Significance
Kawasaki disease (KD), the leading cause of acquired heart disease in children worldwide, has remained a mystery for more than 40 y. No etiological agent has yet been identified. By using simulations with the flexible particle dispersion model from different Japanese cities from each single high (low) KD incidence day, the source region KD is retrieved in cereal croplands in northeastern China. We infer the incubation time for KD ranges from 6 h to 2 d, thus favoring an antigenic or toxic exposure as the trigger. Candida sp. is reported as the dominant fungal species collected aloft (54% of all fungal DNA clones) demonstrating the potential for human disease in aerosols transported by wind currents traveling long distances.
Keywords: northeastern China source, agriculture, heart disease, FLEXPART, cereal croplands
Abstract
Evidence indicates that the densely cultivated region of northeastern China acts as a source for the wind-borne agent of Kawasaki disease (KD). KD is an acute, coronary artery vasculitis of young children, and still a medical mystery after more than 40 y. We used residence times from simulations with the flexible particle dispersion model to pinpoint the source region for KD. Simulations were generated from locations spanning Japan from days with either high or low KD incidence. The postepidemic interval (1987–2010) and the extreme epidemics (1979, 1982, and 1986) pointed to the same source region. Results suggest a very short incubation period (<24 h) from exposure, thus making an infectious agent unlikely. Sampling campaigns over Japan during the KD season detected major differences in the microbiota of the tropospheric aerosols compared with ground aerosols, with the unexpected finding of the Candida species as the dominant fungus from aloft samples (54% of all fungal strains). These results, consistent with the Candida animal model for KD, provide support for the concept and feasibility of a windborne pathogen. A fungal toxin could be pursued as a possible etiologic agent of KD, consistent with an agricultural source, a short incubation time and synchronized outbreaks. Our study suggests that the causative agent of KD is a preformed toxin or environmental agent rather than an organism requiring replication. We propose a new paradigm whereby an idiosyncratic immune response, influenced by host genetics triggered by an environmental exposure carried on winds, results in the clinical syndrome known as acute KD.
After more than four decades of intensive research on Kawasaki disease (KD) (1, 2), no agreement regarding its cause has yet emerged (3). In fact, it is not even clear whether KD should be considered an infectious disease or an abnormal host response in genetically susceptible children to one or more noninfectious environmental triggers (4). Although progress has been made in defining the genetic influence on KD susceptibility and disease outcome, the complex genetic variants that are responsible have only partially been identified. Based on both the clinical presentation and histopathology of tissues, it has been postulated that the trigger enters through the mucosa of the upper respiratory tract (5). A recent study demonstrated a role for both regional winds and large-scale atmospheric circulation in transporting an etiologic agent responsible for the seasonal and nonseasonal anomalous variation in the number of KD patients in both Japan and the United States (6). Certain patterns of winds in the troposphere above the earth’s surface flowing from Asia were associated with the times of the annual peak in KD cases and with days having anomalously high numbers of KD patients. It was demonstrated that these regional wind patterns first impact Japan and, when large-scale conditions are favorable, extend across the North Pacific to the West Coast of the United States (6, 7). In advance of peak KD occurrences, a regional pattern with decreased atmospheric pressure forms over Japan that pumps winds to the archipelago (Fig. S1). Further, this same study (7) demonstrated capabilities of predicting anomalous KD activity in Japan and in the West Coast of the United States with time leads of a few months, based on coupled ocean and atmosphere processes developing in regions of the tropical and north Pacific oceans. However, the nature of the etiologic agent, the source region from which the putative KD agent is lofted into the atmosphere, and the incubation time between exposure and onset of fever remained unknown. Previous studies of KD dynamics suggested a potential source region in northeastern Asia (6, 7). In the present study, we accurately define this region, with the sharpened temporal resolution afforded by the best record of KD available worldwide, that of daily case occurrences in individual prefectures in Japan. The exposure mechanism and incubation time are addressed using two different approaches: an air parcel trajectory analysis of the time taken by wind to travel from the inferred source region and an analysis of the population dynamics of the disease using a simple ordinary differential equation model. Finally, toward a detailed understanding of KD etiology, we also investigate possible KD agent(s) by means of direct airborne sampling conducted over Japan, followed by a detailed analysis of nucleic acids extracted from the aerosolized atmospheric samples trapped on filters collected at selected altitudes during the 2011 KD season.
Results
Inference of the Source Region.
A core element in addressing these questions is the high-resolution daily KD case record for the 47 Japanese prefectures (8). The Japanese Epidemiologic Surveillance System for KD provided the KD onset dates for all cases during the interval 1970–2010 (Data and Methods; see also the clinical definition of KD cases in ref. 6). Dates contained solely within the three KD epidemics in Japan in 1979, 1982 and 1986 were selected for the first analysis, to capitalize on the larger signal-to-noise ratios due to large bursts of KD cases that occurred during those epidemics. For each epidemic, days having a number of cases at least equal to the 95% value of the distribution of KD cases within that epidemic were selected as peak (p95) days. Similarly, for assessing possible differences with the rising phase in each epidemic (Data and Methods), days above the 70th percentile of KD cases within each epidemic that were also separated in time at least 12 d before the identified sequence of p95 d, were selected as rising (p70) KD days. We then generated ensembles of backward (in time) air parcel trajectories with the flexible particle dispersion model (FLEXPART) Version 8.23 (9) at a sequence of 3-h intervals before each of these peak and rising dates. An ensemble of such trajectories was generated for each of a number of selected locations in Japan (Data and Methods). Each backward trajectory traced the geographical and vertical location of an air parcel in its upstream transit 10 d before it crossed over the selected location. Within the air parcel trajectories associated with peak KD events, we postulated that there is a common precursory source region where the air preferentially resided (higher-residence time regions) that should show up in the analyses. It is in these high-residence time regions that there is the greatest chance that the potential agent would be lofted from the land surface into the air mass; this air parcel would then be subsequently borne via the wind currents to Japan. Therefore, we searched the set of peak KD-associated back trajectories to identify common upstream regions having maximal residence times within the swarm of trajectories at all times in a season when both abnormal and extreme numbers of KD cases occurred in Japan.
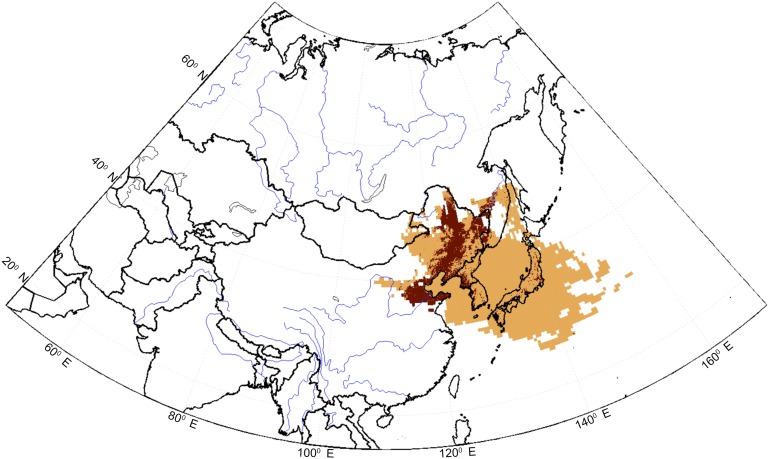
Fig. 1 displays the largest residence times over land (>95%) inferred from simulations for the composite average of all peak days within the three epidemics for Tokyo, Japan’s most populated city. Potential source areas associated with epidemic peak events (orange areas in Fig. 1 denote grid points with largest residence times; see SI Data and Methods for details) reproducibly pointed to the same constrained region, between northeastern China and southwestern Russia, as the most likely source region for the KD agent. The remarkable stability in the location of the overlapping region obtained at all lag times analyzed strongly supports the hypothesis that the northeastern winds pick up the etiological agent of epidemic KD in that region. Furthermore, when simulations were performed including all peak days calculated from the period excluding the epidemic years (hereafter p95 for the whole 1987–2010 interval), the same area over land with maximum residence times was identified, indicating the historical stability of the inferred results (Fig. S2). Both the high resolution and calibration of the reanalysis used to run the FLEXPART model, the stability of the region obtained when using different extreme percentiles for KD (ranging from 1% to 5%), and the fact that the simulations are obtained in a hindcast mode, adds strong confidence to the inferred region. Because epidemics occurred in different months (March to May in 1979, May 1982, and March 1986) (6), the comparison was made between the residence times of the upstream wind trajectories associated with epidemic peaks and others similarly derived from trajectories for nonepidemic peaks, the latter therefore outside of the epidemic months but contained within the winter calendar months (October–March, Fig. S2). Confining the sampling to these calendar months and comparing the two situations effectively removed the potentially confounding effects of seasonality in air mass exposure from either the same or different months.
Fig. 1.
Upstream grid cell locations registering residence times over 30 s for the ensemble of FLEXPART 10-d backward simulations (light brown) for dates within the three epidemics (1979, 1982, and 1986) when KD cases were at or above the 95% threshold of cases (threshold calculated for the entire timespan, 1977–2010). The ensemble represents a total of 257 dates. A 0.5° grid scale was used (latitude, longitude). Brown dots denote crops according to the land cover type yearly climate modeling grid (CMG) datasets with 0.05° resolution from the NASA Land Processes Distributed Active Archive Center (LP DAAC, Sioux Falls, SD), ASTER L1B (32). Grid cells with dots have at least 50% or more subgrids as crops or 100% subgrids as mosaic (mosaic representing crops + natural vegetation).
In an attempt to assess whether there were differences in regions where the air resided preferentially during the initial stages of the epidemics (rising, green in Fig. S3A) in comparison with those at the peak of the epidemic (peak, blue in Fig. S3A), we compared residence times from ensembles of similar backward simulations for the two situations. Results therein clearly indicate that the southernmost regions in NE and East China, from Fig. 1, also contribute during the rising stages compared with the more northern areas in northeastern China, which dominate during the epidemic peaks indicating that most of the etiologic agent load must come from this region. When the temporal difference between the two situations, rising and peak phases are maximized (e.g., for ensembles at lag 15 d), the source regions strongly differ further clarifying this separation (Fig. S3B). The large number of simulated days involved provides consistency to these results (902 in total, corresponding to KD days in Fig. 1 and in Figs. S2–S4).
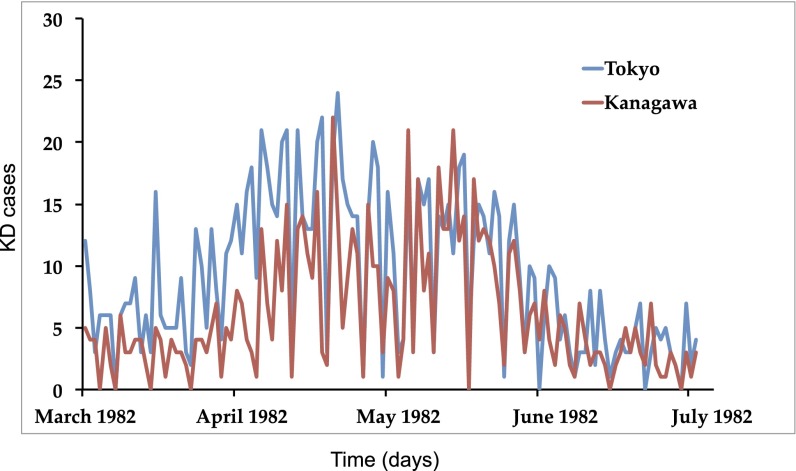
Following the assertion that the KD agent is carried by the wind, it is expected that cases would occur simultaneously in neighboring locations because wind patterns operate over regional scales. In fact, for the largest KD epidemics shown in Fig. 2 (1982; 1986 in Fig. S4), a striking covariation was seen in the daily number of cases in the two major cities in Japan (Tokyo and Yokohama, 28 km apart). Such covariation between the anomalous KD occurrences in these same two prefectures and between Tokyo and Saitama also during the nonepidemic period from 1987 to 2010 is shown in Fig. S5. Delving further, a scale-dependent correlation (SDC) (10, 11) analysis was used to investigate possible commonalities in KD behavior over short time scales (only a few days) among the most populated cities of the Tokyo metropolitan area (Greater Tokyo, comprising Tokyo, Yokohama, Saitama, and Chiba; SI Data and Methods and Table S1). SDC analysis revealed surprisingly high in-phase correlations during epidemics occurring in the same year (see also Table S1). Results demonstrate that maximum numbers of KD cases occurred in these two cities on exactly the same days [Monte Carlo randomization test (MCR), P < 0.001]. Applying the same analysis to all major cities in the Tokyo metropolitan area resulted in similar temporal coherence. Table S1 reports the results of two-way SDC analyses among Tokyo, Kanagawa, Saitama, and Chiba, showing that in all city pairwise comparisons, maximum correlations consistently occurred at zero lag times, attaining correlation values between 0.56 and 0.86 (MCR, 0.04 < P < 0.000). The total variability accounted for by the daily covariation among cases between cities is indeed remarkable (on average above 50%, in a range between 31% and 75%), as it represents a portion of variability not fully resolved in the seasonal–interannual scales addressed in previous studies (6, 7). The present analysis reveals that the two cities had their KD cases fully synchronized for a period of at least 3 wk during the 1982 epidemic. Furthermore, results from an SDC analysis of wind and Greater Tokyo KD fluctuations attained values above 0.6 (MCR, P < 0.05) during the same intervals and with maxima around lags of 2–3 d, indicating that this is the approximate time between the agent leaving the source, traveling for 2 d, arriving in Japan, and rapidly causing the onset of fever in a KD patient. Previous studies of KD examined temporal scales ranging from seasonality to the interannual variability (6, 7), whereas here we focus on the daily variation. In the present analysis, fluctuations in daily KD cases are shown to be coherently associated with regional winds and their previous upstream history, an unprecedented result even compared with other well-known infectious diseases (12). The fact that exactly the same dates are associated with maximal numbers of KD cases in different locations and that the peak in KD cases is rapid after the arrival of the wind suggests that there is a short incubation time for KD.
Fig. 2.
Daily time series of KD date of onset for patients in Tokyo (blue) and Kanagawa (red) during the epidemics of 1982 (see Fig. S4 for 1986). Axes display cases (Y) and day since epidemic onset (X). See SDC (11) analysis between the KD datasets for Tokyo and Kanagawa, as well as for all cities in the Greater Tokyo Area in Data and Methods and Table S1. Maximum overall correlation is attained in this 1982 epidemic, with over 75% of the total KD variability synchronized between Tokyo and Kanagawa during at least 3 wk.
Incubation Time for KD.
To obtain an independent estimate of the incubation time (here time between exposure to the presumed agent and the onset of fever) from that suggested by the SDC analysis, an experiment was conducted using the ensemble of 10-d backward FLEXPART simulations for the 532 d cataloged as high KD occurrences (Movie S1). For comparison, we ran a second experiment with the ensemble of FLEXPART simulations for all 3,206 d with low KD occurrences (having 0 to 1 cases per day) in the postepidemic interval (1987–2010; Fig. S6A). As for Movie S1, average residence times for simulations shown in Fig. S6B mark the time between the moment a particle leaves the source area and the onset of fever in patients to be in the range of 6 h to 2.5 d (see SI Data and Methods for details). This value matches the predicted incubation time inferred from the SDC analysis above to likely be less than 1 d, beyond the 2-d time necessary for the wind to traverse the distance from northeastern China to Japan. Although the confidence interval extends this incubation time to around 2 d more, this longer duration is less likely, and therefore these results imply that there is rapid host response following exposure to the KD agent. Similar results were obtained when days were selected for either Tokyo or the composite of cities in the entire Greater Tokyo Area (Tokyo, Kanagawa, Saitama, and Chiba, which includes over 35 million people).
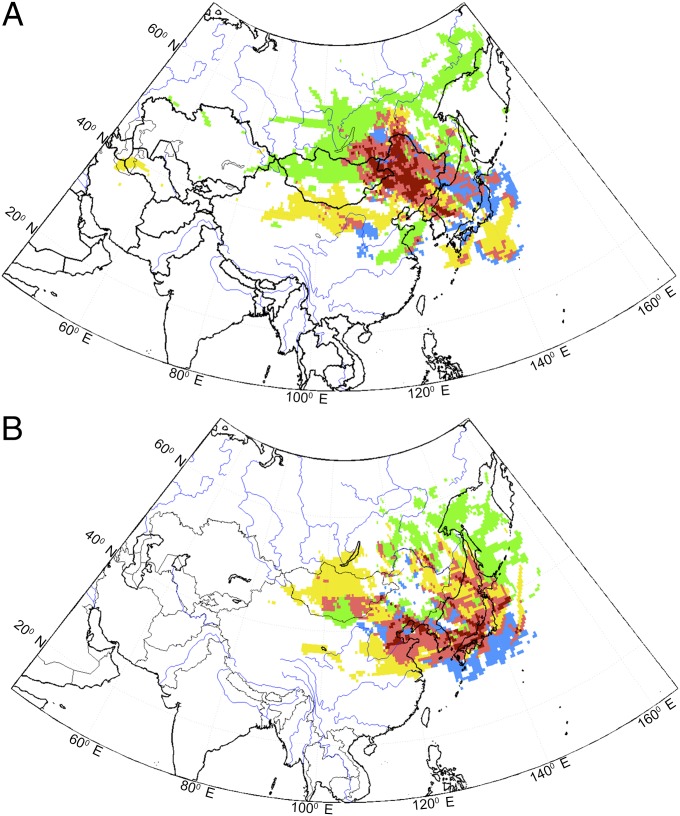
Apart from the three main KD epidemics in Japan, there are seasonal peaks and troughs in disease occurrence (8). We therefore investigated coincidence in potential source regions for KD during periods out of epidemics, and conducted backward trajectory analyses also for high KD activity after the last big epidemic (in the postepidemic interval, 1987–2010). In this case, dates were selected with individual days displaying high (≥95%) numbers of KD cases (around 2 SDs above the mean of the normalized distribution) in locations spanning Japan from north to south (Sapporo, Tokyo, and Nagoya). We used data for the entire 1970–2010 records (epidemic values were included also but only used to set the threshold for the selection of high KD days) and compared with days when KD occurrences were unusually low (days displaying zero KD cases having no high KD event in the preceding 10 d; SI Data and Methods). To properly characterize the time lag between an air parcel leaving the presumed source region and having an influence on KD in the selected location, the air parcel crossing times were varied from the same day to 1 mo in advance of the date associated with the peak in KD cases. We allowed such a variety of lag times to give equal chance to any value in the range of the suspected lag times for the potential KD incubation time, both according to previous hypotheses and known times for airway diseases (13–16). Average residence times were therefore computed for high and low KD dates and areas with residence times greater than the threshold for a high-residence time of 30 s in the preliminary high–low maps retained for further analyses (this threshold corresponds to 95% of the average simulated residence time; Data and Methods). Fig. 3A shows the average residence time map composited for days 1–3 to conform to results obtained above from the trajectory mapping and the SDC analysis of the KD epidemics. Furthermore, we sought for residence times maxima within the nonepidemic trajectories between 1 d and 1 mo, but, similar to the epidemic trajectory results, maximum residence times were centered around 1–2 d. Residence times were retrieved at each grid point for each of the three Japanese locations, namely Sapporo (green), Tokyo (blue), and Nagoya (yellow). Red areas in Fig. 3A indicate the overlap between plumes of ensemble FLEXPART simulations for two cities (light) or three cities (dark). These areas largely coincide with the ones identified in Fig. 1 and Fig. S2 in northeastern China that were associated with the source for the three KD epidemics in Japan. Remarkably, other smaller areas highlighted by the same analysis were associated with positive high–low residence times at lag times from 10 to 30 d but were instead residual and not coherently positioned in the spatial analysis when percentiles and intervals for data selection were varied. Therefore, striking similarities exist between Figs. 1 and 3 that identify the same region as being linked to high KD case numbers in and out of epidemic periods.
Fig. 3.
Areas of (A) high–low or (B) low–high differences in the residence time between sets of cases with high KD and low KD occurrences in the postepidemic interval. Residence time differences higher than 30 s are shown for Tokyo (blue), Sapporo (green), and Nagoya (yellow). The ensembles included have been generated averaging residence time data corresponding to lags of 1–3 d, which appears to capture all of the potential range of values for the KD incubation time. The total number of dates used in the simulations was 1,344. Red areas indicate overlapping high-residence time differences from two cities (light) or three cities (dark) residence time anomaly areas.
As a comparison, a similar analysis to determine upstream regions with the high-residence time from trajectories associated with anomalously low numbers of KD cases among the three cities was conducted. In contrast to the preferred upstream regions associated with peak KD events, the high-residence time regions associated with low KD incidence appear to lie mostly in the sea between China and Japan (Fig. 3B). Importantly, this observation demonstrates that there are other preferential air mass sources that supply wind flow when KD occurrence is low in Japan, and reinforces our conclusion that there is a KD source region over northeastern China. Also, any local influence on KD is at most small compared with the northeastern China region. This result underscores the need to identify possible KD agent(s) in land regions and appears to eliminate the possibility of a sea-related origin.
A surprisingly short interval between exposure and onset of fever is indicated by the trajectory analysis. In searching for candidate KD agents, such a short interval makes many infectious agents unlikely, which leads us to consider that the trigger may be a plant or microbial toxin or an inhaled antigen that would, therefore, not replicate inside the host. Under this model, a dose-dependent reaction may account for variance in disease severity among patients. As an alternative hypothesis, we explored the relationship of daily time series of atmospheric pollutants and pollen species with KD cases in Tokyo. Time series for SO2, OX, NO, NO2, NOX, CO, and NMHC from three stations in the Tokyo Metropolitan area and pollen counts measured in Tokyo were compared with local wind and KD records, but no consistent relationship emerged. Neither seasonality nor trends in emissions of atmospheric mercury were consistent either with fluctuations in KD case occurrences (SI Data and Methods).
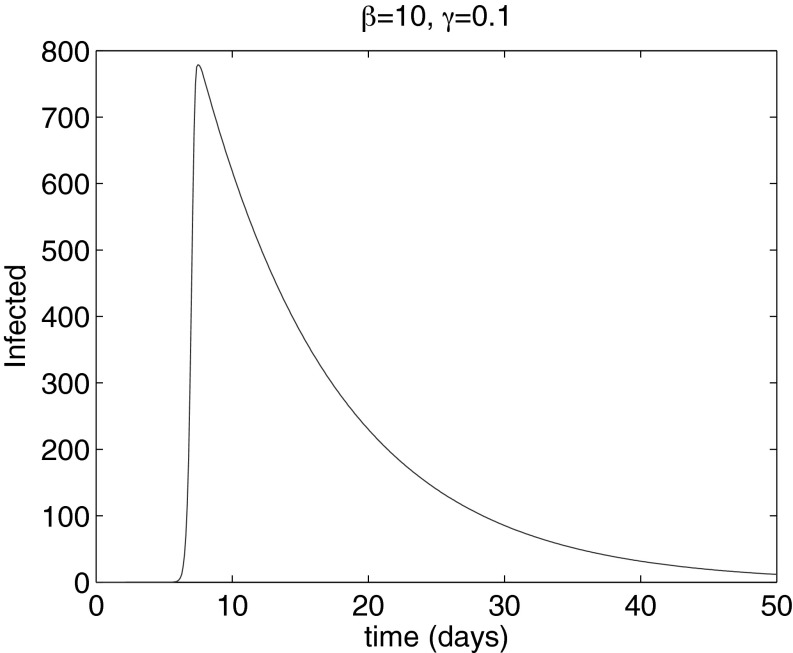
To investigate whether person-to-person spread of an infectious agent could fit the observed data, a simple susceptible–exposed–infected–recovered (SEIR) population disease model was built (SI Data and Methods). Results show that not even an extremely rapidly replicating infectious disease could propagate that quickly (even diseases with idealized incubation times of only 2 h, e.g., 10 times shorter than the fastest respiratory viruses known, e.g., influenza B and rhinovirus) (15, 16). No known infectious agent would be able to produce synchronous (same day) infections over distances between cities, such as those in the Tokyo metropolitan area on the basis of secondary infections only (SI Data and Methods and Fig. S7). In fact, at least 5.6 d would be needed for an infectious agent with only a 2-h incubation time and with the current city populations in the Greater Tokyo metropolitan area to cover the distance between any two cities using only secondary infections, even if there were up to 1% asymptomatic individuals in the population (Fig. 4). In this experiment, children younger than 6 mo were considered not susceptible due to protection from maternal antibodies (17). Indeed, the rarity of KD in very young infants suggests that there may be the acquisition of a transplacental antibody that is protective (18). The possibility that a known infectious agent is responsible for these dynamics is therefore remote. Instead, an immediate response in the form of an idiosyncratic immune reaction in genetically susceptible children that takes place within 24 h after inhalation of the etiologic trigger is further reinforced by our results.
Fig. 4.
Epidemic evolution 30 km from the initial site of infection, for β = 10 d−1 and γ = 0.1 d−1. The maximum number of infected denotes the time at which the disease reaches the next town (on average after 5.66 d).
Tropospheric Microbiome Campaign.
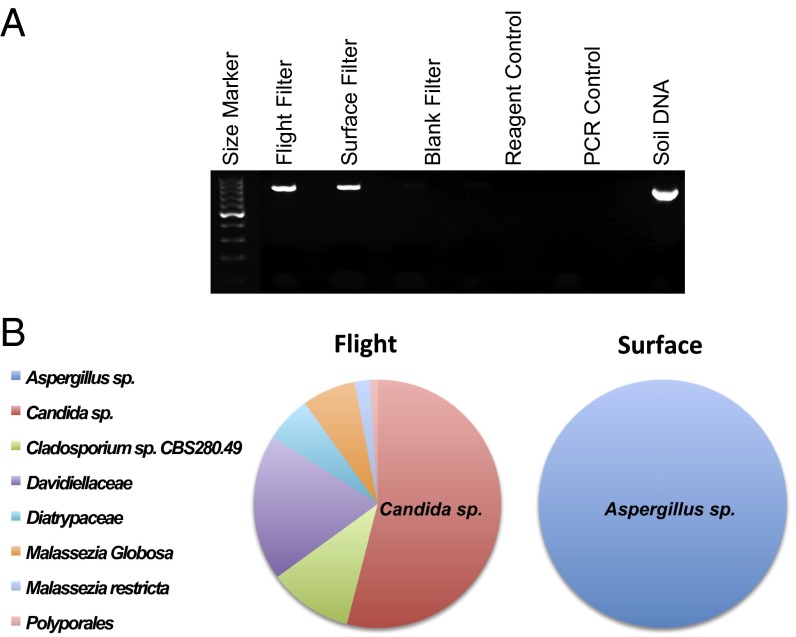
To investigate the microbial diversity of aerosols carried by these tropospheric winds, an air sampling campaign was conducted during a period of high KD activity in the first week of March 2011 over Japan, and aerosol samples were collected at heights between 2,000 and 3,000 m only. The research aircraft flew in a northwesterly direction into the winds toward northeastern China (see Fig. S8 and SI Data and Methods for an explanation of the aircraft campaigns conditions). Flight direction was designed to optimally filter air far from any surface contamination and on selected days when the air came exclusively from the region suspected as a source for the KD agent (compare Fig. 1 with Fig. S8). Major differences in the mycobiome of the tropospheric aerosols were detected compared with ground aerosols with the intriguing finding of Candida species as the dominant fungus aloft (Fig. 5). Filters collected on the same day of the flight included the collection of aerosols at ground level (Fig. 5 A and B, surface filter) and a negative control filter placed in the air sampling device and removed without filtering air (Fig. 5A, blank filter). Quartz filters were handled in a sterile manner to avoid contamination with extraneous nucleic acid and shipped on dry ice for analysis. Methods for profiling microbes were optimized using nucleic acids extracted from the filters. Approximately 2 ng of DNA were obtained from one-fourth of a filter and used as template in 16S rRNA and 18S rRNA gene-specific PCR amplification to identify bacterial and fungal signatures, respectively. The 16S rRNA products were detected only inconsistently on flight and surface filters. In contrast, 18S rRNA gene products were amplified in both the flight filter sample (Fig. 5 A and B, flight filter) and the surface filter sample, whereas the unsampled filter (blank filter), extraction reagents, and PCR controls were negative (Fig. 5A). Differences in the consistency of bacterial vs. fungal signal in these assays may be a consequence of differences in the size of fungal spores (1–40 μm) vs. bacterial cells (1–5 μm) that influences the distribution of fungi and bacteria in the troposphere or entrapment in the filter matrix.
Fig. 5.
Differences in the mycobiome distribution from tropospheric and surface-level aerosols. (A) Fungal 18S rRNA gene PCR demonstrates amplification products in flight and surface filters. DNA extracted from soil was used as a positive control for fungal amplification. (B) Pie charts demonstrating the percent abundance of fungal taxa identified from clone library sequencing (100 clones per filter) of 18S rRNA gene amplification products from flight and surface filters. Candida sp. sequences accounted for 54% of total flight filter sequences, whereas Aspergillus sp. sequences accounted for 100% of surface filter sequences.
Large differences in the distribution of fungi amplified with primers for the 18s rRNA gene were obtained from the surface and flight filters based on the sequencing of 100 clones containing PCR product from each filter. Whereas Candida species sequences were abundant in the flight filter sample, accounting for 54% of all fungal sequences, only Aspergillus species sequences were identified from the surface filter sample (Fig. 5B). Although the 723-nt Candida sequence matched several different species with only a single mismatch and 99% identity, including Candida tropicalis, we were unable to pursue further species discrimination due to the limited yield of nucleic acid from the filter. Despite the abundance of marine and agricultural soil Candida species,† to our knowledge, Candida has not been previously reported from surveys of microbes collected from aircraft sampling over Asia, the Mediterranean, or the Caribbean (19, 20). The presence of Candida sequences is intriguing in light of murine models of coronary arteritis based on exposure to Candida glucans comprising mannose, β-1,3-glucan and β-1,6-glucan. Vulnerability to disease varies by mouse strain and is associated with elevated levels of IL-6, TNF-α, and IFN-γ (21, 22). Similar to KD, vasculitis can be abrogated by use of i.v. Ig (23) and immunosuppressive drugs including the imidazole nucleoside mizoribine (24) and the TNF-α antagonist etanercept (25). However, the finding of Candida sp. aloft can only be reported here as providing support for the concept and feasibility of a windborne pathogen rather than implicating any particular organism.
Discussion
A closer inspection of the geographic area identified by the FLEXPART air parcel trajectory residence times from both nonepidemic intervals and large epidemics of KD in Japan reveals a strikingly uniform source landscape with intensively cultivated croplands and farms. Cropland coverage is denoted by dark dots in Fig. 1 (also Figs. S2 and S3). In contrast to the preferred upstream regions associated with peak KD events, the high-residence time regions associated with low KD incidence appear to lie mostly in the sea between China and Japan (Fig. 3B). The principle crops in the region uncovered include cereals such as corn, rice, and spring wheat in a region known to be the main grain area of China (26) [Fig. 1, with brown dots in denoting at least 50% total coverage by crops according to the land cover type yearly climate modeling grid (CMG) datasets with 0.05° resolution from the NASA Land Processes Distributed Active Archive Center (LP DAAC).]. Among other crops, the Heilongjiang province is the principal corn-growing region and accounts for over 29% of total Chinese corn grain production (27). Given the clear link to croplands/agricultural soils indicated by a variety of our results at a time and in a region where the ground is frozen throughout the high KD season, a whole range of new possibilities emerges regarding the nature of the KD etiological agent. Possibilities include environmental toxins linked to crops or to plant decay by-products (e.g., bacterial/fungal toxins), as well as a combination of the former with agricultural practices, atmospheric chemistry, and an idiosyncratic immune reaction in genetically susceptible children (28, 29). The lack of an industrial signature in the inferred region and the lack of correlation with pollutant levels in the winds associated with increased KD activity argue against a chemical pollutant as the relevant causative agent. A new calculation of the impact of KD in northeastern China when the population density is taken into account might also point to an underestimation of the impact of this disease in the region (SI Data and Methods).
Many crop species harbor fungal spores and inhalation of spore-associated mycotoxins is a common route of exposure. Fungal toxins (mycotoxins) are a diverse group of molecules that are difficult to classify because they are synthesized through diverse biochemical pathways and cause a diverse array of symptoms in humans with the liver, kidneys, and brain being prominent targets. What mycotoxins share is a rapid onset of symptoms following exposure (30). Some notable examples of crop-associated mycotoxins are the aflatoxins, difuranocoumarin derivatives synthesized by a polyketide pathway by many strains of Aspergillus, the citrinins produced by Penicillium and Aspergillus species, the fumonisins produced by Fusarium species, and the ochratoxins produced by various Aspergillus species. None of the known mycotoxins, however, produce a vasculitis similar to the clinical picture in KD.
Results in this study on source regions and incubation times, as well as detailed analysis of nucleic acids in atmospheric samples provide important clues regarding both the potential source regions and the nature of the etiologic agent for KD in Japan. Avenues for future research should include detailed characterization of the organic and inorganic compounds and microbiome of tropospheric winds collected aloft over northeastern China and Japan during the annual peak in disease activity in Japan. These air sampling campaigns will need to be further expanded in the future in close connection with a simultaneous detailed surveillance of the occurrence of KD in Japan to further explore the role that microorganisms or their toxins may play in triggering KD. Study of the transport and circulation patterns for air masses before peaks and troughs in KD activity in other regions across the globe with substantial numbers of KD cases may further inform on their sources and the role that tropospheric winds and their chemistry play in the transport of the KD agent. Finally, future studies will be needed to directly test the capacity for microbes, antigens, or toxins contained in those aerosol samples to elicit a humoral and cellular immune response in KD patients.
Data and Methods
KD Datasets.
Approved written consent was obtained from all participants. Human population epidemiology studies were approved by the Bioethics Committee for Epidemiologic Research, Jichi Medical University. Data used in this study for Japan are total daily counts of KD patients admitted to hospitals for each of the 47 prefectures of Japan. To account for possible effects of different population sizes in cities, incidence was also calculated for comparison in the case of major Japanese cities from population censuses interpolated from 10-y national demographic records and only small differences emerged with results obtained for cases. KD cases were then weighed against the total pediatric population in that prefecture, with data covering the interval 1977–2010 (see KD case ascertainment protocol for Japan sites) (6). KD dates used in this study were in all case onset dates. The Japanese datasets were derived from 21 separate questionnaire surveys of hospitals in Japan spanning the period 1970–2010. For subjects with multiple admissions, only the first hospitalization date was used.
Backtrajectory Ensemble Simulations.
Trajectories traced back in time 10 d for each dataset and location were generated using the FLEXPART Lagrangian particle dispersion model Version 8.23 in backward mode. FLEXPART, a Lagrangian model suitable for the simulation of a large range of atmospheric transport processes (9), was originally designed for calculating the long-range and mesoscale dispersion of air pollutants from point sources but has evolved into a comprehensive tool for atmospheric transport modeling and analysis. FLEXPART can be run in backward or forward mode. FLEXPART uses 3D atmospheric wind data, in this case supplied from the European Centre for Medium-Range Weather Forecasts (ECMWF) Re-Analysis (ERA) 40 (31) for dates until June 1989 and after that to December 2010 from the ECMWF global ERA-Interim reanalysis at 1° resolution, 60 vertical levels and time resolution of 3 h. The particles modeled were air tracers, with 10,000 particles used on each model run. Residence time is the collective amount of time that a particular area upstream of, or including, the selected location was overlain by any of the air parcels from the trajectories in the sample set. Model output used was residence time, with an output grid of 0.5° latitude × longitude and a time resolution of 3 h (SI Data and Methods for further description).
Supplementary Material
Acknowledgments
We thank Emiliano Gelati for the data and preliminary maps of cropland areas from MODIS. We especially thank the Institut Català de Ciències del Clima Foundation and its administration for its flexibility in the use of research funds for projects of exceptional interest. Thanks are also given to the anonymous reviewers for their insightful comments. This study was funded by Kawasaki Disease: Disentangling the Role of Climate in the Outbreaks Project 081910 from La Marató de TV3 Foundation 2008 (through a grant to X.R.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
†Kang H-Y, Ryu Y-W, Xylitol production by flocculating yeast, Candida sp. HY200. 25th Symposium on Biotechnology for fuels and chemicals, National Renewable Energy Laboratory, May 4–7, 2003, Breckenridge, CO, poster presentation 2-23.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400380111/-/DCSupplemental.
References
- 1.Kawasaki T, Kosaki F, Okawa S, Shigematsu I, Yanagawa H. A new infantile acute febrile mucocutaneous lymph node syndrome (MLNS) prevailing in Japan. Pediatrics. 1974;54(3):271–276. [PubMed] [Google Scholar]
- 2.Rowley AH. Kawasaki disease: Novel insights into etiology and genetic susceptibility. Annu Rev Med. 2011;62:69–77. doi: 10.1146/annurev-med-042409-151944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns JC. Finding Kawasaki disease. Ann Pediatr Cardiol. 2012;5(2):133–134. [PMC free article] [PubMed] [Google Scholar]
- 4.Cimaz R, Falcini F. An update on Kawasaki disease. Autoimmun Rev. 2003;2(5):258–263. doi: 10.1016/s1568-9972(03)00032-6. [DOI] [PubMed] [Google Scholar]
- 5.Rowley AH, et al. IgA plasma cell infiltration of proximal respiratory tract, pancreas, kidney, and coronary artery in acute Kawasaki disease. J Infect Dis. 2000;182(4):1183–1191. doi: 10.1086/315832. [DOI] [PubMed] [Google Scholar]
- 6.Rodó X, et al. Association of Kawasaki disease with tropospheric wind patterns. Sci Rep. 2011;1:152. doi: 10.1038/srep00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ballester J, et al. Kawasaki disease and El Niño-Southern Oscillation. Geophys Res Lett. 2013;40(10):2284–2289. [Google Scholar]
- 8.Nakamura Y, et al. Epidemiologic features of Kawasaki disease in Japan: Results of the 2009-2010 nationwide survey. J Epidemiol. 2012;22(3):216–221. doi: 10.2188/jea.JE20110126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stohl A, Forster C, Frank A, Seibert P, Wotawa G. Technical note: The Lagrangian particle dispersion model FLEXPART version 6.2. Atmos Chem Phys. 2005;5:2461–2474. [Google Scholar]
- 10.Rodó X. Reversal of three global atmospheric fields linking changes in SST anomalies in the Pacific, Atlantic and Indian oceans at tropical latitudes and midlatitudes. Clim Dyn. 2001;18:203–217. [Google Scholar]
- 11.Rodríguez-Arias MA, Rodó X. A primer on the study of transitory dynamics in ecological series using the scale-dependent correlation analysis. Oecologia. 2004;138(4):485–504. doi: 10.1007/s00442-003-1464-4. [DOI] [PubMed] [Google Scholar]
- 12.Koelle K, Rodó X, Pascual M, Yunus M, Mostafa G. Refractory periods and climate forcing in cholera dynamics. Nature. 2005;436(7051):696–700. doi: 10.1038/nature03820. [DOI] [PubMed] [Google Scholar]
- 13.Fujita Y, et al. Kawasaki disease in families. Pediatrics. 1989;84(4):666–669. [PubMed] [Google Scholar]
- 14.Rowley AH, Baker SC, Orenstein JM, Shulman ST. Searching for the cause of Kawasaki disease—cytoplasmic inclusion bodies provide new insight. Nat Rev Microbiol. 2008;6(5):394–401. doi: 10.1038/nrmicro1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lessler J, et al. Incubation periods of acute respiratory viral infections: A systematic review. Lancet Infect Dis. 2009;9(5):291–300. doi: 10.1016/S1473-3099(09)70069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reich NG, Perl TM, Cummings DA, Lessler J. Visualizing clinical evidence: Citation networks for the incubation periods of respiratory viral infections. PLoS ONE. 2011;6(4):e19496. doi: 10.1371/journal.pone.0019496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura Y, Yoshinaga M, Masuda K, Takei S, Miyata K. Maternal antibody against toxic shock syndrome toxin-1 may protect infants younger than 6 months of age from developing Kawasaki syndrome. J Infect Dis. 2002;185(11):1677–1680. doi: 10.1086/340513. [DOI] [PubMed] [Google Scholar]
- 18.Burns JC, Glodé MP. Kawasaki syndrome. Lancet. 2004;364(9433):533–544. doi: 10.1016/S0140-6736(04)16814-1. [DOI] [PubMed] [Google Scholar]
- 19.Griffin DW, et al. Airborne desert dust and aeromicrobiology over the Turkish Mediterranean coastline. Atmos Environ. 2007;41:4050–4062. [Google Scholar]
- 20.Kakikawa M, Kobayayshi F, Maki T, Yamada M, Higashi T, Chen B, Shi G, Hong C, Tobo Y, Iwasaka Y. Dustborne microorganisms in the atomosphere over an Asian dust source region, Dunhuang. Air Quality Atmos Health. 2008;1:195–202. [Google Scholar]
- 21.Nagi-Miura N, et al. Lethal and severe coronary arteritis in DBA/2 mice induced by fungal pathogen, CAWS, Candida albicans water-soluble fraction. Atherosclerosis. 2006;186(2):310–320. doi: 10.1016/j.atherosclerosis.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 22.Oharaseki T, et al. Susceptibility loci to coronary arteritis in animal model of Kawasaki disease induced with Candida albicans-derived substances. Microbiol Immunol. 2005;49(2):181–189. doi: 10.1111/j.1348-0421.2005.tb03708.x. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi K, et al. Administration of human immunoglobulin suppresses development of murine systemic vasculitis induced with Candida albicans water-soluble fraction: An animal model of Kawasaki disease. Mod Rheumatol. 2010;20(2):160–167. doi: 10.1007/s10165-009-0250-5. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, et al. Mizoribine provides effective treatment of sequential histological change of arteritis and reduction of inflammatory cytokines and chemokines in an animal model of Kawasaki disease. Pediatr Rheumatol Online J. 2011;9(1):30. doi: 10.1186/1546-0096-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi R, et al. Etanercept suppresses arteritis in a murine model of kawasaki disease: A comparative study involving different biological agents. Int J Vasc Med. 2013;2013:543141. doi: 10.1155/2013/543141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Qian C, Deng A, Zhang W. Progressive and active adaptations of cropping system to climate change in Northeast China. Eur J Agron. 2012;38:94–103. [Google Scholar]
- 27.Low D. 2003. Crop Farming in China. PhD dissertation (Swiss Fed Inst Tech, Zurich), No. 15209.
- 28.Kuijpers TW, et al. Kawasaki disease: A maturational defect in immune responsiveness. J Infect Dis. 1999;180(6):1869–1877. doi: 10.1086/315111. [DOI] [PubMed] [Google Scholar]
- 29.Lee K-Y, Han J-W, Lee J-S. Kawasaki disease may be a hyperimmune reaction of genetically susceptible children to variants of normal environmental flora. Med Hypotheses. 2007;69(3):642–651. doi: 10.1016/j.mehy.2006.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev. 2003;16(3):497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kållberg P, Simmons S, Uppala S, Fuentes M. The ERA-40 Archive, ERA-40 Project Report Series No. 17. 2004. (European Centre for Medium-Range Weather Forecasts, Reading, UK) [Google Scholar]
- 32. (2001) Climate Modeling Grid (CMG) datasets. NASA Land Processes Distributed Active Archive Center (LP DAAC), ASTER L1B. US Geological Survey/Earth Resources Observation and Science (EROS) Center. Available at https://lpdaac.usgs.gov/about/citing_lp_daac_and_data. Accessed Spring 2013.