Significance
Plants use the hormone jasmonic acid (JA) to modulate plant:microbe interactions. Disease-causing microbes use proteins to alter host JA signaling to aid their growth in plant tissues. Beneficial symbiotic fungi, which colonize plant tissues and provide essential ecosystem services such as carbon sequestration and plant fertilization, can also alter JA signaling in plant cells to promote colonization. Here, we demonstrate that the MiSSP7 (Mycorrhiza-induced small secreted protein-7) protein of the beneficial fungus Laccaria bicolor interacts with host plant JA signaling repressors and, in contrast to biotrophic pathogens, promotes symbiosis by blocking JA action. These results shed new light on how beneficial and pathogenic microbes have evolutionarily diverged in the mechanisms by which they overcome plant defenses.
Keywords: ectomycorrhizal fungus, defense, tree nutrition, host immunity, plant hormone
Abstract
Ectomycorrhizal fungi, such as Laccaria bicolor, support forest growth and sustainability by providing growth-limiting nutrients to their plant host through a mutualistic symbiotic relationship with host roots. We have previously shown that the effector protein MiSSP7 (Mycorrhiza-induced Small Secreted Protein 7) encoded by L. bicolor is necessary for the establishment of symbiosis with host trees, although the mechanistic reasoning behind this role was unknown. We demonstrate here that MiSSP7 interacts with the host protein PtJAZ6, a negative regulator of jasmonic acid (JA)-induced gene regulation in Populus. As with other characterized JASMONATE ZIM-DOMAIN (JAZ) proteins, PtJAZ6 interacts with PtCOI1 in the presence of the JA mimic coronatine, and PtJAZ6 is degraded in plant tissues after JA treatment. The association between MiSSP7 and PtJAZ6 is able to protect PtJAZ6 from this JA-induced degradation. Furthermore, MiSSP7 is able to block—or mitigate—the impact of JA on L. bicolor colonization of host roots. We show that the loss of MiSSP7 production by L. bicolor can be complemented by transgenically varying the transcription of PtJAZ6 or through inhibition of JA-induced gene regulation. We conclude that L. bicolor, in contrast to arbuscular mycorrhizal fungi and biotrophic pathogens, promotes mutualism by blocking JA action through the interaction of MiSSP7 with PtJAZ6.
Plants are constantly confronted by different organisms that seek to colonize their tissues in an effort to gain the nutrients stored therein. Plants use hormones, such as jasmonic acid (JA), to mediate defense signaling during microbial colonization. JA is ubiquitous throughout the plant kingdom and is involved in the control of cell development and cycling, of vegetative growth, and in the mediation of plant defensive responses (1, 2). In Arabidopsis, the active form of JA, JA-Ile, is perceived by CORONATINE-INSENSITIVE 1 (COI1), which then forms a complex with JASMONATE ZIM-DOMAIN (JAZ)-transcriptional regulators and targets them for ubiquitination and degradation (3–5). Because JAZ proteins are negative regulators of the JA signaling network, their degradation leads to transcriptional activation of plant defense networks within affected cells (6–8). Biotrophic pathogens have evolved methods of favoring JA signaling through the use of effector proteins because the outcomes of this hormonal pathway is considered less detrimental to their growth within plant tissues compared with defenses induced by other plant hormones [e.g., salicylic acid (SA)] (9–12). Not all organisms attempting to colonize plant tissues, however, have adverse affects on plant health. In particular, roots of most trees in temperate and boreal forests form a nutrient-acquiring symbiosis with mutualistic ectomycorrhizal (ECM) fungi (13). Trees that form a relationship with ECM fungi generally benefit from these relationships through an increase in growth rate and via elevated tolerance to biotic and abiotic stresses (13, 14). Although ECM fungi are thought to have evolved from wood- and litter-decaying fungal ancestors, their lifestyle more closely reflects that of biotrophic pathogens (15–17). Like colonization of plant tissues by biotrophic pathogens, the colonization of roots by ECM fungi is also disruptive to plant tissues because fungal hyphae use both hydrolytic enzymes and mechanical force to colonize the apoplastic space of the root (18). The invasive ECM hyphae that reside within the host apoplastic space form a hyphal network encasing the epidermal cells, the so-called Hartig net. The formation of the Hartig net is necessary to maximize the benefits of this mutualistic relationship because it is at the fungal:plant cell interface within the Hartig net that the fungus provides different growth-limiting nutrients to the plant (e.g., nitrogen, phosphorus) in exchange for photosynthetically derived sugars. It is interesting to note that formation of the Hartig net, as opposed to colonization of tissues by the hyphae of biotrophic fungi, is inhibited by increased levels of JA (19). How the ECM fungus is able to aggressively colonize plant tissues and not be repulsed by plant defenses controlled by JA and other plant hormones is not well understood.
Because pathogenic organisms favor the use of secreted “effector” proteins to subvert host immunity, a great deal of research has focused on the role of pathogenic effectors and how they are used to modify the signaling of defense pathways controlled by plant hormones (10–12, 20–24). For example, the pathogenic bacteria Pseudomonas syringae and the oomycete Hyaloperonospora arabidopsidis secrete a number of effectors that target either JAZ proteins (10, 11, 23) or ethylene signaling (24), whereas Phytophthora infestans produces different effectors that interfere with SA-related signaling (21, 22). Like pathogenic organisms, the ECM fungus Laccaria bicolor also produces effector proteins called MiSSPs (Mycorrhiza-induced Small Secreted Proteins) (25). The first MiSSP of L. bicolor to be characterized, MiSSP7, was found to enter host cells and localize to the nucleus where it altered host cell transcription. Localization of MiSSP7 in host cell nuclei was found to be essential for the formation of the Hartig net, although the mechanistic reasoning behind this effect was unknown (26). Here we demonstrate that MiSSP7 interacts with the transcriptional repressor protein PtJAZ6 in the nuclei of the host plant Populus trichocarpa, where it protects PtJAZ6 from JA-induced degradation. Furthermore, MiSSP7 is able to counter the negative impacts of JA on fungal colonization of host tissues by repression of JA-induced gene trancription, likely through its interaction with JAZ proteins. Our results further the concept that, like pathogenic organisms, mutualistic fungi use effectors to target plant host hormone pathways to foster fungal colonization.
Results
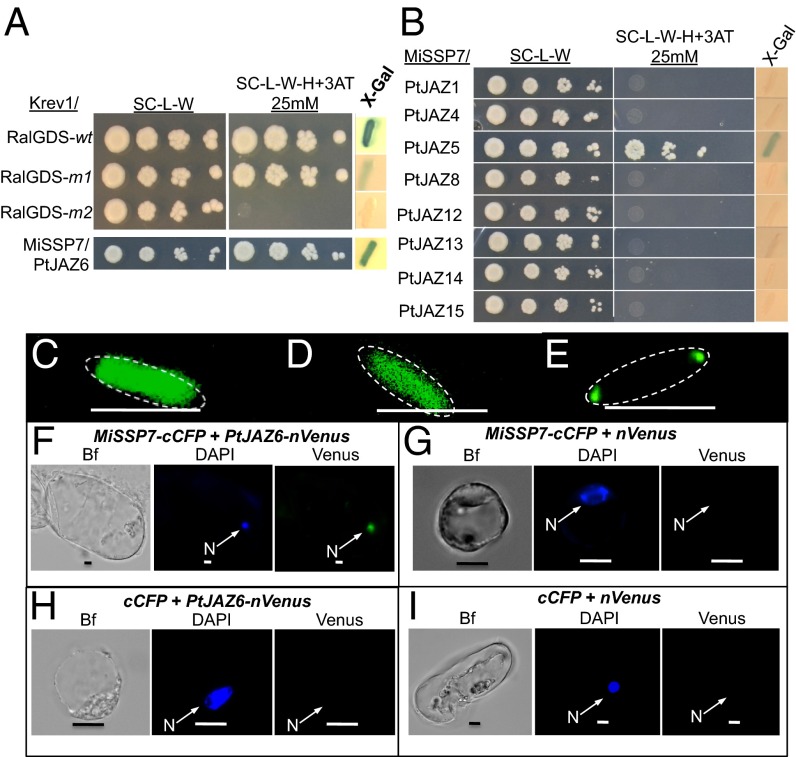
As reported previously, L. bicolor MiSSP7 is able to enter roots cells of its host P. trichocarpa and alter transcription therein (26). Using a yeast reporter assay (27), we tested if the mature version of MiSSP7 (without a signal peptide) could bind to DNA and induce gene transcription. We found that MiSSP7 alone was not able to induce expression of the reporter genes (Fig. S1), demonstrating that the MiSSP7 protein itself does not directly activate the differential gene transcription previously observed (26). Therefore, we used yeast two-hybrid assays (YTH) (27) to determine the protein target of MiSSP7 in poplar root cells. In this particular version of YTH, a positive interaction between two proteins permits growth on medium lacking histidine and the production of β-galactosidase. We screened ∼1.8 × 107 yeast zygotes from a cDNA library of poplar ECM roots and identified one plant protein that interacted with MiSSP7: a JAZ domain containing protein encoded by P. trichocarpa (POPTR_0003s06670) (Fig. 1A). In plant genomes, JAZ proteins form a large multigene family (Fig. S2). The JAZ protein identified as interacting with MiSSP7 shows closest homology to AtJAZ6 in Arabidopsis (Fig. S2); therefore, we will henceforth refer to this protein as PtJAZ6. Although no other JAZ proteins were identified as interacting with MiSSP7 in the general YTH screens, we wished to test if MiSSP7 was able to interact with the other Populus JAZ-domain containing proteins found in the P. trichocarpa gene repertoire (Fig. S2) and expressed in the ectomycorrhizal roots of Populus colonized by L. bicolor (Fig. 1B and Fig. S3A). Of the other JAZ-domain–containing proteins tested, only PtJAZ5 (POPTR_0001s16640), the paralogous gene to PtJAZ6 (Fig. S2), which shares 73% amino acid identity with PtJAZ6, was also found to interact with MiSSP7 (Fig. 1B). Transcription of both PtJAZ6 and PtJAZ5 was found to be significantly regulated by MeJA treatment of poplar roots and, to a lesser extent, during the normal course of L. bicolor colonization of poplar root tips (Fig. S3A).
Fig. 1.
MiSSP7 interacts with PtJAZ6 and PtJAZ5. (A) The control colonies for the ProQuest YTH system are shown. (Upper) Krev1/RalGDS-wt is a strong positive interaction, Krev1/RalGDS-m1 is a weak interacting pair, and Krev1/RalGDS-m2 is a pairing of proteins that show no detectable interaction. (Lower) Growth of yeast colonies because of the interaction between MiSSP7 and PtJAZ6, as demonstrated by the activation of two reporter genes (histidine biosynthesis, β-Gal activity). (B) MiSSP7 also interacts with PtJAZ5 but no other tested root-expressed JAZ protein in YTH system. SC-L-W: synthetic complete medium without both leucine and tryptophan; SC-L-W-H+3AT: synthetic complete medium without leucine, tryptophan, and histidine with the addition of 3-aminotriazol. (C) MiSSP7-GFP expressed in E. coli remains diffused within the cytosol before the induction of PtJAZ6-DivIVa. (D) Coexpression of DivIVa with MiSSP7-GFP exhibits maintenance of the GFP signal in the cytosol. (E) Coexpression of PtJAZ6-DivIVa with MiSSP7-GFP causes relocalization of the GFP signal to the poles of the E. coli, indicating a positive physical interaction between the two proteins. Scale bar (C–E) = 3 μm. (F) PtJAZ6-nVenus interacts with MiSSP7-cCFP in the nuclei of Populus protoplast cells and reconstitutes the Venus fluorescence in a BiFC interaction assay. Nuclear localization was determined by the colocalization of the Venus signal with DAPI fluorescence. (G) MiSSP7-cCFP does not interact with nVenus alone to reconstitute the green fluorescent signal. (H) cCFP and PtJAZ6-nVenus do not interact to reconstitute the green fluorescent signal. (I) cCFP and nVenus alone do not interact to reconstitute the green Venus fluorescent signal. Bf, brightfield image; n = nucleus. (Scale bars, 10 μm.)
Because the interaction between MiSSP7 and PtJAZ6 was the stronger interaction (Fig. 1 A and B), we proceeded with an in-depth characterization of PtJAZ6’s role during the root colonization process by L. bicolor. To further verify this interaction, we used two other model interaction systems: the in vivo DivIVa interaction system used to confirm direct physical interactions between proteins as reported in Edwards et al. (28) (Fig. 1 C–E) and the in planta bimolecular fluorescence complementation method (BiFC) (Fig. 1 F–I). In the former test, we found no relocation of MiSSP7 to the poles of the Escherichia coli in cells expressing only MiSSP7-GFP (i.e., without induction of PtJAZ6-DivIVa) (Fig. 1C), or in cells coexpressing MiSSP7-GFP and the DivIVa protein (Fig. 1D), demonstrating that MiSSP7-GFP does not interact with the DivIVa protein itself. When we expressed MiSSP7-GFP with PtJAZ6-DivIVa, we found that the GFP signal relocated to the poles of the cell thus confirming the physical interaction between these two proteins (Fig. 1E). As a further level of confirmation concerning the MiSSP7:JAZ interaction in a physiologically relevant context, we also used BiFC in poplar cells. We observed fluorescence only in cells coexpressing MiSSP7 with PtJAZ6 (Fig. 1 F–I). This fluorescence was observed in the nuclei of the poplar cells, the localization of both MiSSP7 and JAZ proteins (Fig. S3 B and C). Therefore, through the proof of three separate methods in E. coli, Saccharomyces cerevisiae, and the homologous system Populus, we have demonstrated that MiSSP7 interacts with the JAZ-domain protein PtJAZ6.
AtCOI1 is a receptor of JA and is a known interacting protein of most JAZ proteins in Arabidopsis (4–7). Interaction between AtCOI1 and a JAZ protein in the presence of the active form of JA (i.e., JA-Ile) is necessary for induction of JA responsive genes through the degradation of JAZ proteins (5, 7). Unlike Arabidopsis, Populus encodes two homologs to AtCOI1, which will be denoted here as PtCOI1 (POPTR_0008s06460) and PtCOI2 (POPTR_0010s20030). To determine if the identified PtJAZ6 mirrored the biology of its ortholog AtJAZ6, we performed both a directed YTH and DivIVa interaction assay between PtJAZ6 and either PtCOI1 or PtCOI2 with and without the JA-Ile mimic coronatine (Fig. 2). In both assays only PtCOI1 was found to interact with PtJAZ6 and only in the presence of coronatine (Fig. 2 A–D). Furthermore, the strength of the interaction between PtJAZ6:PtCOI1 was increased in yeast cells grown under higher levels of coronatine as determined by a concomitant increase in the levels of β-galactosidase activity (Fig. 2 E and F). Because coronatine did not affect the growth rate of these cells (Fig. S4A) nor did it promote the interaction between two proteins from a pathway that is unrelated to JA-Ile/coronatine (i.e., negative control for coronatine promotion of protein:protein interactions) (Fig. S4B), these results indicate that, like the Arabidopsis model, PtJAZ6 interacts with PtCOI1 but only in the presence of a JA-Ile mimic. Because JAZ proteins are transcriptional repressors of the JA pathway, we tested the ability of PtJAZ6 to repress the expression of JA marker genes (26). Transgenic poplar roots with increased expression of PtJAZ6 resulted in the repression of gene transcription associated with JA signaling (Fig. S5 and Table S1). Therefore, PtJAZ6 negatively regulates part of the JA pathway in poplar roots.
Fig. 2.
PtJAZ6 interacts with PtCOI1 in the presence of coronatine. (A) The control colonies for the ProQuest YTH system are shown in the upper part of the panel: Krev1/RalGDS-wt is a strong positive interaction, Krev1/RalGDS-m1 is a weak interacting pair, and Krev1/RalGDS-m2 is a pairing of proteins that show no detectable interaction. The test of PtCOI1 and PtCOI2 interaction with PtJAZ6 using the YTH system in the lower section of this panel demonstrates that a positive interaction is only achieved between PtCOI1 and PtJAZ6 when coronatine is present in the growth medium, as indicated by the activation of two reporter genes: histidine biosynthesis and β-Gal activity (blue coloration after treatment with X-gal). (B) PtJAZ6-GFP expressed in E. coli remains diffused within the cytosol before induction of PtCOI1-DivIVa. (C) Coexpression of DivIVa with PtJAZ6-GFP exhibits GFP signal in the cytosol. (D) Coexpression of PtJAZ6-GFP with PtCOI1-DivIVa in medium supplemented with coronatine causes relocalization of the GFP signal to the poles of the E. coli, indicating a positive interaction between the two proteins. Scale bar (B–D) = 3 μm. (E) The strength of the interaction between PtJAZ6 and COI1 in yeast cells improves with increasing quantities of coronatine in the growth medium (as determined by an increase in β-galactosidase activity in the yeast cultures; superscript letters indicate significant differences between treatments as determined by one-way ANOVA followed by a Tukey HSD (honestly significant difference) multiple comparison test (P < 0.05), whereas no amount of coronatine tested induced an interaction between PtCOI2 and PtJAZ6 (F).
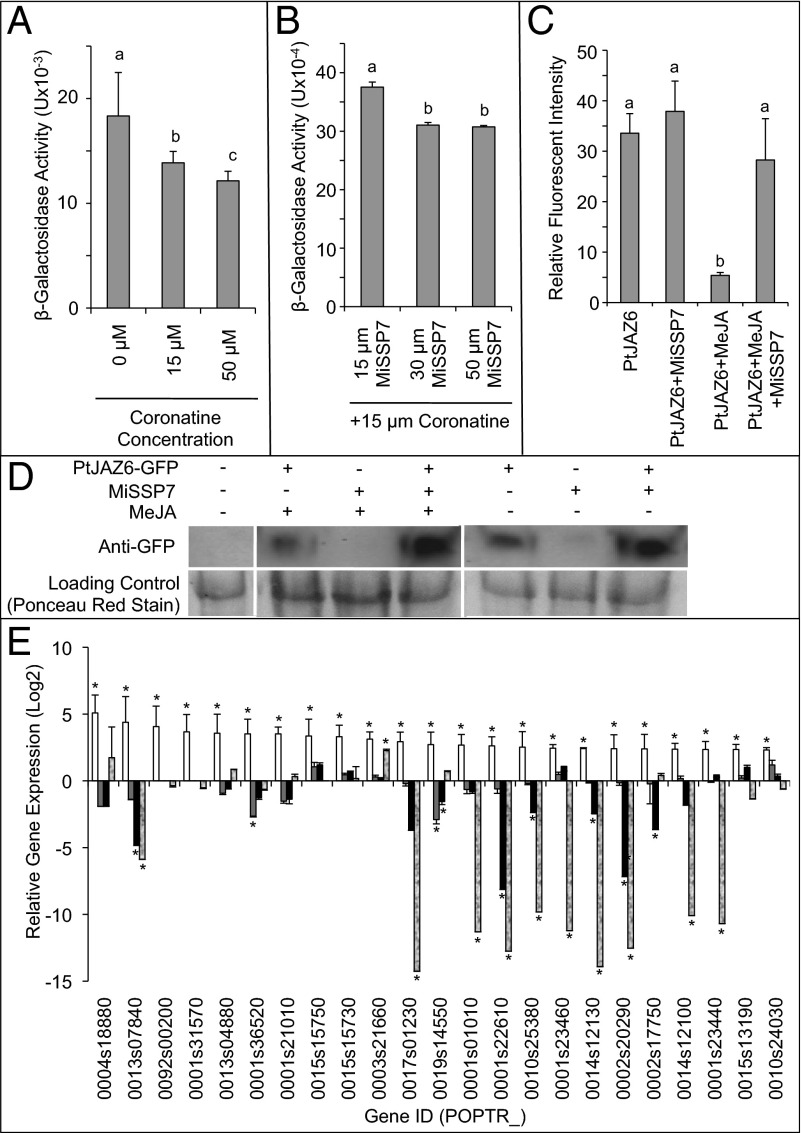
The result that coronatine induces the interaction between PtCOI1 and PtJAZ6 led us to test whether or not coronatine could also increase the strength of the interaction between MiSSP7 and PtJAZ6 as quantified by β-galactosidase activity of the yeast cells (Fig. 3A). At all concentrations tested, however, coronatine had no visible effect on promoting the interaction between these proteins. Rather, as concentrations increased, coronatine significantly disrupted the interaction between MiSSP7 and PtJAZ6 as demonstrated by a significant decrease in β-galactosidase activity (Fig. 3A). Thus, the interaction between MiSSP7 and PtJAZ6 is not stimulated by a mimic of JA-Ile. Furthermore, we tested the ability of MiSSP7 to alter coronatine-induced binding between PtCOI1 and PtJAZ6. We found that MiSSP7, which is taken up by yeast cells and localizes to the nucleus as in plants (Fig. S4C), significantly decreased the ability of PtCOI1 to interact with PtJAZ6 in the presence of coronatine (Fig. 3B). Because our findings above demonstrate that MiSSP7 interferes with coronatine-induced binding of PtCOI1 to PtJAZ6, we investigated whether MiSSP7 could inhibit JA induced degradation of PtJAZ6 in planta. In this experiment we expressed PtJAZ6-GFP and MiSSP7 in Nicotiana benthamiana and treated the leaves with MeJA or buffer. GFP fluorescence in nuclei of epidermal cells was quantified. Treatment with MeJA alone resulted in the almost complete loss of nuclear fluorescence indicating degradation of the protein PtJAZ6-GFP (Fig. 3C), but leaves coexpressing MiSSP7 inhibited the loss of PtJAZ6-GFP fluorescence (Fig. 3C). These results were confirmed by Western blotting (Fig. 3D). Taken together, these data would suggest that MiSSP7 is interfering with the JA-induced degradation of the PtJAZ6-GFP fusion protein in the nucleus thereby resulting in an increased lifetime for PtJAZ6.
Fig. 3.
MiSSP7 alters PtJAZ6 cycling in planta, blocks MeJA activity in N. benthamiana leaves and represses the transcription of JA-inducible genes in planta. (A) Treatment with increasing concentrations of coronatine significantly reduced the β-galactosidase activity in yeast colonies expressing MiSSP7 and PtJAZ6, as determined by reduced formation of chlorophenol red. (B) Treatment with increasing concentrations of MiSSP7 and a fixed quantity of coronatine significantly reduced the β-galactosidase activity in yeast colonies expressing COI1 and PtJAZ6; superscript letters in the first two panels indicate significant differences between treatments as determined by one-way ANOVA followed by a Tukey HSD multiple comparison test (P < 0.05 ± SEM). (C) MiSSP7 treatment of N. benthamiana leaves reduces MeJA induced nuclear degradation of PtJAZ6-GFP as determined by decrease in GFP fluorescence. Bars labeled with identical superscript letters indicate significant similarity in PtJAZ6-GFP fluorescence (P < 0.05) ± SEM. (D) Representative Western blots quantifying the presence of the PtJAZ6-GFP construct in N. benthamiana leaves treated with water (control) MeJA or MeJA + MiSSP7. The lower portion of the panel shows the Ponceau red stain of the protein loading control. (E) Relative fold-change in transcript abundance of JA-marker genes, as defined by Major and Constable (29), in roots treated with 10−8 M MeJA (white bars), in two independent transgenic lines expressing MiSSP7 (gray and black bars) and in roots colonized by L. bicolor (stippled bars). Ratio of gene expression in MeJA-treated tissues was obtained from RNA-seq data; the other datapoints were obtained by comparing expression ratios from whole genome oligoarrays. Gene annotations can be found in Table S1. The asterisk represents significant difference from untreated/uncolonized poplar roots (fold-change > 2.5; P < 0.05), ± SEM.
Because MiSSP7 interacts with PtJAZ6, a repressor of JA-induced gene expression, and as MiSSP7 increases the lifetime of PtJAZ6 in the nucleus during JA treatment, it would thus follow that MiSSP7 treatment of poplar roots should result in the repression of JA-inducible genes. We tested this hypothesis by analyzing the expression of known JA-inducible genes (29) in the roots of poplar heterologously overexpressing MiSSP7. We found that, of the 23 JA-inducible genes tested, 11 were significantly repressed in at least one of the transgenic lines expressing MiSSP7 and mainly consisted of genes coding for cell wall active enzymes (e.g., extensin, pectinesterase, chitinase) (Fig. 3E and Table S1). It was not expected that all of the genes tested would be repressed because MiSSP7 is only able to interact with two JAZ proteins of poplar and JAZ proteins have been demonstrated to have discrete functions (8). We further tested the expression of these JA marker genes in roots colonized by L. bicolor (Fig. 3E). We found that of the 23 genes tested, 10 were significantly repressed in colonized root tissues. Six of these genes were also down-regulated by MiSSP7 and were found to be homologous to extensin, peroxidase, and pectinesterase genes (Fig. 3E and Table S1). Therefore, MiSSP7 is able to repress the expression of several JA marker genes related to cell wall remodeling.
Because MiSSP7 is able to reduce JA-induced degradation of PtJAZ6 and partly repress JA-induced gene transcription (Fig. 3), we wished to determine if MiSSP7 application could block the previously described inhibitory effect of JA on L. bicolor Hartig net development (19). The inhibition of Hartig net formation induced by exogenous MeJA is dose-dependent, with lower concentrations of MeJA (10−14 M) having no significant effect on the establishment of the Hartig net and concentrations higher than 10−12 M significantly inhibiting Hartig net formation (Fig. 4A). The inhibition of Hartig net establishment was not a result of reduced fungal growth rate caused by MeJA addition (Fig. S6). When 15 μM synthetic MiSSP7 was exogenously added to the root system undergoing colonization by L. bicolor in the presence of MeJA, however, the development of a Hartig net was reinstated at concentrations of MeJA that would normally be inhibiting this process (i.e., 10−10 M) (Fig. 4A). Because MiSSP7 is able to inhibit JA activity during Hartig net formation, and as lack of MiSSP7 expression during root colonization stops Hartig net formation (26), it is possible that the lack of MiSSP7 expression during the colonization process could be complemented by either increasing the expression of PtJAZ6 or by inhibiting JA-induced gene transcription. To test this hypothesis, we generated transgenic roots with increased or decreased production of PtJAZ6 and tested the ability of L. bicolor to form a Hartig net in these roots (Fig. 4B and Figs. S6 and S7). We found that in PtJAZ6 RNAi lines, the development of the Hartig net was significantly impeded in roots being colonized by wild-type L. bicolor (Fig. 4B). Conversely, formation of a Hartig net in L. bicolor Δmissp7 lines was found to be reinstated if PtJAZ6 was overexpressed in root tissues (>24–fold) (Fig. 4C and Fig. S7). The observed establishment of the Hartig net in 35S::PtJAZ6 roots colonized L. bicolorΔmissp7 was abolished by the external application of MeJA, indicating that the phenotype was a result of altered JA signaling rather than an artifact of the transformation process (Fig. 4C). To complement these transgenic assays, we tested whether the previously described inhibitors of JA-induced gene expression, SHAM and aspirin (30), could complement the L. bicolor missp7 RNAi mutant. Because these inhibitors have not been verified as repressors of the JA pathway in poplar, we analyzed the ability of both compounds to repress JA-induced gene transcription. We found that both inhibitors significantly repressed JA-induced gene expression patterns (Fig. S8). We also found that both SHAM and aspirin application to roots led to the formation of a Hartig net by L. bicolor Δmissp7 RNAi lines. Therefore, an increase in the expression of PtJAZ6 or the pharmacological suppression of JA-induced gene expression are each able to replace the role of MiSSP7 during the development of the Hartig net.
Fig. 4.
MiSSP7 activity in ECM root tips is complemented by pharmacological repression of JA-induced genes or transgenic transgenic overexpression of PtJAZ6. (A) Growth of L. bicolor hyphae between epidermal roots cells (= Hartig net formation) of P. tremula × P. alba 717–1B4 with simple application of differing concentrations of MeJA (white bars; 10−8 to 10−14 M) or concurrent application of 15 μM MiSSP7 with differing concentrations of MeJA (gray bars). Bars labeled with identical superscript letters indicate significant similarity in Hartig net development (P < 0.05) ± SEM. (B) Depth of hyphal penetration during colonization by wild-type L. bicolor S238N (= Hartig net formation) into the apoplast between epidermal root cells of either wild-type P. tremula × P. alba 717–1B4 or transgenic lines overexpressing PtJAZ6 (35S::PtJAZ6 Line x) or lines with RNAi reduction of PtJAZ6 (PtJAZ6-RNAi Line x) under untreated colonization conditions, with external addition of JA (+JA) or MeJA (+MeJA), or external addition of JA-signaling inhibitors (+Aspirin or +SHAM). (C) The effect that either variation in JA-induced gene transcription or that transgenic overexpression of PtJAZ6 (35S::PtJAZ6 Line x) or RNAi reduction of PtJAZ6 (PtJAZ6-RNAi Line x) have on the ability of wild-type L. bicolor (S238N) or two mutant lines of L. bicolor unable to produce MiSSP7 (L. bicolor Δmissp7;CL03, Cl19) to form a Hartig net. Additionally, we tested the ability of externally applied MeJA (10−8 M) to saturate the phenotype of 35S::PtJAZ6 roots (35S::PtJAZ6 + MeJA Line x). Bars labeled with identical superscript letters indicate significant similarity in Hartig net development (P < 0.05) ± SEM. Each transgenic line (denoted by a number of 1 − n) is an independent transformation event.
Discussion
Plants have developed a complex defensive response system to protect themselves against invasion by detrimental organisms, often mediated by plant hormones. In turn, invading organisms have developed various methods to circumvent the plant’s defenses or control plant cell function to their benefit. Pathogens, such as P. syringae and H. arabidopsidis, attempt to manipulate the plant response by producing effectors that target different components of the JA (i.e., JAZ proteins) and ET signaling pathways (i.e., ERF proteins) in such a fashion that colonization is favored (10, 11, 23, 24). Like pathogenic bacteria, mutualistic fungi affect plant hormone signaling cascades to achieve colonization (31–35), although the knowledge of the mechanistic reasoning behind most of these differences is in its infancy. We demonstrate here that MiSSP7, an effector protein produced by the mutualistic ECM fungus L. bicolor, targets plant-encoded JAZ proteins and interacts with them in the nucleus of the plant. MiSSP7 is able to block the activity of MeJA and promote the proliferation of L. bicolor in plant tissues. This effect is likely a result of the ability of MiSSP7 to reduce the JA-induced degradation of the JAZ protein, thereby repressing JA-induced gene transcription. Finally, we demonstrate that the activity of MiSSP7 during the colonization process can be replaced by either inhibiting JA signaling in planta or through transgenic overexpression of the PtJAZ6 gene.
It is interesting that MiSSP7 acts to repress the expression of JA-induced genes in tissues as both biotrophic pathogens and mutualistic arbuscular mycorrhizal (AM) fungi promote JA accumulation in plant tissues to favor colonization (35). More recently, we made the observation that mutualistic ECM fungi are distinct from other biotrophic organisms in that JA inhibits their proliferation within root tissues (19). This finding is consistent with the earlier demonstration that colonization by another ECM fungus, Paxillus involutus, results in accumulation of the JA antagonist SA in root tissues and a decrease in JA biosynthesis (31). To our knowledge, our results give the first mechanistic insight into how an ECM fungus may negatively regulate plant JA-induced gene regulation in cells through the use of an ECM fungal effector protein. MiSSP7, by reducing the JA-induced degradation of the PtJAZ6 protein (Fig. 3 C and D) and through the competitive exclusion of COI1 binding with PtJAZ6 (Fig. 3B), likely results in the observed repression of JA-induced gene regulation in poplar roots during colonization by L. bicolor. Of the JA marker genes repressed by MiSSP7, the majority have annotated functions relating to cell wall modification (e.g., extensin, pectin esterase, chitinase) (Fig. 3E and Table S1), suggesting that rather than inhibiting chemical defense induced by the JA pathway, MiSSP7 is affecting cell wall chemistry. The effect on cell wall modification suggested by these data is consistent with our earlier findings regarding the role of MiSSP7 during L. bicolor colonization of poplar roots (26). Within root rhizodermal cells this alteration to cell wall dynamics would then be sufficient to allow for the penetration of fungal hyphae into the root and establishment of the Hartig net. It is interesting to note, however, that some JA-inducible genes normally repressed during the L. bicolor colonization process of poplar roots (Fig. 3E) are not affected by MiSSP7 application. This finding would suggest that there are other effectors or mechanisms at play that target other aspects of the JA signaling cascade during the establishment of the ECM root tip. Taken together, these findings have a fundamental impact upon our understanding of how L. bicolor “negotiates” a symbiotic relationship by altering a plant’s ability to respond to JA. It remains to be seen if other mutualistic fungi similarly target JAZ-domain proteins and, thereby, affect the JA responsiveness of their host plant to foster symbiotic interactions.
Materials and Methods
Template cDNA for all cloning and YTH procedures was generated from RNA extracted using P. trichocarpa roots undergoing colonization by L. bicolor S238N after 0, 2, 4, 6, and 12 wk postcontact with the fungus. Yeast DNA reporter assay and YTH screens were performed as outlined previously (27). DivIVa and BiFC interaction tests were used to verify each interaction found in the YTH as per previous reports (28, 36).
An in vitro assay was used to determine the effects of different chemicals on the ability of L. bicolor S238N to colonize poplar roots after 2 wk of contact between the two organisms as per Felten et al. (33). To test if transgenically altering the transcription of PtJAZ6 could complement the loss of MiSSP7 in L. bicolor Δmissp7 RNAi mutants during mycorrhization, we used Agrobacterium rhizogenes strain 15834 to generate stably transformed roots of P. tremula × Populus alba 717–1B4 using a technique similar to that described in Chabaud et al. (37).
Full methods and any associated references can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank M. Buée and Y. Dessaux for kindly supplying the strain 15834 of Agrobacterium rhizogenes; T. Kazmierzack for supplying Nicotiana benthamiana plants; and B. Pêtre and M. Ouassou for their technical assistance. We thank the Arabidopsis Biological Resource Center at The Ohio State University for the bimolecular fluorescence complementation vectors. We would also like to thank Dr. M. Doktycz for helpful discussions. This work was supported in part by the European Commission within the Project ENERGYPOPLAR (FP7-211917), the Laboratory of Excellence Advanced Research on the Biology of Tree and Forest Ecosystems (ANR-11-LABX-0002-01), and the Agence Nationale de Recherche project FungEffector (to F.M.); the Genomic Science Program (project ‘Plant-Microbe Interactions’) funded by the US Department of Energy, Office of Science, Biological and Environmental Research under the Contract DE-AC05-00OR22725.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE56865, GSE56863, GSE56864, and GSE53475).
This article contains Supporting Information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322671111/-/DCSupplemental.
References
- 1.McDowell JM, Dangl JL. Signal transduction in the plant immune response. Trends Biochem Sci. 2000;25(2):79–82. doi: 10.1016/s0968-0004(99)01532-7. [DOI] [PubMed] [Google Scholar]
- 2.Thomma B, et al. Different micro-organisms differentially induce Arabidopsis disease response pathways. Plant Physiol Biochem. 2001;39(7):673–680. [Google Scholar]
- 3.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5(5):344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 4.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21(8):2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA. 2008;105(19):7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 7.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 8.Pauwels L, Goossens A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–3100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiler EW, et al. The Pseudomonas phytotoxin coronatine mimics octadecanoid signalling molecules of higher plants. FEBS Lett. 1994;345(1):9–13. doi: 10.1016/0014-5793(94)00411-0. [DOI] [PubMed] [Google Scholar]
- 10.Jiang S, et al. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLOS Path. 2013;9(10):e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gimenez-Ibanez S, et al. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014;12(2):e1001792. doi: 10.1371/journal.pbio.1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He P, et al. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 2004;37(4):589–602. doi: 10.1111/j.1365-313x.2003.01986.x. [DOI] [PubMed] [Google Scholar]
- 13.Korkama T, Fritze H, Pakkanen A, Pennanen T. Interactions between extraradical ectomycorrhizal mycelia, microbes associated with the mycelia and growth rate of Norway spruce (Picea abies) clones. New Phytol. 2007;173(4):798–807. doi: 10.1111/j.1469-8137.2006.01957.x. [DOI] [PubMed] [Google Scholar]
- 14.Bellion M, Courbot M, Jacob C, Blaudez D, Chalot M. Extracellular and cellular mechanisms sustaining metal tolerance in ectomycorrhizal fungi. FEMS Microbiol Lett. 2006;254(2):173–181. doi: 10.1111/j.1574-6968.2005.00044.x. [DOI] [PubMed] [Google Scholar]
- 15.Hibbett DS, Matheny PB. The relative ages of ectomycorrhizal mushrooms and their plant hosts estimated using Bayesian relaxed molecular clock analyses. BMC Biol. 2009;7:13. doi: 10.1186/1741-7007-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Floudas D, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336(6089):1715–1719. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 17.Plett JM, Martin F. Blurred boundaries: lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet. 2011;27(1):14–22. doi: 10.1016/j.tig.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Massicotte HB, Peterson L, Melville LH. Hartig net structure of ectomycorrhizae synthesized between Laccaria bicolor (Tricholomataceae) and two hosts: Betula alleghaniensis (Betulaceae) and Pinus resinosa (Pinaceae) Am J Bot. 1989;76:1654–1667. [Google Scholar]
- 19.Plett JM, et al. Ethylene and jasmonic acid act as negative modulators during mutualistic symbiosis between Laccaria bicolor and Populus roots. New Phytol. 2014;202(1):270–286. doi: 10.1111/nph.12655. [DOI] [PubMed] [Google Scholar]
- 20.Martin F, Kamoun S. 2012. in Effectors in Plant-Microbe Interactions, eds Martin F, Kamoun S (John Wiley & Sons, Oxford, UK)
- 21.Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem. 2004;279(25):26370–26377. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- 22.Tian M, Benedetti B, Kamoun S. A Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 2005;138(3):1785–1793. doi: 10.1104/pp.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhtar MS, et al. European Union Effectoromics Consortium Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333(6042):596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohn JR, Martin GB. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 2005;44(1):139–154. doi: 10.1111/j.1365-313X.2005.02516.x. [DOI] [PubMed] [Google Scholar]
- 25.Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452(7183):88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 26.Plett JM, et al. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr Biol. 2011;21(14):1197–1203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 27.Plett JM, Montanini B, Kohler A, Ottonello S, Martin F. Tapping genomics to unravel ectomycorrhizal symbiosis. Methods Mol Biol. 2011;722:249–281. doi: 10.1007/978-1-61779-040-9_19. [DOI] [PubMed] [Google Scholar]
- 28.Edwards AN, et al. An in vivo imaging-based assay for detecting protein interactions over a wide range of binding affinities. Anal Biochem. 2009;395(2):166–177. doi: 10.1016/j.ab.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Major IT, Constabel CP. Shoot-root defense signaling and activation of root defense by leaf damage in poplar. Botany. 2007;85(12):1171–1181. [Google Scholar]
- 30.Pena-Cortes H, et al. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191(1):123–128. [Google Scholar]
- 31.Luo Z-B, et al. Upgrading root physiology for stress tolerance by ectomycorrhizas: Insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol. 2009;151(4):1902–1917. doi: 10.1104/pp.109.143735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 33.Felten J, et al. The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol. 2009;151(4):1991–2005. doi: 10.1104/pp.109.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Splivallo R, Fischer U, Göbel C, Feussner I, Karlovsky P. Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 2009;150(4):2018–2029. doi: 10.1104/pp.109.141325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Ráez JA, et al. Hormonal and transcriptional profiles highlight common and differential host responses to arbuscular mycorrhizal fungi and the regulation of the oxylipin pathway. J Exp Bot. 2010;61(10):2589–2601. doi: 10.1093/jxb/erq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee LY, et al. Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell. 2012;24(5):1746–1759. doi: 10.1105/tpc.112.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chabaud M, et al. 2003. Agrobacterium rhizogenes-mediated root transformation. The Medicago truncatula Handbook, eds Mathesius U, Journet EP, Sumner LW. (The Samuel Roberts Noble Foundaton, Ardmore, OK)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.