenilin S3/NICD cleavage. In the absence of Kuz both S2 and S3 cleavage were impaired consistent with the sequential cleavage model.48 Whereas in mammalian cells ligand independent Notch molecules function independently of Kuz48 in flies Kuz was absolutely required for S2. Interestingly, in flies exogenous ADAM17/TACE can partly rescue the Kuz cleavage defect in vitro.74
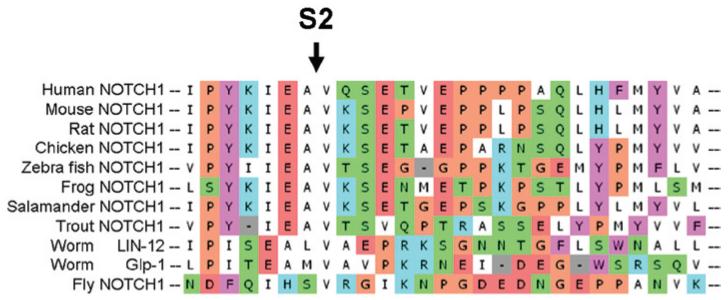
Figure 3. Conservation of the S2 ADAM cleavage site.
Partial alignment of NOTCH1 proteins from different species, with the arrow indicating the S2 cleavage site. Image of alignment was constructed using the program UTOPIA. Aminoacides, in green: Asparagine (N), Glutamine (Q), Serine (S) and Threonine (T); in red: Aspartic acid (D) and Glutamic acid (E); in purple: Phenylalanine (F) and Tyrosine (Y); in blue: Histidine (H), Lysine (K) and Arginine (R); in orange: Glycine (G) and Proline (P); in white: Alanine (A), Isoleucine (I), Leucine (L), Methionine (M) and Valine (V).
The discrepancies between the Kuz requirement in flies and Tace requirement in mammalian cells can now be reconciled by recent work. Both the Weinmaster and Vooijs labs conclusively demonstrated that ADAM10 has an essential role in ligand dependent Notch1 signaling but that ADAM17/TACE or other ADAM proteases may function redundantly with Kuz/ADAM10 particularly in ligand-independent Notch signaling proteins such as those found mutated in cancer.25, 45 In vitro and in flies both Kuz/ADAMIO and TACE are sufficient to cleave Notch in the absence of ligand, a process that may be important in tumors overexpressing ADAM proteins74,84,85 and van Tetering et al., in preparation). Also in worms inactivation of either SUP-17/ADAM10 or ADM-4/ADAM17 has no dramatic effect on LIN-12/Glp-1/Notch signaling however their combined inactivation produces a Notch loss of function phenotype suggesting that these proteases act redundant. Many tissues in SUP-17/ADM-4 mutants however appeared normal suggesting that Notch activation in other tissues requires the action of other unknown (ADAM-like) proteases in C.elegans to regulate LIN-12/Glp-1 activity of which there are several.76
Mice lacking Notch1 are embryonic lethal at E9.5 with severe defects in haematopoiesis and neurogenesis.33,86,87 Whereas ADAM17/TACE co-purifies with Notchl processing activity in HeLa cells and can cleave Notch1 in vitro, Adam17/Tace-deficient mice or flies do not phenocopy the Notch-deficient phenotype suggesting that it is either redundant with another protease or has a more spatial or temporal restricted role in Notch signaling.74,88 TACE may have more tissue-restricted roles in regulating Notch1 cleavage however, for example during differentiation of monocyte progenitors into macrophages.47 In line with the kuz phenotypes in flies, mice lacking AdamlO die at E9.5 with reduced neuronal Hes5 expression, a Notch target gene, resembling Nofch1-null embryos.89 Moreover, T-cell-specific deletion/disruption of Adam 10 in vivo phenocopied the Notch1 null phenotype during thymocyte development.90-92 Recently by using ADAM10 silencing by siRNA or Adam lO KO fibroblasts it was finally demonstrated that ADAM10 is required for ligand-dependent endogenous Notch1 signaling and that it executes S2 cleavage at Val171 1.25,45 In contrast to flies overexpression of ADAM17 could not compensate for ADAM10 loss.45 This finding was recently confirmed by the analysis of brain-specific Adam 10 deletion showing a severe defect in Notch cleavage and signaling in mice.93 Some residual S2/S3 cleavage could be detected in the absence of Adam10 indicating a different protease is capable of cleaving Notch1 as well.93
ADAM10 however may also be involved in ligand independent signaling and appears to act redundant with TACE/ADAM17 in the activation of T-ALL derived Notch proteins.25, 45 The Meltrins ADAM9, ADAM12, and ADAM15, all have been implicated in Notch signaling directly or indirectly by activating ADAM1094,95 however appear not to be required for Notch1 proteolysis.25 As mentioned these findings were somewhat unanticipated because earlier no defects were observed in Notch cleavage.48,89 This can now be reconciled by the demonstration that pathological (ligand independent) and physiological Notch signaling may engage distinct sheddases.45
A tantalizing new finding is the involvement of the non-membrane bound Matrix Metalloproteinase-7 (MMP-7) in Notch1 activation. Sawey and colleagues convincingly demonstrated that in pancreatic explants derived from Mmp7−/− mice acinar transdifferentiation -a Notch1 dependent process-is defective due to impaired Notch cleavage.96 Direct proof for the involvement of Mmp7 in ligand-independent Notch1 cleavage at S2 awaits further analysis but would be remarkable given the “protected” nature of the S2 scissile bond deep within the juxtamembrane LNR module.24 This study further illustrates the diversity of proteolytic cascade involved in Notch activation of particular relevance to pancreatic cancer but may apply to other cancer types as well. Until now there is little insight into the proteases and mechanism involved in shedding of Notch2, Notch3 and Notch4. ADAM10 has been implicated in marginal zone B-cell development; a Notch2 dependent process however no direct effect on Notch2 cleavage was shown.97 It is likely their activation follows a similar paradigm as for Notch1.
NOTCH LIGAND CLEAVAGE BY ADAMS
A series of papers in 2003 reported that mammalian Notch ligands are also subject to RIP. Delta and Jagged are both cleaved by multiple ADAMs including ADAM10 ADAM17/TACE, ADAM12 and ADAM9 followed by γ-secretase cleavage.94,98-101. These findings support earlier work on the involvement of Delta cleavage in vivo in flies.102, 103 Kuz is both necessary and sufficient to induce D1 cleavage and sensitive to metalloprotease inhibitors.102 Although the exact nature and role of ligand cleavage in Notch receptor signaling remains unresolved, a role for Kuz in Notch ligand cleavage would be consistent with a role for soluble ligands in Notch/LIN-12 activation in worms.104 However in flies and mammals soluble Notch ligands are inactive or act antagonistically/competitively with membrane bound forms.105,106 Furthermore, Delta intracellular domain is required for some ligand mediated Notch activation in flies and mammals and promotes ligand multimerization thought to be necessary for efficient Notch activation.107,108
Kuz cleavage of Delta may also explain the non-cell autonomous phenotypes observed for Kuz in fliesm80 although only cis-cleavage by Kuz has been shown.109 Delta cleavage would be consistent with models of ligand transendocytosis in receiving cells.101,109 Similar to Notch1 the P-P1’ cleavage sites are conserved between mammals but mutation of S2 cleavage sites does not prevent cleavage whereas pharmacological and genetic protease inhibition does.25,101 Whereas Notch1 S2 cleavage occurs at the cell surface (Fig. 1)25 the subcellular location of ligand cleavage has not been determined but may—depend on and—occur during endocytosis.110,111 A comprehensive review on ligand cleavage has been published elsewhere.112
Kuz has also been postulated to act in cis-inhibition whereby ligands in Notch receptor expressing cells prevent Notch receptors binding to ligands and signaling in trans to adjacent cells.113,114 Thus a general picture emerges whereby RIP regulates the activities of Notch receptors and ligands to achieve a delicate balance and directionality of signaling needed for context dependent fine-tuning including in the maintenance of stem cells in the Drosophila mid gut 115 as well as in the mammalian epidermis.116 An interesting twist to the story is emerging by the identification of Kul (Kuz-like) the Kuzbanian homolog in flies. Interestingly no Kul homolog has been identified in mammals. Kul protease appears to be critical to maintain signaling directionality during lateral specification in flies.73 An important role for Kul has been demonstrated in wing margin specification where Kuz had already been demonstrated to be dispensable for Dl signaling/processing.75 Whether Kul’s role extends to other invertebrate tissues as well is not known and why flies have evolved to have an additional protease for Delta processing is intriguing.
NOTCH S2-PROTECTION FROM RIP
Under physiological conditions the Notch receptor requires binding of DSL ligands from adjacent cells in trans to EGF repeats 10-12 on Notch expressing cells.117 In the absence of ligand the Notch juxtamembrane region or negative control region (NRR) comprising the LNR and HD domain (Fig. 1) acts to inhibit Notch proteolysis.34,40,41,118 The role of ligand binding and endocytosis in triggering Notch receptor activation by RIP has emerged as a key step in the Notch activation cascade.46,48 Both genetic and biochemical studies supported a model in which receptor oligomerization would regulate receptor proteolysis.40,48 Notch receptors at the cell surface are mostly in their monomeric form with or without ligand. Although Notch proteins can hetero- and homodimerize mediated by the EGF repeats this appears not to regulate shedding of the extracellular domain, since ligand independent active and inactive proteins do not differ in oligomerization.119 Dimerization of the Notch extracellular domain was recently confirmed by electron microscopy.120 It will be interesting to see whether Notch receptors carrying T-ALL mutations have altered dimerization properties. Although from mammalian studies it appears unlikely that dimerization regulates Notch receptor function119 in Drosophila, where furin cleavage is infrequent, dimerization of Notch receptors may play a more prominent role in regulating activity.
The finding that in about 25% of human T-ALL cases mutations are found in the NRR that cause destabilization, ligand-independent Notch cleavage and activation underscores the central importance NRR unfolding in the activation cascade.26,44,121 X-ray structural analysis and epitope-specific antibodies have helped us understand how this activation occurs. Ligand binding to Notch dissolves the “closed” NRR structure leading to access to the Val1711 scissile bond.24,25 Importantly both physiological and ligand independent Notch molecules are cleaved at the same position. These results suggest that conformational changes induced in the extracellular domain by DSL binding or by cancerous mutation are similar but engage different sheddases.25 Further support for this comes from the analysis of Notch proteins with heterologous (CD4ΔIg) extracellular domains.48 These constitutively active proteins are also cleaved at Val1711119 (and Vooijs et al., unpublished).
Currently two models prevail that explain access to S2. Both rely on the force generated between membrane bound ligands and Notch receptors on adjacent cells to liberate the NECD that has been correlated with the extent of adhesion between receptor and ligands.122 One based on X-ray structural analysis predicts that destabilization of the NRR “lifts” the LNR modules to allow S2 protease access.24 The other proposes that ligand binding leads to physical dissociation of the non-covalently held NRR and after which S2 proteolysis occurs.43 It is clear that for cancer prone ligand-independent Notch receptors, NRR relaxation is sufficient to induce S2 as opposed to ligand dependent signaling whereby ligand endocytosis may be needed to pull apart the non-covalent heterodimer. Irrespective of both models the S2 cleavage remains a limiting step for S3 proteolysis. More detailed analysis is needed and X-ray structures from mutated Notch receptors are eagerly awaited. How then is the specificity of S2 proteolysis regulated?
SUBSTRATE REQUIREMENTS FOR S2
Regulation of S2 proteolysis by ADAMs may occur at transcriptional levels by gene expression and alternative splicing, subcellular distribution and posttranslational modification. One of these is prodomain removal by furin in the secretory pathway, which ensures cell surface expression and activity. For example furin and PC7 are critical for ADAM10 maturation and cell surface activity.123 Thus defects in furin may not only affect Notch1 processing at S1 but also S2 processing by ADAM10.25
To date little is known on what determines substrate specificity. Overall cleavage sites are highly variable and a clear consensus sequence is lacking, the secondary structure and the distance of the juxtamembrane stalk (for Notchl, APP, TNFα and EGF ligands are all located within 20 amino-acids proximal to the TMD) seem to be of critical importance for efficient substrate recognition.53,124 Elegant experiment from Arribas demonstrated that swapping the juxtamembrane domains from ADAM permissive substrates TGFa and APP with a substrate normally not shed, could convert this chimeric protein into an efficient ADAM substrate.125 Early work from the Struhl and Kopan labs48, 126 had already demonstrated that the extracellular domain of Notch inhibited S2 cleavage in vitro and in vivo. Using heterologous extracellular domains trimming of the extracellular domain of Notch increased signaling activity in vivo. Engineered dimerization/oligomerization of the extracellular domain also inhibited S2 and S3 cleavage of Notch.48 These experiments established for the first time that S2 was an intermediate step between S1 and S3 and tightly (negatively) regulated by NECD. Although γ-secretase binds to membrane bound Notch during maturation in the secretory pathway the generation of a free N-terminus close to the transmembrane domain is required to become a RIP substrate. Shah provided evidence that Nicastrin an essential component of the γ-secretase activity functions as a docking receptor to haul S2 cleaved Notch into the RIP-S3 processing machinery.127
Interestingly like for S3 Presenilin cleavage of Notch1 the substrate requirements for S2 seem to be independent of the intracellular domain. Furthermore, NEXT-like Notchl proteins (N1ΔE) that are highly active are still S2 cleaved. Cleavage at Val1711 can be blocked by pharmacological or genetic ADAM10 inhibition but has no consequence for γ-secretase cleavage or transcriptional activity. This shows that, at least with truncated artificial substrates like N1ΔE, ADAM proteins will produce Val1711 even with only several residues at the C terminal- cleavage site and further question the substrate requirements for S2 cleavage.25 Moreover, these data suggest that cleavage by ADAM10, under some conditions, may not require binding to Notch. This opens up the hypothesis that the S2/S3 RIP complex is already pre-associated at the cell surface and that the substrate and protease transmembrane domains may interact in the lipid bilayer.
By using randomized peptides libraries the in vitro cleavages specificities of TACE/ADAM17 versus ADAM10 has revealed distinct amino-acid preferences.128 The most important difference maps to the P1’ position of the substrate where TACE is selective for smaller aliphatic residues and ADAM10 can accommodate aromatic amino acids. Whereas the Pl-Pl’ (Ala-Val) amino acids in the mNotchl S2 cleavage site are conserved Notch1 cleavage at Val1711 still has to be demonstrated in other species. Remarkably, whereas the Pl-Pl’ A-V motif in Notchl is common in multiple TACE substrates it is unique among known ADAM10 substrates.128 In view of this and the finding that recombinant murine Notchl proteins/peptides are preferred by ADAM17/TACE47 whereas in vivo mNotchl proteins require ADAM10 these findings suggest that protease activity in vitro may not always be extrapolated.25 In mammals the S2 cleavage site of Notch1 YxIEA-VxSE is highly conserved between species between Notch proteins of different species only AV is conserved (Fig. 3). Based on this analysis NOTCH receptors 2, 3 and 4 are also predicted to be substrates for TACE, but no results have been reported on this yet.
Mutation of the Notch1 S2 cleavage site does not abolish cleavage but generates a novel epitope, which has not been determined.25 P1-mutants of other ADAM10 substrates such as lysine (P1-K|L-P1’) in APP also does notabolish cleavage.129 These findings further support the notion that structural alterations induced by mutation or ligand-binding expose scissile residues that enable access of the sheddase and cleavage. Direct confirmation for this has come from the analysis of T-ALL mutations that affect the S2 position as well as direct mutation of the S2 cleavage site, which produces a gain of function protein.25,130
ADAM10 and TACE/ADAM17 function in multiple RIP processes such as cleavage of the extracellular domain of Alzheimer’s precursor protein (APP), TNFa and EGF ligands among many other Type I integral membrane proteins.131 ADAM10 has up to 40 substrates including Type II membrane proteins that are SPP substrates.132 Although ligand-independent activity induced by ADAM expression has been described this is probably not a physiological mechanism.84 A common feature of ectodomain shedding is constitutive versus regulated shedding. For example ADAM10 is required for constitutive α-secretase cleavage of APP133 whereas both ADAM17/TACE and ADAM10 are involved in PMA-stimulated (regulated) shedding.134,135 Thus distinct proteases are involved in constitutive versus regulated cleavage of RIP substrates Notch and APP. Whereas Notch gain of function mutations are constitutively cleaved, this cleavage can be further induced by PMA, the mercuric compound APMA and well as by EDTA, the latter by disrupting the non-covalently associated heterodimer.25 If during constitutive and regulated cleavages of Notch different proteases are recruited that cleave Notch at different positions has to be determined. The only evidence for this so far stem from the analysis of Adam17/Tace requirement to block PMA stimulated differentiation of monocyte precursors into macrophages a process that depends on Notch-Jagged signaling.47 It should be noted that it is not clear whether PMA/APMA regulated shedding of Notch receptors has any physiological role.
LOCATION OF S2 CLEAVAGE
The best characterized substrates for ADAM proteases are membrane bound signaling molecules such EGF ligands, TNFα, APP, Ephrins and Notch136 Most ADAM proteases are produced as inactive zymogens and activated by prodomain removal in the secretory pathway. Prodomain removal is necessary for proper cell surface expression. The a-secretase cleavage of APP by ADAM10 predominantly occurs at the plasma membrane,129 but also in the trans-Golgi network upon stimulation with PMA, where it may compete with β-secretase cleavage.137 Intracellular cleavage of APP induced by PMA is consistent with the perinuclear expression of TACE.137,138 Using S2-epitope specific antibodies van Tetering showed by immunofluorescence and biochemically that Notch S2 cleavage by ADAM10 predominantly occurred at the cell surface. Interestingly this analysis also demonstrated intracellular vesicles that were decorated with the S2 antibody. As this analysis was performed in the presence of γ-secretase inhibitor this indicated that S2 cleaved Notch from the cell surface may be internalized for further processing. At present the identity and fate of these S2-positive vesicles is not known but of great interest. Outstanding questions are whether these vesicles contain γ-secretase competent Notch molecules and whether these are recycled to the plasma membrane or follow other intracellular routes for activation or degradation.25 Endocytosis is a conserved biological mechanism regulating the location and activity of membrane bound signaling molecules.139 Also in the activation of Notch signaling, ligand endocytosis plays a critical role presumably to pull apart the NRR to expose the S2 site.46 Furthermore, asymmetric cell division and unequal distribution of Notch receptors and ligands among daughter cells controls important cell fate decisions in several tissues.140 There is compelling evidence that demonstrates that receptor endocytosis controls Notch cleavage and activity in signal receiving cells independent of ligand endocytosis in signaling cells in flies and mammals.26,141-143 Several years ago it was already established that γ-secretase cleavage sites on Notch1 and APP shifted in the more acidic environments of endocytic vesicles. NICD species with different NH2-residues (V, L, S) have different stability and therefore signaling strength, which highlighted an additional level of regulation.144-146 At least under some conditions receptor endocytosis is required for γ-secretase cleavage.146-148 From this it is clear that γ-secretase activity occurs both at the plasma membrane as well as in endocytic vesicles. For example the E3 ubiquitin ligase Deltex that directly binds to Notch promotes signaling independent of ligand via receptor internalization in endocytic compartments. Depending on the co-factors AP-3/HOPS and type of endosome (early vs late) the fate of Notch receptors is degradation or signaling.143, 149 Under conditions of signaling the Notch ectodomain is removed however whether this is as a consequence of ectodomain shedding at S2 (or an alternative position) or non-specific degradation is unknown.149 Whether similar roles for Deltex can be extended to other tissues and species is not known but ligand-independent Notch activation via Deltex may occur through heterodimer destabilization due to changes in pH/ionic environments in endocytic organelles.150 Exciting new data is emerging that show that ligand dependent Notch signaling may also occur in multivesicular endosomes so-called Sara endosomes.151 Whether Sara endosomes present a conserved intracellular Notch signaling node is not known but exiting new insights are expected. The availability of antibodies for S2 cleaved forms of Notch1 was long awaited and may be used to address the origin and fate of S2 activation step in mammalian cells as well.25
TARGETING S2 PROTEOLYSIS IN CANCER
In the last decade it has become apparent that deregulation of the Notch signaling pathway is a common event in human cancer. This research was fueled by the discovery that Notch mutations and alterations underlie the pathogenesis of T-cell acute lymphocytic leukemias (T-ALL).26, 152 Missense mutations in the extracellular HD of Notch1 occur in about 25% of all human T-ALL cases26 and lead to ligand independent activation.44 Less frequent are insertions that duplicate the S2 site and lead to ligand independent Notch activity.130 Finally alterations in the endocytic pathway as discussed above may provide another oncogenic pathway for Notch activation. In addition to T-ALL, Notch activation is common in solid cancers as well. For example in breast cancer one of the most frequent malignancies in the western world Notch1 activation is a common event and correlates with poor prognosis.153-156 There is ample evidence for crosstalk with other frequently deregulated pathways in breast cancer such as RAS/EGFR/HER2157 activation and Estrogen pathway158 that are common drug targets for cancer intervention. New data is emerging that activation of Notch3 and Notch4 as well may also be important in breast cancer. In contrast Notch2 activity seems to correlate with better prognosis in breast cancer.159 Finally, Notch receptor signaling is critical in normal mammary stem cell160, 161 as well as in tumor initiating cells or breast cancer stem cells.162 Finally an important new role of Notch signaling is emerging in maintaining normal and tumor angiogenesis (reviewed in ref. 163). The role of Notch signaling in cancers will be discussed in detail elsewhere. Given the widespread role and importance of NOTCH activation in human malignancies therapeutic targeting of Notch may allow for disease control. Most if not all Notch signaling requires S2 and S3 cleavage making Notch proteolysis an attractive drug target. At the same time this provides challenges for cancer drug development, as these drugs will also target physiological Notch activation. Currently 15 clinical trials are underway that evaluate the efficacy of γ-secretase inhibitors (GSIs) as anti-cancer drugs (web link 4). While targeting γ-secretase using has shown encouraging results it has many pitfalls as well. Among these is the lack of specificity and the mechanism based toxicity caused by attenuating physiological Notch function. One of the best-illustrated side-effects is gastrointestinal toxicity caused by precocious secretory differentiation of intestinal epithelial cells limiting its long term use because of intestinal stem cell depletion.17,164,165 Both pharmacological and genetic approaches have provided insight into the normal role of Notch in suppressing secretory differentiations and have revealed KLF4 as a key regulator. Significant progress was made by Real and colleagues who demonstrated that combined glucocorticoid and GSI treatment increased anti-leukemic activity in T-ALL by reversing glucocorticoid resistance while suppressing gut toxicity by inhibition of KLF4.166 Since γ-secretase has many different substrates pleiotropic effects are unavoidable when targeting this enzyme, although modulators have been identified that may show selectivity for specific substrates.167 The fact that GSI have been studied intensely the past decade a safe-drug has yet to enter clinical practice.168 Recently, monoclonal antibody based approaches have been developed specifically targeting the Notch NRR that show promising results in preclinical studies.169-171 Strategies to target the nuclear transcription complex are also promising.172
An alternative approach could encompass targeting S2 cleavage: the rate-limiting step in the Notch activation cascade in physiological and pathological conditions.25 Two approaches not mutually exclusive could be envisaged. One based on targeting the metalloprotease, another based on blocking proteolysis of S2 cleaved Notch. Multiple ADAMs have causal roles in cancer formation and progression but whether these ADAM deregulate Notch activation in these tumors is unknown (reviewed in).173 Despite enormous efforts and investment the implementation of metalloprotease inhibitors in anticancer medicine has largely failed.174 For example, the hydroxamic acid Zn2+ chelators GM6001 (Gelardin), Batimastat (BB94) and Marimastat have all failed for clinical application because of undesirable side-effects. Recently progress has been made with the development of new inhibitors more specific for ADAM10 and ADAM17. One of these INCB3619 is designed to target EGFR/HER shedding a frequent and causal alteration in breast and lung cancer.175-177 Since Notch activity is frequently reported in both cancers it will be interesting to see whether inhibition of Notch shedding occurs and whether this is of therapeutic value.
While investigating the requirement for ADAM proteases on Notch S2 cleavage we discovered that S2 cleavage and transcriptional activity of activated Notch1 could neither be blocked using broad-spectrum metalloprotease inhibitors (GM6001, BB94) nor with more specific ADAM17/TACE or ADAM10 specific inhibitors.178 Interestingly whereas these inhibitors efficiently blocked Vall711 cleavage residual S2 cleavage occurred at another unknown position, S2b. Thus whereas ligand dependent Notch signaling requires ADAM10, ADAM/MP inhibition in ligand independent signaling does not abrogate activity. The S2b cleavage is insensitive to broad-spectrum aspartyl, cysteine and serine protease inhibitors as well as to BACE inhibitors. These results strongly point to the existence of additional S2 proteases insensitive to these mechanism-based metalloprotease inhibitors.25 One likely outcome from these and other studies is that pathological (ligand-independent) and physiological Notch signaling may engage distinct sheddases.45 While this hypothesis awaits further validation it suggests the possibility of targeting disease-specific Notch proteases while leaving normal Notch signaling intact.
Another potential strategy to block Notch activity in tumors is to prevent conversion from S2 cleaved Notch into a γ-secretase substrate using S2 specific antibodies.25 Such capping antibodies would be highly specific and could interfere with efficient recruitment of S2 cleaved into the γ-secretase complex. Such antibodies would target receptors and are not hampered by membrane diffusion or cellular uptake. Experimental data suggests that such an approach may be feasible.127 It is important to note that completely blocking Val1711 pharmacologically however does not abrogate signaling and physiological and pathological Notch signaling both proceed through Val1711 cleavage.
It is clear that many challenges still lie ahead before drugs will be developed that are effective in controlling Notch dependent disease while leaving normal Notch signaling intact.
CONCLUSION
The highly conserved Notch signaling pathway controls developmental patterning and homeostasis in animal tissues. A conserved proteolytic cascade leading to liberation of the Notch intracellular domain and transcription governs the rate-limiting step in Notch activation: ectodomain shedding at S2. Loss of function studies point to a dominant role for ADAM10 in the cleavage and activation of Notch1 after ligand binding. Under malignant conditions shedding of Notch1 receptors is induced by different proteases that eagerly await identification. Exciting biology lies ahead of us in revealing the many facets of NOTCH proteolysis controlling Notch activation in normal and injured tissues providing new insights that may be applied for therapeutic intervention.
ACKNOWLEDGMENTS
A.G and M.V. were supported by the European Research Council under the European Community Seventh Framework Programme (FP7/2007-2013)/ERC Grant 208259 (to M. V.) and the Dutch Cancer Society Grant KWF UU2006-3623 (to M. V. )
Footnotes
MEROPS Protease database (REF): http://merops.sanger.ac.uk
Protein families and Domains: http://pfam.sanger.ac.uk
GSI clinical trials: http://clinicaltrials.gov/ct2/results?term=NOTCH+and+CancerGSIclinicaltrials
REFERENCES
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100(4):391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Urban S, Freeman M. Intramembrane proteolysis controls diverse signalling pathways throughout evolution. Curr Opin Genet Dev. 2002;12(5):512–518. doi: 10.1016/s0959-437x(02)00334-9. [DOI] [PubMed] [Google Scholar]
- 3.Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes Dev. 2009;23(22):2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107(2):173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 6.Weihofen A, Binns K, Lemberg MK, et al. Identification of signal peptide peptidase, apresenilin-type aspartic protease. Science. 2002;296(5576):2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 7.De Strooper B, Saftig P, Craessaerts K, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391(6665):387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 8.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopan R, Goate A. A common enzyme connects notch signaling and Alzheimer’s disease. Genes Dev. 2000;14(22):2799–2806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 10.Feng L, Yan H, Wu Z, et al. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318(5856):1608–1612. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 11.Golde TE, Eckman CB. Physiologic and pathologic events mediated by intramembranous and juxtamembranous proteolysis. Sci STKE. 2003;2003(172):RE4. doi: 10.1126/stke.2003.172.re4. [DOI] [PubMed] [Google Scholar]
- 12.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 13.Gazave E, Lapebie P, Richards GS, et al. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 2009;9:249. doi: 10.1186/1471-2148-9-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson A, Radtke F. Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett. 2006;580(12):2860–2868. doi: 10.1016/j.febslet.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Vooijs M, Ong CT, Hadland B, et al. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007 Feb;134(3):535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3(10):756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 17.van Es JH, van Gijn ME, Riccio O, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 18.De Strooper B, Annaert W, Cupers P, et al. A presenilin-1 -dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 19.Stifani S, Blaumueller CM, Redhead NJ, et al. Human homologs of a Drosophila Enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2(2):119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 20.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393(6683):382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 21.Christensen S, Kodoyianni V, Bosenberg M, et al. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122(5):1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 22.Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79(2):273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 23.Fryer CJ, Lamar E, Turbachova I, et al. Mastermind mediates chromatin-specific transcription and turnover of the Notch enhancer complex. Genes Dev. 2002;16(11):1397–1411. doi: 10.1101/gad.991602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon WR, Vardar-Ulu D, Histen G, et al. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14(4):295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 25.van Tetering G, van Diest P, Verlaan I, et al. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem. 2009;284(45):31018–31027. doi: 10.1074/jbc.M109.006775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 27.Kimble J, Simpson P. The LIN-12/Notch signaling pathway and its regulation. Annu Rev Cell Dev Biol. 1997;13:333–361. doi: 10.1146/annurev.cellbio.13.1.333. [DOI] [PubMed] [Google Scholar]
- 28.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387(6636):908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 29.Okajima T, Irvine KD. Regulation of notch signaling by o-linked fucose. Cell. 2002;111(6):893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 30.Moloney DJ, Panin VM, Johnston SH, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406(6794):369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 31.Haines N, Irvine KD. Glycosylation regulates Notch signalling. Nat Rev Mol Cell Biol. 2003;4(10):786–797. doi: 10.1038/nrm1228. [DOI] [PubMed] [Google Scholar]
- 32.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398(6727):522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 33.Huppert SS, Le A, Schroeter EH, et al. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405(6789):966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 34.Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93(4):1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90(2):281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 36.Logeat F, Bessia C, Brou C, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci USA. 1998;95(14):8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bush G, diSibio G, Miyamoto A, et al. Ligand-induced signaling in the absence of furin processing of Notch1. Dev Biol. 2001;229(2):494–502. doi: 10.1006/dbio.2000.9992. [DOI] [PubMed] [Google Scholar]
- 38.Kidd S, Lieber T. Furin cleavage is not a requirement for Drosophila Notch function. Mech Dev. 2002;115(1-2):41–51. doi: 10.1016/s0925-4773(02)00120-x. [DOI] [PubMed] [Google Scholar]
- 39.Gordon WR, Vardar-Ulu D, L’Heureux S, et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009;4(8):e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenwald I. Structure/function studies of lin-12/Notch proteins. Curr Opin Genet Devel. 1994;4(4):556–562. doi: 10.1016/0959-437x(94)90072-b. [DOI] [PubMed] [Google Scholar]
- 41.Lieber T, Kidd S, Alcamo E, et al. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Devel. 1993;7(10):1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Irizarry C, Carpenter AC, Weng AP, et al. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24(21):9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nichols JT, Miyamoto A, Olsen SL, et al. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176(4):445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malecki MJ, Sanchez-Irizarry C, Mitchell JL, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26(12):4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bozkulak EC, Weinmaster G. Selective use of ADAM10 and ADAM17 in activation of Notch1 signaling. Mol Cell Biol. 2009;29(21):5679–5695. doi: 10.1128/MCB.00406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127(7):1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- 47.Brou C, Logeat F, Gupta N, et al. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5(2):207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 48.Mumm JS, Schroeter EH, Saxena MT, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5(2):197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 49.Jarriault S, Brou C, Logeat F, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377(6547):355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 50.Sanalkumar R, Dhanesh SB, James J. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell Mol Life Sci. 2010;67(17):2957–2968. doi: 10.1007/s00018-010-0391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolfsberg TG, Primakoff P, Myles DG, White JM. ADAM, a novel family of membrane proteins containing A Disintegrin and Metalloprotease domain: multipotential functions in cell-cell and cell-matrix interactions. J Cell Biol. 1995;131(2):275–278. doi: 10.1083/jcb.131.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murphy G. Regulation of the proteolytic disintegrin metalloproteinases, the ‘Sheddases’. Semin Cell Dev Biol. 2009;20(2):138–145. doi: 10.1016/j.semcdb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 54.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38(Database issue):D227–233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386(Pt 1):15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howard L, Maciewicz RA, Blobel CP. Cloning and characterization of ADAM28: evidence for autocatalytic pro-domain removal and for cell surface localization of mature ADAM28. Biochem J. 2000;348(Pt 1):21–27. [PMC free article] [PubMed] [Google Scholar]
- 57.Schlomann U, Wildeboer D, Webster A, et al. The metalloprotease disintegrin ADAM8. Processing by autocatalysis is required for proteolytic activity and cell adhesion. J Biol Chem. 2002;277(50):48210–48219. doi: 10.1074/jbc.M203355200. [DOI] [PubMed] [Google Scholar]
- 58.Gomis-Ruth FX. Catalytic domain architecture of metzincin metalloproteases. J Biol Chem. 2009;284(23):15353–15357. doi: 10.1074/jbc.R800069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Georgiadis D, Yiotakis A. Specific targeting of metzincin family members with small-molecule inhibitors: progress toward a multifarious challenge. Bioorg Med Chem. 2008;16(19):8781–8794. doi: 10.1016/j.bmc.2008.08.058. [DOI] [PubMed] [Google Scholar]
- 60.Van Wart HE, Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc Natl Acad Sci USA. 1990;87(14):5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Becker JW, Marcy AI, Rokosz LL, et al. Stromelysin-1: three-dimensional structure of the inhibited catalytic domain and of the C-truncated proenzyme. Protein Sci. 1995;4(10):1966–1976. doi: 10.1002/pro.5560041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaultier A, Cousin H, Darribere T, Alfandari D. ADAM13 disintegrin and cysteine-rich domains bind to the second heparin-binding domain offibronectin. J Biol Chem. 2002;277(26):23336–23344. doi: 10.1074/jbc.M201792200. [DOI] [PubMed] [Google Scholar]
- 63.Hoiruchi K, Blobel CP. Studies from ADAM knockout mice. In: Hooper NM, Lendeckel U, editors. The ADAM Family of Proteases. Springer; New York: 2005. pp. 29–64. [Google Scholar]
- 64.Cho C, Bunch DO, Faure JE, et al. Fertilization defects in sperm from mice lacking fertilin beta. Science. 1998;281(5384):1857–1859. doi: 10.1126/science.281.5384.1857. [DOI] [PubMed] [Google Scholar]
- 65.Nishimura H, Cho C, Branciforte DR, Myles DG, et al. Analysis of loss of adhesive function in sperm lacking cyritestin or fertilin beta. Dev Biol. 2001;233(1):204–213. doi: 10.1006/dbio.2001.0166. [DOI] [PubMed] [Google Scholar]
- 66.Weskamp G, Cai H, Brodie TA, et al. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol Cell Biol. 2002;22(5):1537–1544. doi: 10.1128/mcb.22.5.1537-1544.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahin U, Weskamp G, Kelly K, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164(5):769–779. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huovila AP, Turner AJ, Pelto-Huikko M, et al. Shedding light on ADAM metalloproteinases. Trends Biochem Sci. 2005;30(7):413–422. doi: 10.1016/j.tibs.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 69.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803(1):55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amour A, Slocombe PM, Webster A, et al. TNF-alpha converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett. 1998;435(1):39–44. doi: 10.1016/s0014-5793(98)01031-x. [DOI] [PubMed] [Google Scholar]
- 71.Amour A, Knight CG, Webster A, et al. The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett. 2000;473(3):275–279. doi: 10.1016/s0014-5793(00)01528-3. [DOI] [PubMed] [Google Scholar]
- 72.Smookler DS, Mohammed FF, Kassiri Z, et al. Tissue inhibitor of metalloproteinase 3 regulates TNF-dependent systemic inflammation. J Immunol. 2006;176(2):721–725. doi: 10.4049/jimmunol.176.2.721. [DOI] [PubMed] [Google Scholar]
- 73.Sapir A, Assa-Kunik E, Tsruya R, et al. Unidirectional Notch signaling depends on continuous cleavage of Delta. Development. 2005;132(1):123–132. doi: 10.1242/dev.01546. [DOI] [PubMed] [Google Scholar]
- 74.Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16(2):209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klein T. kuzbanian is required cell autonomously during Notch signalling in the Drosophila wing. Dev Genes Evol. 2002;212(5):251–255. doi: 10.1007/s00427-002-0233-4. [DOI] [PubMed] [Google Scholar]
- 76.Jarriault S, Greenwald I. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev Biol. 2005;287(1):1–10. doi: 10.1016/j.ydbio.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sotillos S, Roch F, Campuzano S. The metalloprotease-disintegrin Kuzbanian participates in Notch activation during growth and patterning of Drosophila imaginal discs. Development. 1997;124(23):4769–4779. doi: 10.1242/dev.124.23.4769. [DOI] [PubMed] [Google Scholar]
- 78.Pan D, Rubin GM. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90(2):271–280. doi: 10.1016/s0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 79.Wen C, Metzstein MM, Greenwald I. SUP-17, a Caenorhabditis elegans ADAM protein related to Drosophila KUZBANIAN, and its role in LIN-12/NOTCH signalling. Development. 1997;124(23):4759–4767. doi: 10.1242/dev.124.23.4759. [DOI] [PubMed] [Google Scholar]
- 80.Rooke J, Pan D, Xu T, Rubin GM. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science. 1996;273(5279):1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 81.Janes PW, Saha N, Barton WA, et al. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123(2):291–304. doi: 10.1016/j.cell.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 82.Fambrough D, Pan D, Rubin GM, Goodman CS. The cell surface metalloprotease/disintegrin Kuzbanian is required for axonal extension in Drosophila. Proc Natl Acad Sci USA. 1996;93(23):13233–13238. doi: 10.1073/pnas.93.23.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hattori M, Osterfield M, Flanagan JG. Regulated cleavage of a contact-mediated axon repellent. Science. 2000;289(5483):1360–1365. doi: 10.1126/science.289.5483.1360. [DOI] [PubMed] [Google Scholar]
- 84.Delwig A, Rand MD. Kuz and TACE can activate Notch independent of ligand. Cell Mol Life Sci. 2008;65(14):2232–2243. doi: 10.1007/s00018-008-8127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borrell-Pages M, Rojo F, Albanell J, et al. TACE is required for the activation of the EGFR by TGF-alpha in tumors. EMBO J. 2003;22(5):1114–1124. doi: 10.1093/emboj/cdg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121(5):1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 87.Swiatek PJ, Lindsell CE, del Amo FF, et al. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8(6):707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 88.Peschon JJ, Slack JL, Reddy P, et al. An Essential Role For Ectodomain Shedding in Mammalian Development. Science. 1998;282(5392):1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 89.Hartmann D, De Strooper B, Serneels L, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;ll(21):2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 90.Radtke F, Wilson A, Stark G, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10(5):547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 91.Manilay JO, Anderson AC, Kang C, Robey EA. Impairment of thymocyte development by dominant-negative Kuzbanian (ADAM-10) is rescued by the Notch ligand, delta-1. J Immunol. 2005;174(11):6732–6741. doi: 10.4049/jimmunol.174.11.6732. [DOI] [PubMed] [Google Scholar]
- 92.Tian L, Wu X, Chi C, et al. ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int Immunol. 2008;20(9):1181–1187. doi: 10.1093/intimm/dxn076. [DOI] [PubMed] [Google Scholar]
- 93.Jorissen E, Prox J, Bernreuther C, et al. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30(14):4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dyczynska E, Sun D, Yi H, et al. Proteolytic processing of delta-like 1 by ADAM proteases. J Biol Chem. 2007;282(1):436–444. doi: 10.1074/jbc.M605451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tousseyn T, Thathiah A, Jorissen E, et al. ADAM10, the rate-limiting protease of regulated intramembrane proteolysis of Notch and other proteins, is processed by ADAMS-9, ADAMS-15, and the gamma-secretase. J Biol Chem. 2009;284(17):11738–11747. doi: 10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sawey ET, Johnson JA, Crawford HC. Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc Natl Acad Sci USA. 2007;104(49):19327–19332. doi: 10.1073/pnas.0705953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gibb DR, El Shikh M, Kang DJ, et al. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med. 2010;207(3):623–635. doi: 10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Six E, Ndiaye D, Laabi Y, et al. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc Natl Acad Sci USA. 2003;100(13):7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278(36):34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 100.Ikeuchi T, Sisodia SS. The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent "gamma-secretase" cleavage. J Biol Chem. 2003;278(10):7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- 101.Bland CE, Kimberly P, Rand MD. Notch-induced proteolysis and nuclear localization of the Delta ligand. J Biol Chem. 2003;278(16):13607–13610. doi: 10.1074/jbc.C300016200. [DOI] [PubMed] [Google Scholar]
- 102.Qi H, Rand MD, Wu X, et al. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283(5398):91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 103.Klueg KM, Parody TR, Muskavitch MAT. Complex Proteolytic Processing Acts On Delta, a Transmembrane Ligand For Notch, During Drosophila Development. Mol Biol Cell. 1998;9(7):1709–1723. doi: 10.1091/mbc.9.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell. 2004;6(2):183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- 105.Sun X, Artavanis-Tsakonas S. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development. 1997;124(17):3439–3448. doi: 10.1242/dev.124.17.3439. [DOI] [PubMed] [Google Scholar]
- 106.Noguera-Troise I, Daly C, Papadopoulos NJ, et al. Blockade of Dll4 inhibits tumour growth by promoting nonproductive angiogenesis. Nature. 2006;444(7122):1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- 107.Shimizu K, Chiba S, Saito T, et al. Integrity of intracellular domain of Notch ligand is indispensable for cleavage required for release of the Notch2 intracellular domain. EMBO J. 2002;21(3):294–302. doi: 10.1093/emboj/21.3.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Varnum-Finney B, Wu L, Yu M, et al. Immobilization of Notch ligand, Delta-1, is required for induction of Notch signaling. J Cell Sci. 2000;113(23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- 109.Mishra-Gorur K, Rand MD, Perez-Villamil B, Artavanis-Tsakonas S. Down-regulation of Delta by proteolytic processing. J Cell Biol. 2002;159(2):313–324. doi: 10.1083/jcb.200203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Delwig A, Bland C, Beem-Miller M, Kimberly P, Rand MD. Endocytosis-independent mechanisms of Delta ligand proteolysis. Exp Cell Res. 2006;312(8):1345–1360. doi: 10.1016/j.yexcr.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 111.Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131(21):5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- 112.Zolkiewska A. ADAM proteases: ligand processing and modulation of the Notch pathway. Cell Mol Life Sci. 2008;65(13):2056–2068. doi: 10.1007/s00018-008-7586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.de Celis JF, Bray SJ. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development. 2000;127(6):1291–1302. doi: 10.1242/dev.127.6.1291. [DOI] [PubMed] [Google Scholar]
- 114.Jacobsen TL, Brennan K, Arias AM, Muskavitch MAT. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 1998;125(22):4531–4540. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- 115.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315(5814):988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 116.Lowell S, Jones P, Le Roux I, et al. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10(9):491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 117.de Celis JF, Barrio R, del Arco A, Garcia-Bellido A. Genetic and molecular characterization of a Notch mutation in its Delta- and Serrate-binding domain in Drosophila. Proc Natl Acad Sci USA. 1993;90(9):4037–4041. doi: 10.1073/pnas.90.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kimble J, Henderson S, Crittenden S. Notch/Lin-12 Signaling - Transduction By Regulated Protein Slicing. Trends Biochem Sci. 1998;23(9):353–357. doi: 10.1016/s0968-0004(98)01263-8. [DOI] [PubMed] [Google Scholar]
- 119.Vooijs M, Schroeter EH, Pan Y, et al. Ectodomain shedding and intramembrane cleavage of mammalian Notch proteins is not regulated through oligomerization. J Biol Chem. 2004;279(49):50864–50873. doi: 10.1074/jbc.M409430200. [DOI] [PubMed] [Google Scholar]
- 120.Kelly DF, Lake RJ, Middelkoop TC, et al. Molecular structure and dimeric organization of the Notch extracellular domain as revealed by electron microscopy. PLoS ONE. 2010;5(5):e10532. doi: 10.1371/journal.pone.0010532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sanchez-Irizarry C, Malecki M, Lee W, et al. Functional Analysis of Leukemia-Associated Mutations Involving the Heterodimerization Domain of NOTCH1. Am Soc Hematol. 2005 [Google Scholar]
- 122.Ahimou F, Mok LP, Bardot B, Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J Cell Biol. 2004;167(6):1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anders A, Gilbert S, Garten W, et al. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15(10):1837–1839. doi: 10.1096/fj.01-0007fje. [DOI] [PubMed] [Google Scholar]
- 124.Blobel CP. ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol. 2005;6(1):32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 125.Arribas J, Lopez-Casillas F, Massague J. Role of the juxtamembrane domains of the transforming growth factor-alpha precursor and the beta-amyloid precursor protein in regulated ectodomain shedding. J Biol Chem. 1997;272(27):17160–17165. doi: 10.1074/jbc.272.27.17160. [DOI] [PubMed] [Google Scholar]
- 126.Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6(3):625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 127.Shah S, Lee SF, Tabuchi K, et al. Nicastrin functions as a gamma-secretase-substrate receptor. Cell. 2005;122(3):435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 128.Caescu CI, Jeschke GR, Turk BE. Active-site determinants of substrate recognition by the metalloproteinases TACE and ADAM10. Biochem J. 2009;424(1):79–88. doi: 10.1042/BJ20090549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sisodia SS. Beta-amyloid precursor protein cleavage by a membrane-bound protease. Proc Natl Acad Sci USA. 1992;89(13):6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sulis ML, Williams O, Palomero T, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112(3):733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20(2):164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 132.Endres K, Fahrenholz F. Upregulation of the alpha-secretase ADAM10--risk or reason for hope? FEBS J. 2010;277(7):1585–1596. doi: 10.1111/j.1742-4658.2010.07566.x. [DOI] [PubMed] [Google Scholar]
- 133.Kuhn PH, Wang H, Dislich B, et al. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. EMBO J. 2010;29(17):3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lammich S, Kojro E, Postina R, et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci USA. 1999;96(7):3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Buxbaum JD, Liu KN, Luo YX, et al. Evidence That Tumor Necrosis Factor Alpha Converting Enzyme Is Involved in Regulated Alpha-Secretase Cleavage of the Alzheimer Amyloid Protein Precursor. J Biol Chem. 1998;273(43):27765–27767. doi: 10.1074/jbc.273.43.27765. [DOI] [PubMed] [Google Scholar]
- 136.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91(4):439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 137.Skovronsky DM, Moore DB, Milla ME, et al. Protein kinase C-dependent alpha-secretase competes with beta-secretase for cleavage of amyloid-beta precursor protein in the trans-golgi network. J Biol Chem. 2000;275(4):2568–2575. doi: 10.1074/jbc.275.4.2568. [DOI] [PubMed] [Google Scholar]
- 138.Schlondorff J, Becherer JD, Blobel CP. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE) Biochem J. 2000;347(Pt 1):131–138. [PMC free article] [PubMed] [Google Scholar]
- 139.Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463(7280):464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- 140.Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132(8):1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- 141.Vaccari T, Lu H, Kanwar R, et al. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J Cell Biol. 2008;180(4):755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jaekel R, Klein T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev Cell. 2006 Nov;ll(5):655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 143.Wilkin MB, Carbery AM, Fostier M, et al. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr Biol. 2004 Dec 29;14(24):2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 144.Pasternak SH, Bagshaw RD, Guiral M, et al. Presenilin, Nicastrin, Amyloid precursor protein, and γ-secretase activity are co-locelized in the lysosomal membrane. J Biol Chem. 2003;278:26687–26694. doi: 10.1074/jbc.m304009200. [DOI] [PubMed] [Google Scholar]
- 145.Tagami S, Okochi M, Yanagida K, et al. Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1. Mol Cell Biol. 2008 Jan;28(1):165–176. doi: 10.1128/MCB.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fukumori A, Okochi M, Tagami S, et al. Presenilin-dependent gamma-secretase on plasma membrane and endosomes is functionally distinct. Biochemistry. 2006 Apr 18;45(15):4907–4914. doi: 10.1021/bi052412w. [DOI] [PubMed] [Google Scholar]
- 147.Seugnet L, Simpson P, Haenlin M. Requirement For Dynamin During Notch Signaling In Drosophila Neurogenesis. Developmental Biology. 1997;192(2):585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 148.Gupta-Rossi N, Six E, LeBail O, et al. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004 Jul 5;166(1):73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wilkin M, Tongngok P, Gensch N, et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev Cell. 2008 Nov;15(5):762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 150.Rand MD, Grimm LM, Artavanis-Tsakonas S, et al. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20(5):1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Coumailleau F, Furthauer M, Knoblich JA, Gonzalez-Gaitan M. Directional Delta and Notch trafficking in Sara endosomes during asymmetric cell division. Nature. 2009 Apr 23;458(7241):1051–1055. doi: 10.1038/nature07854. [DOI] [PubMed] [Google Scholar]
- 152.Ellisen LW, Bird J, West DC, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991 Aug 23;66(4):649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 153.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006 Feb 1;66(3):1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 154.Reedijk M, Odorcic S, Chang L, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005 Sep 15;65(18):8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 155.Reedijk M, Pinnaduwage D, Dickson BC, et al. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008 Oct;111(3):439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 156.Mittal S, Subramanyam D, Dey D, et al. Cooperation of Notch and Ras/MAPK signaling pathways in human breast carcinogenesis. Mol Cancer. 2009;8:128. doi: 10.1186/1476-4598-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weijzen S, Rizzo P, Braid M, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002 Sep;8(9):979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 158.Rizzo P, Miao H, D’Souza G, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008 Jul 1;68(13):5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004 Nov;14(5):779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- 160.Bouras T, Pal B, Vaillant F, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008 Oct 9;3(4):429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 161.Raouf A, Zhao Y, To K, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008 Jul 3;3(1):109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 162.Harrison H, Farnie G, Howell SJ, et al. Regulation of breast cancer stem cell activity by signaling through the Notch4 receptor. Cancer Res. 2010 Jan 15;70(2):709–718. doi: 10.1158/0008-5472.CAN-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Thurston G, Noguera-Troise I, Yancopoulos GD. The Delta paradox: DLL4 blockade leads to more tumour vessels but less tumour growth. Nat Rev Cancer. 2007 May;7(5):327–331. doi: 10.1038/nrc2130. [DOI] [PubMed] [Google Scholar]
- 164.Milano J, McKay J, Dagenais C, et al. Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004 Nov;82(1):341–358. doi: 10.1093/toxsci/kfh254. [DOI] [PubMed] [Google Scholar]
- 165.Barten DM, Meredith JE, Jr., Zaczek R, et al. Gamma-secretase inhibitors for Alzheimer’s disease: balancing efficacy and toxicity. Drugs R D. 2006;7(2):87–97. doi: 10.2165/00126839-200607020-00003. [DOI] [PubMed] [Google Scholar]
- 166.Real PJ, Tosello V, Palomero T, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009 Jan 1;15(1):50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Weggen S, Eriksen JL, Das P, et al. A subset of NSAIDs lower amyloidogenic A beta 42 independently of cyclooxygenase activity. Nature. 2001;414(6860):212–216. doi: 10.1038/35102591. [DOI] [PubMed] [Google Scholar]
- 168.Bergmans BA, De Strooper B. gamma-secretases: from cell biology to therapeutic strategies. Lancet Neurol. 2010 Feb;9(2):215–226. doi: 10.1016/S1474-4422(09)70332-1. [DOI] [PubMed] [Google Scholar]
- 169.Aste-Amezaga M, Zhang N, Lineberger JE, et al. Characterization ofNotch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 2010;5(2):e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li K, Li Y, Wu W, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008 Mar 21;283(12):8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 171.Wu Y, Cain-Hom C, Choy L, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 172.Moellering RE, Cornejo M, Davis TN, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462(7270):182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Duffy MJ, McKiernan E, O’Donovan N, McGowan PM. Role of ADAMs in cancer formation and progression. Clin Cancer Res. 2009;15(4):1140–1144. doi: 10.1158/1078-0432.CCR-08-1585. [DOI] [PubMed] [Google Scholar]
- 174.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 175.Fridman JS, Caulder E, Hansbury M, et al. Selective inhibition of ADAM metalloproteases as a novel approach for modulating ErbB pathways in cancer. Clin Cancer Res. 2007;13(6):1892–1902. doi: 10.1158/1078-0432.CCR-06-2116. [DOI] [PubMed] [Google Scholar]
- 176.Zhou BB, Peyton M, He B, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10(1):39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Witters L, Scherle P, Friedman S, et al. Synergistic inhibition with a dual epidermal growth factor receptor/HER-2/neu tyrosine kinase inhibitor and a disintegrin and metalloprotease inhibitor. Cancer Res. 2008;68(17):7083–7089. doi: 10.1158/0008-5472.CAN-08-0739. [DOI] [PubMed] [Google Scholar]
- 178.Ludwig A, Hundhausen C, Lambert MH, et al. Metalloproteinase inhibitors for the disintegrin-like metalloproteinases ADAM10 and ADAM17 that differentially block constitutive and phorbol ester-inducible shedding of cell surface molecules. Comb Chem High Throughput Screen. 2005;8(2):161–171. doi: 10.2174/1386207053258488. [DOI] [PubMed] [Google Scholar]