Significance
Iron-sulfur clusters are ubiquitous cofactors of proteins intervening in disparate biological processes. Iron-sulfur cluster biosynthesis pathways are tightly regulated in Gram-negative bacteria. One of the participating transcription factors, iron-sulfur cluster pathway (ISC) regulator (IscR), can itself bind an iron-sulfur cluster. Depending on its ligation status, IscR recognizes and binds to distinct promoters, therefore modulating cluster biosynthesis. This unique protein at the crossroad between the ISC and sulfur assimilation (SUF) iron-sulfur cluster biosynthetic pathways was thought to be restricted to Gram-negative bacteria. We demonstrated the existence of a functional IscR in the unique Gram-positive bacterium Thermincola potens. Structural and functional analysis of T. potens and Escherichia coli IscR unveiled a conserved mechanism of promoter discrimination, along with subtle structural differences that explain their distinct DNA sequence recognition specificity.
Keywords: Rrf2-like regulator, transcription regulation, helix-turn-helix motif, DNA recognition, specificity modulation
Abstract
Iron-sulfur clusters function as cofactors of a wide range of proteins, with diverse molecular roles in both prokaryotic and eukaryotic cells. Dedicated machineries assemble the clusters and deliver them to the final acceptor molecules in a tightly regulated process. In the prototypical Gram-negative bacterium Escherichia coli, the two existing iron-sulfur cluster assembly systems, iron-sulfur cluster (ISC) and sulfur assimilation (SUF) pathways, are closely interconnected. The ISC pathway regulator, IscR, is a transcription factor of the helix-turn-helix type that can coordinate a [2Fe-2S] cluster. Redox conditions and iron or sulfur availability modulate the ligation status of the labile IscR cluster, which in turn determines a switch in DNA sequence specificity of the regulator: cluster-containing IscR can bind to a family of gene promoters (type-1) whereas the clusterless form recognizes only a second group of sequences (type-2). However, iron-sulfur cluster biogenesis in Gram-positive bacteria is not so well characterized, and most organisms of this group display only one of the iron-sulfur cluster assembly systems. A notable exception is the unique Gram-positive dissimilatory metal reducing bacterium Thermincola potens, where genes from both systems could be identified, albeit with a diverging organization from that of Gram-negative bacteria. We demonstrated that one of these genes encodes a functional IscR homolog and is likely involved in the regulation of iron-sulfur cluster biogenesis in T. potens. Structural and biochemical characterization of T. potens and E. coli IscR revealed a strikingly similar architecture and unveiled an unforeseen conservation of the unique mechanism of sequence discrimination characteristic of this distinctive group of transcription regulators.
Iron-sulfur (Fe/S) proteins play crucial roles for the functioning of both prokaryotic and eukaryotic cells, being required for biological functions ranging from electron transport to redox and nonredox catalysis, and from DNA synthesis and repair to sensing in regulatory processes (1). The main role of the Fe/S cluster assembly machineries is to mobilize iron and sulfur atoms from their storage sources, assemble the two components into an Fe/S cluster, and then transfer the newly formed cluster to the final protein acceptors (2). In Escherichia coli, there are two of these Fe/S cluster ‘‘factories,’’ the ISC (iron-sulfur cluster) and SUF (sulfur assimilation) systems, whose corresponding genes are organized in two operons, iscSUA-hscBA-fdx and sufABCDSE, respectively (2, 3). Deletion mutants of the ISC system display a variety of growth defects due to loss of Fe/S cluster-containing enzyme activity and disruption of sulfur metabolism whereas failure of both the ISC and SUF systems leads to synthetic lethality (4, 5).
In E. coli, the ISC machinery is considered the housekeeping system responsible for the maturation of a large variety of Fe/S proteins whereas the SUF system is triggered under stress conditions, such as oxidative stress or iron starvation (6). ISC pathway regulator (IscR) is a [2Fe-2S] cluster-containing transcription factor with a single predicted helix-turn-helix motif, first identified for its role in regulating expression of the ISC biogenesis pathway (7) and subsequently found to control the expression of more than 40 genes in E. coli (7, 8). According to the currently accepted model for Fe/S cluster biogenesis, under conditions unfavorable for Fe/S cluster formation, the labile IscR cluster is lost, and IscR-mediated repression of the isc (iron-sulfur cluster) operon is alleviated. At the same time, apo-IscR activates the suf (sulfur assimilation) operon to further compensate for damage or loss of Fe/S clusters (9, 10). Once the demand for Fe/S biogenesis is met, higher levels of cluster-containing holo-IscR exist, causing an increased repression of the ISC pathway. Moreover, under iron limitation, the ISC and SUF machineries are unable to maintain the levels of holo-IscR, and therefore this feedback mechanism allows IscR to sense Fe/S demand and enables E. coli to respond appropriately to stress conditions (11).
There are two classes of IscR binding sites in the E. coli genome: a type-1 site deduced from iscR, yadR, and yhgI promoter regions and a type-2 site compiled from the IscR sites upstream of the hyaA, ydiU, and sufA promoters (8). Interestingly, IscR binds type-1 promoters solely in its holo-form whereas binding to type-2 promoters was shown to be independent of the presence of the Fe/S cluster (12). In E. coli, IscR mutation E43A enabled specific recognition of type-1 promoters by apo-IscR, likely mimicking the interaction mode of the cluster-bound form of the protein (13).
Although a molecular-level understanding of the complex processes of Fe/S cluster biosynthesis in several organisms is now emerging from the combination of in vivo and in vitro approaches, these machineries are still poorly understood in Gram-positive bacteria. Although homologs of the E. coli ISC or SUF systems are present in several organisms, some species exhibit unusual Fe/S cluster biosynthetic machineries. Most Gram-positive bacteria carry only a suf operon, containing genes coding for SufU and the SufBCD complex (14, 15), but no sufE or sufA-related genes, even if in some cases sufA can be found elsewhere in the genome (14, 16).
Thermincola potens (strain JR) is an anaerobic, thermophilic, Gram-positive dissimilatory metal-reducing bacterium (DMRB), isolated from a thermophilic microbial fuel cell (17). It is of the first Gram-positive DMRB for which there is a complete genome sequence, which revealed an unusual abundance of multiheme c-type cytochromes (17, 18). Using homology searches, we identified a series of genes with sequence similarity to both E. coli SUF and ISC machineries in the T. potens genome, including a gene locus coding for a putative IscR protein. Taken together, our results both identify and characterize a unique Fe/S biogenesis regulator in Gram-positive bacteria. Through structural and biochemical analysis of both T. potens and E. coli apo-IscR proteins and their E43A mutants, we were able to unveil subtle structural features important for DNA recognition and binding specificity.
Results
Unique Fe/S Cluster Biogenesis in T. potens.
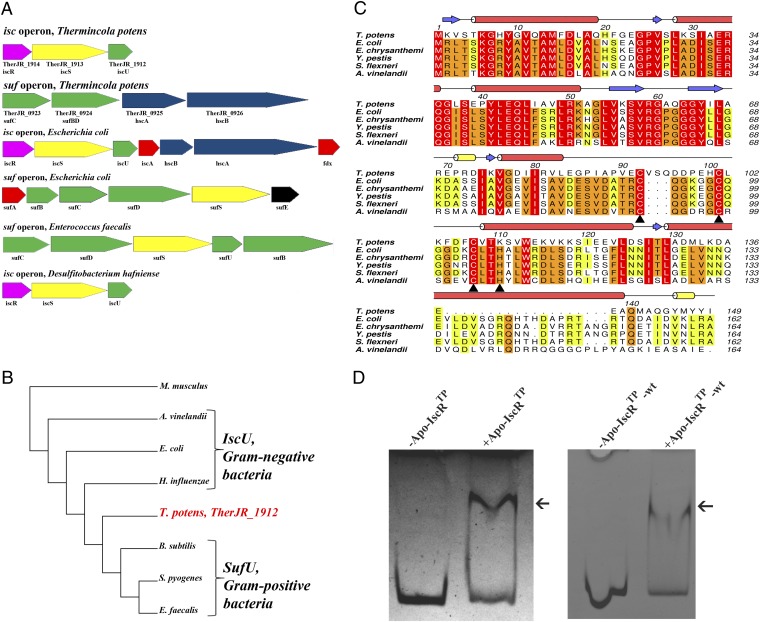
In Gram-positive bacteria, there is conservation of the suf operon, often present as sufCDSUB, which is the only machinery for Fe/S cluster biosynthesis in the majority of these organisms (14, 19). Surprisingly, homology searches on the Gram-positive DMRB T. potens JR genome (17) allowed identifying two gene loci with sequence similarity to E. coli SUF and ISC machineries (Fig. 1A). In T. potens, there are ORFs coding for homologs of the transcription factor IscR (TherJR_1914, 37% identical to the E. coli protein) (7), the cysteine desulfurase IscS (TherJR_1913) (4, 20), and the scaffold IscU (TherJR_1912) (21) from the ISC pathway. An additional suf-like operon in T. potens comprises homologs of sufC (TherJR_0923), sufB, and sufD (TherJR_0924) from the E. coli SUF pathway, and the chaperones hcsA (TherJR_0925) and hcsB (TherJR_0926) from the E. coli isc operon (22–24).
Fig. 1.
Identification of an Fe/S cluster biosynthesis regulator in T. potens. (A) T. potens possesses a unique organization of genes involved in Fe/S cluster biosynthesis, with both isc and suf operons. Colors denote gene function conservation between Gram-negative (E. coli), Gram-positive (E. faecalis), and DMRB Gram-positive bacteria (T. potens and D. hafniense). (B) The amino acid sequence of T. potens scaffold protein reveals features characteristic of the IscU-proteins from Gram-negative bacteria. Neighbor-joining phylogenetic analysis of conserved protein sequences of putative IscU-type or SufU-type proteins in both Gram-positive (T. potens, Streptococcus pyogenes, E. faecalis, B. subtilis) and Gram-negative bacteria (E. coli, Azotobacter vinelandii, Haemophilus influenza), using Mus musculus as outgroup. The sequences were aligned with three distinct alignment algorithms as implemented in ADOPS (57). The resulting cladogram places the T. potens scaffold protein between IscU proteins from Gram-negative bacteria and SufU proteins from other Gram-positive bacteria. (C) The T. potens TherJR_1914 gene codes for a protein that is highly homologous to IscR from Gram-negative bacteria. Strictly conserved amino acids are highlighted in red, and increasing residue conservation is represented by a color gradient from green to red. Alignment prepared with ClustalW (58) and colored with Aline (59). (D) T. potens apo-IscR recognizes the upstream suf operon region between genes TherJR_0922 and TherJR_0923. Incubation of either apo-IscR Cys-to-Ser mutant (apo-IscRTp) or its as-purified wild-type version (apo-IscRTp-wt) with the putative suf promoter region (suf sequence) (Table S1) resulted in a mobility-shift (arrow).
Compared with other Gram-positive bacteria, namely from the Firmicutes phylum, some unique features of the T. potens suf operon become evident. In T. potens, the suf operon does not code for cysteine desulfurase (SufS) homologs although there are elsewhere in the T. potens genome two additional genes coding for putative cysteine desulfurases (TherJR_0460 and TherJR_3003) homologous to CsdA/SufS, which can function as complementary sulfur sources for Fe/S cluster biogenesis, possibly through the recruitment of the SUF machinery (25). The T. potens suf operon is also devoid of homologs of SufU, recently reported to be a zinc-dependent sulfurtransferase in Bacillus subtilis (26), but encodes a sufBD protein, which together with sufC was shown to act as scaffold in Gram-negative bacteria (27, 28). Furthermore, the suf operon in T. potens includes the hscA and hscB genes coding for the chaperones responsible for transferring preformed clusters from the scaffold IscU to final acceptors and that, in E. coli, are cotranscribed with the isc and not with the suf operon (4). Additionally, genes coding for A-type carriers are absent from the T. potens genome.
IscU is a highly conserved protein that functions as scaffold for cluster assembly and subsequent transfer. Preserved features include the cluster ligands (three cysteines and one histidine), an aspartate residue that plays a critical role in cluster transfer to apo-proteins and the LPPVK motif recognized by the chaperone HscA (29) (Fig. S1). Some Gram-positive bacteria (e.g., Enterococcus faecalis) were shown to possess an IscU homolog, SufU, which does not contain the HscA recognition site and has a 19-residue insertion between the first two conserved cysteines (14). The T. potens scaffold protein displays conservation of the characteristic IscU LPPVK motif and does not contain the insertion signature specific of SufU-type proteins. Accordingly, phylogenetic analysis of IscU and SufU protein sequences places the protein encoded by the TherJR_1912 gene between the Gram-negative IscU-type and the SufU-like proteins from Gram-positive bacteria (Fig. 1B).
As previously reported for Clostridium perfringens (30), searches for Fe/S cluster biogenesis operons in Gram-positive bacteria with completely sequenced genomes, namely the DMRBs Desulfitobacterium hafniense and Desulfotomaculum reducens, revealed that they possess a single ISC gene locus (iscRSU) but no SUF apparatus. In contrast, in other Gram-positive bacteria (e.g., E. faecalis), only the SUF pathway can be found. Therefore, contrary to other Gram-positive bacteria described so far, T. potens not only has two gene loci coding for the two Fe/S cluster biosynthesis machineries present in E. coli (ISC and SUF), but these systems display a unique organization.
The T. potens TherJR_1914 Gene Codes for IscR.
IscR is a [2Fe-2S] cluster-containing transcriptional regulator encoded by the first gene of the iscRSUA-hscBA-fdx operon that regulates both ISC and SUF systems in E. coli and other Gram-negative bacteria (10, 11). Apart from the sequence-unrelated SufR found in cyanobacteria (31, 32), no IscR homolog was described in Gram-positive bacteria, with the possible exception of some species of the Clostridium genus for which functional data are still lacking (30). In T. potens, the TherJR_1914 gene encodes a protein of the Rrf2 family of transcriptional regulators, sharing only 37% identity with E. coli IscR but with full conservation of the cysteine residues known to coordinate the [2Fe-2S] cluster (Cys92, 98, 104, E. coli numbering) (Fig. 1C) (10).
Clusterless (apo) IscR from E. coli was shown to activate suf operon expression during stress conditions, such as iron starvation (6). The as-purified apo form of the protein encoded by T. potens gene TherJR_1914 (apo-IscRTp-wt) was found to bind to the upstream region of the putative suf operon, between genes TherJR_0922 and TherJR_0923 (Fig. 1D). A similar behavior was observed for a triple mutant (C92/101/107S) of IscRTp (apo-IscRTp) (Fig. 1D) where all putative cluster-binding cysteine residues (Fig. 1C) were replaced by serine. Given the structural similarity between cysteine and serine and the requirement for homogeneous sample for downstream functional and structural assays, this variant was used in all experiments where the apo form of IscRTp was required. The ability of apo-IscRTp-wt and apo-IscRTp to bind the promoter region of the suf operon suggests that IscRTp can function as an Fe/S cluster regulator in this organism, with the apo form involved in the regulation of the suf operon expression, as observed for E. coli. Although such regulators have been found and characterized in a number of Gram-negative bacteria (7, 8, 33–35), the characterization of orthologous proteins from Gram-positive species has not yet been reported. Therefore, T. potens has a unique organization and regulation of Fe/S cluster assembly genes, among Gram-positive bacteria.
Overall Structure of T. potens IscR.
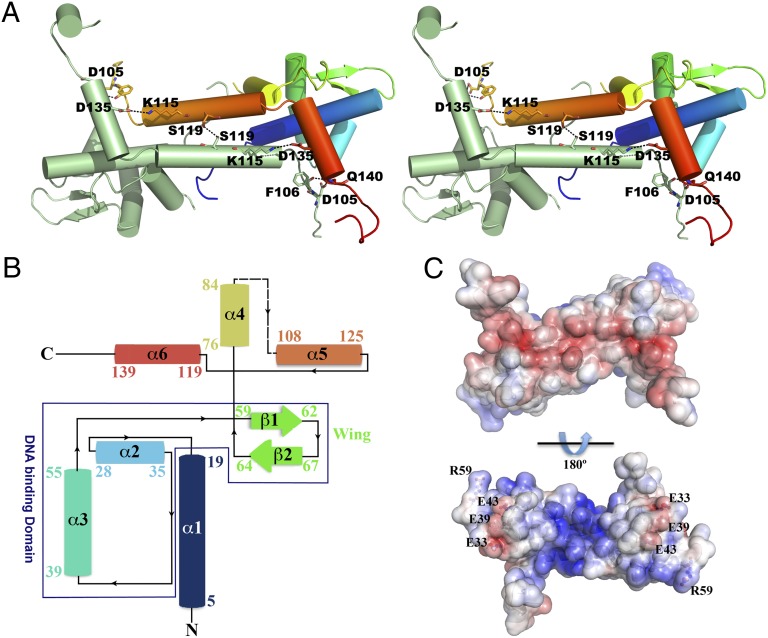
The 3D structure of free apo T. potens IscR, with the putative cluster-binding cysteines mutated to serine (the clusterless IscR triple-mutants C92/101/107S for T. potens or C92/98/104S for E. coli are hereby termed apo-IscRTp and apo-IscREc, respectively) was determined by X-ray crystallography from tetragonal (P41) crystals diffracting to 1.6-Å resolution. The crystallographic asymmetric unit contains the functional IscR homodimer (Fig. 2A). Apo-IscRTp monomers are predominantly α-helical and, whereas the N-terminal region is formed by three consecutive α-helices preceding the characteristic wing β-hairpin, the C-terminal domain is exclusively α-helical (Fig. 2B). Within each monomer, the N-terminal α-helix (α1) interfaces with the wing-helix subdomain (α2-α3-β1-β2) and with the N-terminal end of the dimerization helix (α5) from the adjacent monomer. The dimerization helix, which comprises two of the putative iron-sulfur cluster binding residues (101 and 107), is further stabilized by close contacts with helix α6 from the adjacent monomer. Residues 86–100, encompassing part of the putative iron-sulfur cluster-binding segment (Fig. 2B), are disordered in both monomers and could not be modeled (Fig. 2 A and B). The structures of the monomers are nearly identical, superposing with an rmsd of 0.17 Å for 133 aligned Cα atoms.
Fig. 2.
The 3D structure of T. potens IscR. (A) Stereoscopic view of the biologically active apo-IscRTp dimer, highlighting important residues (represented as sticks) at the dimerization interface. One of the monomers is colored from N- (blue) to C-terminal (red), with highlighted residues color-coded (nitrogen blue, oxygen red). Hydrogen bonds are represented as dashed lines. (B) Topology diagram of the apo-IscRTp monomer. Secondary structure element colors match those of A. (C) Solid-surface representation of the apo-IscRTp dimer, with mapped electrostatic surface potential contoured from +5 (blue) to −5 (red) kbTe−1 [kb, Boltzmann’s constant; T, temperature (K); e, charge of an electron].
Apo-IscRTp dimer formation involves mostly interactions between residues from helix α5 of each monomer, but residues from helices α1 and α4 further stabilize the homodimer. The extensive dimerization interface between the two helices α5 is hydrophobic, except for a single hydrogen bond between the side chains of neighboring Ser119 residues. Further intermonomer polar interactions are established between the side chain of Gln140 at the C terminus of helix α6 and Asp105 OD1 and Phe106 O (Fig. 2A).
In agreement with its DNA-binding function (Fig. 1D), the electrostatic surface potential of apo-IscRTp is highly polarized (Fig. 2C), being predominantly negative at the solvent-exposed face of helices α5 and α6, and positively charged at the opposite, putative DNA-binding side. This asymmetric surface-charge distribution promotes initial DNA positioning by nonspecific electrostatic contacts, before fine-tuning through the establishment of base- and shape-specific interactions. Negatively charged residues (Glu33, Glu39, and Glu43) protrude from this positively charged surface (Fig. 2C), resembling E. coli IscR (13). Among these residues, Glu39 is unique in T. potens IscR (replaced by a leucine residue in closely related molecules) (Fig. 1C).
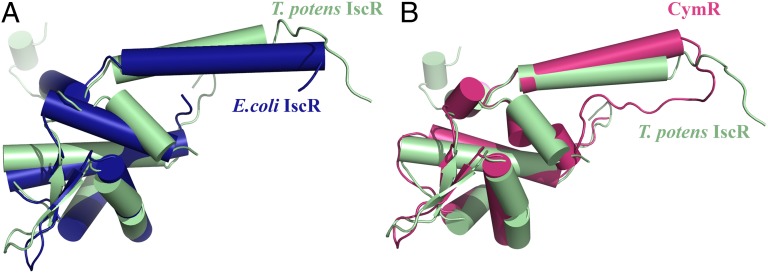
T. potens IscR belongs to the Rrf2-like family of transcriptional regulators and displays highest structural similarity with the global cysteine regulator CymR from B. subtilis (PDB ID code 2Y75) (36) and S. aureus (PDB ID code 3T8T) (37) as well as with the recently determined structure of E. coli IscR (PDB ID code 4HF0) (13), superposing with these models with an rmsd of 1.5–2.3 Å. The conserved DNA-binding helix-turn-helix motif can also be superposed to the corresponding domain of more distantly related proteins, albeit with somewhat higher rmsd values (Table 1). The DNA-binding winged-helix motif is structurally similar in the selected structures (Fig. 3 A and B), and most structural differences occur in the dimerization helix α5 and in the length and orientation of helix α4, which precedes the iron-sulfur cluster-binding region.
Table 1.
Structural similarity between T. potens IscR and other winged-helix transcription regulators
| Protein | PDB ID code | rmsd, Å | No. of aligned Cα atoms | Amino acid sequence identity, %* | Z-score |
| B. subtilis CymR | 2y75 | 1.5 | 119 | 55 | 17.2 |
| E. coli IscR (unliganded) | 4hf0 | 2.0 | 116 | 41 | 16.8 |
| E. coli IscR (DNA complex) | 4hf1 | 2.3 | 120 | 40 | 16.3 |
| S. aureus CymR | 3t8t | 2.3 | 119 | 46 | 15.9 |
| Putative transcriptional regulator from L. innocua | 3lwf | 3.8 | 124 | 48 | 14.9 |
| B. cereus protein BC1842 | 1ylf | 2.7 | 118 | 20 | 13.9 |
S. aureus, Staphylococcus aureus; L. innocua, Listeria innocua; B. cereus, Bacillus cereus.
Structure-based sequence alignment, as calculated by Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server/start) (60).
Fig. 3.
IscR from T. potens is structurally similar to other winged-helix transcriptional regulators. Superposition of apo-IscRTp monomer (green) with (A) free E. coli apo-IscR C92/98/104A (blue; PDB ID code 4HF0) and (B) free B. subtilis CymR (magenta; PDB ID code 2Y75) highlighting the overall structural conservation.
Molecular Details of IscR-DNA Interaction.
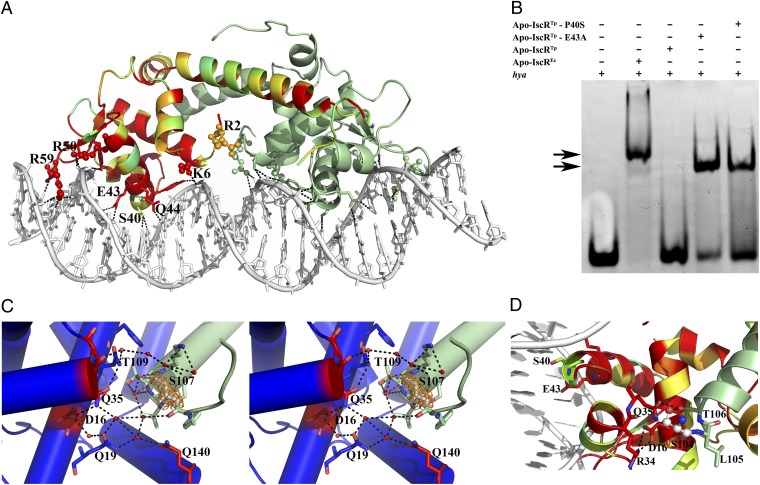
To better grasp the fine molecular details of specific promoter sequence recognition by IscR, we determined the structure of apo-IscR from E. coli (C92/98/104S triple mutant with the putative cluster-binding cysteine residues mutated to serine; apo-IscREc) in complex with the E. coli hya (hydrogenase-1) promoter sequence (8, 12, 31). The asymmetric unit contains the apo-IscREc biological dimer bound to a 26-bp double-stranded oligonucleotide with a single nucleotide overhang at the 5′ end of each strand (Fig. 4A and Table S1). This structure is very similar to the recently reported model of E. coli apo-IscR (C92/98/104A triple mutant) in complex with DNA (PDB ID code 4HF1) (13). Overall, the protein main chains of the two models superpose with an rmsd of 0.5 Å for 124 aligned Cα atoms. DNA binding induces a small concerted movement of the wing-helix motif within each apo-IscREc monomer and of the dimerization helix α5 of the adjacent monomer (Fig. S2 A and B) (13). In the complex, helix α3 of the winged-helix motif from each apo-IscREc monomer is presented to the major groove of the corresponding DNA half-site whereas the β-hairpin inserts into the minor groove (Fig. 4A).
Fig. 4.
Binding of IscR to type-2 promoter sequences. (A) Bidentate binding of apo-IscREc (C92/98/104S) to the hya promoter DNA sequence. In one of the monomers, residues are colored according to conservation, where red corresponds to positions strictly conserved between E. coli and T. potens IscR. Residues at the DNA-interacting interface are highlighted as sticks and basic residues as spheres. Hydrogen bonds between apo-IscREc and DNA are represented as dotted lines. (B) Electrophoretic mobility-shift assay analysis of apo-IscR binding to the E. coli hya promoter. Arrows denote observed band-shifts. (C) Stereoscopic view of the intricate network of hydrogen bonds in apo-IscRTp (one monomer of the functional dimer is colored green and the other one blue) centered on the cluster-binding residue 107 (light gray). The 2Fo − Fc electron density map around residue 107 is represented as an orange mesh. Water molecules and strictly conserved residues in closely related IscR molecules are colored red. (D) In apo-IscREc (C92/98/104S), serine 104 (ball and stick) participates in a network of polar interactions with neighboring residues (sticks), cross-linking helices α1, α2, and α5. The corresponding cysteine residue in the wild-type protein could be part of a sensing mechanism for the presence of the Fe/S cluster.
As observed for apo-IscRTp, the apo-IscREc biological dimer is highly polarized with a clustering of basic residues on the DNA-interacting surface (Fig. S2C). Although the positive electrostatic surface might be required to orient the protein toward productive binding with the DNA duplex, in the apo-IscREc-DNA complex very few protein-DNA interactions involve basic side chains. A notable exception is Arg59 that protrudes from the β-hairpin wing and wedges into the symmetrically related AT-rich regions of the narrow groove (Fig. 4A). Accordingly, the E. coli apo-IscR R59A mutant is unable to bind to either type-1 or type-2 promoter sequences, demonstrating the relevance of this residue for DNA recognition (13). Further interactions with the minor grove are established by residues at the tip of the wing (Gly60-Pro61) that slot between deoxyribose moieties and by Pro27 that packs against phosphodiester linkages.
Apo-IscREc residues contributing to the partial negative charge within the DNA-binding surface (Glu33, Asp30, and Glu43) (Fig. S2C) are structurally equivalent to the acidic residues identified on the equivalent side of apo-IscRTp. However, only Glu43 contacts directly the bound oligonucleotide. Together with Gln44 (conserved) and Ser40 (variable), Glu43 is involved in base-specific recognition within the major groove (Fig. 4A). It has been demonstrated that Glu43 is a crucial selectivity filter that negatively affects binding of E. coli apo-IscR to type-1 promoter sequences, containing thymine at positions 6 and 7, due to lack of suitable hydrogen-bond donors (13).
Subtle Structural Differences Modulate DNA Sequence Recognition Specificity.
Overall, there is a striking conservation of the DNA-binding interface (Figs. 1C and 4A). Apo-IscRTp differs from the E. coli homolog only at four positions within the interaction surface, which could result in altered DNA binding affinity and specificity: Ser27 (Pro27 in apo-IscREc), Pro40 (Ser40 in apo-IscREc), and Ala61-Gln62 (Pro61-Gly62 in apo-IscREc). The replacement of Pro27 by a serine is likely to increase the flexibility of the linker between the first two α-helices although a large change in DNA affinity is not predictable. In contrast, the substitution of Pro61-Gly62 by an Ala-Gln dipeptide can impact the conformation of the wing β-hairpin and interfere with the tight packing of this structural element within the minor groove. In particular, the residue at position 40 is likely to play a key role in sequence-specific recognition of DNA. In the apo-IscREc-DNA complex, the protein packs very tightly within the major groove, leaving limited space for bulkier residues (Fig. 4A). Although a proline could be accommodated at the N terminus of helix α3 without helical disruption, the resulting steric hindrance might prevent the placement and base readout of the conserved Glu43-Gln44 and/or contacts of the residues interacting with the phosphate backbone (Tyr9, Ser38, Tyr41). Substitution of the purines interacting with Ser40 (G20′ and A19) prevents binding of E. coli apo-IscR to the hya promoter sequence, highlighting the importance of this residue for base-specific recognition (12). Further, mutation of Ser40 to alanine in E. coli IscR decreases binding to the hya promoter by 90% compared with the wild-type protein, a decrease that is sequence-dependent and more pronounced for type-2 sites (13).
The influence of Pro40 in apo-IscRTp interaction with DNA is evidenced by its inability to bind the E. coli hya promoter sequence (Fig. 4B and Table S2). Replacement of Pro40 in apo-IscRTp by the structurally equivalent amino acid in E. coli IscR (apo-IscRTp P40S) is sufficient to allow binding to the heterologous promoter (Fig. 4B and Table S2). The presence of a serine residue at position 40 is likely to alleviate the tight packing of IscR within the major groove of DNA, reducing steric hindrance and allowing binding. Accordingly, the IscRTp E43A mutant, where the shorter alanine side chain can provide room for positional adjustments of this region, also recognized the hya sequence (Fig. 4B) with an affinity comparable with that of the E. coli protein, as assessed by microscale thermophoresis (Table S2). Taken together, these results suggest that substitution of Ser40 by a proline in apo-IscRTp prevents base recognition through steric hindrance, an impairment lifted by introducing less bulky residues at either position 40 or 43.
Position of the Cluster-Binding Residues.
In contrast to previous studies with E. coli IscR, where all putative cluster-binding cysteine residues were mutated to alanine to obtain homogeneous clusterless protein (10, 12, 13), in T. potens IscR, the corresponding residues were mutated to serine, which is a closer structural match. In all E. coli and T. potens IscR structures, the region involved in iron-sulfur cluster association is partially disordered, but the serine residues replacing Cys107 in apo-IscRTp and Cys104 in apo-IscREc are clearly visible in the electron density maps (Fig. 4 C and D). In contrast to what is observed for the Cys-to-Ala mutant structure of E. coli free apo-IscR where the two visible cluster ligands (Ala104 and His107) are on the outer face of the longer dimerization helix α5 (13), in apo-IscRTp, the equivalent Ser107 is part of the coil region preceding helix α5, and the crystal structure shows that it participates in a water-mediated network of hydrogen bonds connecting this structural segment to helices α1 and α2 (Fig. 4C). In particular, the Ser107 side-chain is hydrogen-bonded to Thr109 OG1 within each monomer. Both residues also establish polar interactions with ordered solvent molecules that participate in a hydrogen-bond network interfacing the two monomers of the functional dimer and involving the side chains of Gln19, Asp16, and Gln35 from the adjacent monomer (the last two residues strictly conserved across IscR molecules) (Fig. 1C). Of particular relevance is the involvement of Asp16 side chain in a salt bridge with Arg34 within the DNA-binding helix-turn-helix motif. The cluster-binding segment is further stabilized by a polar contact with Gln140 of the adjacent monomer, which also connects the corresponding helices α1 and α6. Altogether, this polar interaction network tightly connects the cluster-binding segment at the N-terminal portion of helix α5 from one monomer with the N-terminal helix α1, helix α6, and helix α2 from the adjacent monomer. Particularly, this network suggests an interconnection between structural changes in the cluster-binding segment and functional effects at the DNA-binding interface.
The geometry of the hydrogen bonds established by the mutated Ser107 in apo-IscRTp, and the rotational freedom of Thr109, evidenced by the two discrete conformations of its side-chain in the current crystal structure, are compatible with the existence of similar hydrogen bonds involving Cys107 in the cluster-free wild-type IscR. Indeed, the cysteine side chain thiol group is a moderately good hydrogen bond donor, sometimes crucial for protein activity and function (38–41).
In the crystal structure of the apo-IscREc-DNA complex, Ser104 (structurally equivalent to Ser107 in apo-IscRTp) is also well defined in the electron density maps. In one of the monomers, Ser104 is part of helix α5, as previously reported for the triple Cys-to-Ala mutant structure (13). However, in the other monomer of the apo-IscREc dimer, this residue hydrogen bonds to the conserved Thr106 (Thr109 in T. potens IscR), which in turn engages in a network of direct polar contacts cross-linking the dimerization helix α5 to the C terminus of the adjacent helix α2 (Arg34, Gln35) and to helix α1 (Asp16) (Fig. 4D). This hydrogen bond network involves direct interactions between the amino acid side chains, in contrast to what is observed in apo-IscRTp, where solvent molecules mediate some of the contacts. In the Cys-to-Ala triple mutant of E. coli IscR, this arrangement of polar contacts is preserved, with the expected exception of residue 104, which is there an alanine (13).
The predicted function of residue 107/104 (in T. potens and E. coli, respectively) in iron-sulfur cluster binding, as well as its location between the dimerization helix of one monomer and the first helix of the helix-turn-helix DNA-binding motif of the neighboring subunit, suggest a possible role as a central nanoswitch, whereby cluster binding-induced movement could trigger a global motion involving both the dimer interface and the DNA-binding region from the opposite monomer. The resulting structural changes could explain the observed alteration in DNA binding specificity upon cluster association (13).
A Single Mutation Allows apo-IscRTp to Recognize Type-1 Promoter Sequences from T. potens and E. coli.
In E. coli, holo-IscR was shown to interact with both type-1 and type-2 DNA motifs in a similar manner whereas apo-IscR bound solely to type-2 promoter sequences (12). Recently, it was also demonstrated that replacement of Glu43 by alanine in E. coli IscR C92/98/104A removed unfavorable interactions with type-1 motifs, allowing recognition of these promoters (13). The T. potens isc promoter region displays cis-regulatory elements similar to those identified in the type-1 E. coli iscRSUA-HscBA-fdx, including the −35 hexamer and the −10 element sequences (7). In fact, it is possible to delimit a segment (iscTp_1) (Fig. 5A and Table S1) displaying 48% identity to the E. coli isc promoter sequence and containing a −10 element and a consensus −35 hexamer of the Eσ70-binding site with a 20-bp spacer region (13).
Fig. 5.
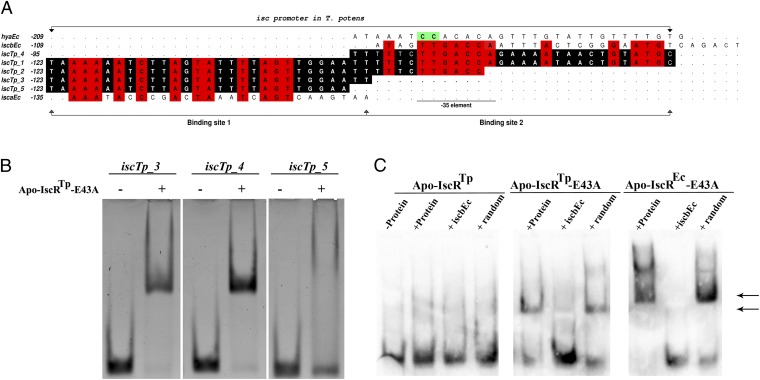
Modulation of apo-IscRTp specificity by a single point mutation. (A) IscR binding sites in type-1 [T. potens isc (iscTp) and E. coli isca (iscaEc) and iscb (iscbEc)] and type-2 [E. coli hya (hyaEc)] promoters. Numbers refer to the most upstream base of each IscR site relative to the corresponding start codon. Conserved bases between the isc promoters are highlighted in red whereas bases conserved between the five T. potens isc promoter sequences are shaded black. The highly conserved CC motif in type-2 promoters is colored green (12). (B) There are two independent binding sites for apo-IscRTp E43A in the T. potens isc promoter. Purified apo-IscRTp E43A (7.5 μM) was incubated with iscTp_3, iscTp_4 and iscTp_5 sequences, analyzed by nondenaturing PAGE, and visualized by ethidium bromide staining. An arrow denotes the DNA band-shift upon complex formation. (C) Cross-recognition of the E. coli isc promoter by T. potens IscR. Purified proteins (apo-IscRTp, apo-IscRTp E43A, or apo-IscREc E43A) were incubated with DIG-labeled iscrb promoter sequence (iscbEc) (Table S1) and analyzed by nondenaturing PAGE. An arrow denotes bands indicative of DNA–IscR complex formation. Where indicated, cold iscrb or a similarly sized random sequence (random) was added in 100-fold molar excess as competitor.
Similar to what is observed for E. coli apo-IscR and isc, apo-IscRTp does not bind iscTp_1 (Fig. S3A). Using an enzymatic system under oxygen-depleted atmosphere (42), an Fe/S cluster could be reversibly reconstituted in wild-type apo-IscRTp, yielding the holo form of the protein (reconstituted IscRTp-wt; R-IscRTp-wt), as judged by the appearance of absorption maxima at 320 and 420 nm (Fig. S3B). A dose-dependent structural change of iscTp_1 DNA could be identified by circular dichroism spectroscopy, upon R-IscRTp-wt binding (Fig. S3C). These results demonstrate that, as expected for a bona fide IscR, the enzymatically reconstituted Fe/S cluster-bound form of IscRTp-wt binds to the T. potens isc promoter region (Fig. S3C). As seen for E. coli IscR (13), the single point mutant apo-IscRTp E43A binds specifically to the iscTp_1 sequence, seemingly forming two distinct complexes—with either one or two IscR dimers binding to the target sequence—as suggested by the two observed DNA band shifts (Fig. S3A). This finding is further supported by the observation of a single complex with the 3′-trimmed iscTp_1 sequence, termed iscTp_2 (Fig. 5A, Table S1, and Fig. S3A). In E. coli, DNase footprinting led to the identification of two IscR binding sites within the isc promoter region, iscra and iscrb (8). Two highly homologous regions could be identified in the T. potens isc promoter, iscTp_3 and iscTp_4 (Fig. 5A and Table S1), to which apo-IscRTp E43A displays specific binding (Fig. 5B and Table S2). Further, removal of the two 3′-end nucleotides of iscTp_3, yielding the shorter iscTp_5 (Fig. 5A and Table S1), effectively prevents binding of apo-IscRTp E43A (Fig. 5B and Table S2), in good agreement with the observed bidentate binding of IscR to the minor groove of AT-rich segments at the termini of its recognition sequence (Fig. 4A).
In line with the structural similarity of E. coli and T. potens IscR proteins and the considerable conservation of isc promoter sequences, there is cross-recognition between the transcriptional regulator of T. potens and the E. coli promoter. Although apo-IscRTp does not bind E. coli iscrb (iscbEc, Table S1), this sequence is specifically recognized by the E43A mutant (Fig. 5C and Table S2), as observed for E. coli apo-IscR (13). Therefore, the unique mechanism of promoter-sequence discrimination by IscR seems to be conserved between these organisms.
Discussion
We performed a detailed analysis of the product of gene TherJR_1914 from T. potens, undoubtedly establishing its functional relationship with the Fe/S cluster-binding transcription regulator IscR, known to control Fe/S cluster biogenesis in several Gram-negative bacteria. The identification of an IscR homolog in T. potens was unprecedented: most other Gram-positive bacteria studied so far do not code for any IscR-like proteins or have an isc operon, and the rare cases where an isc operon is present (e.g., the DMRB D. hafniense or the bacterium C. perfringens) (30) lack the SUF machinery.
The combination of biochemical and structural studies, on T. potens IscR and its homolog from E. coli, revealed also an unforeseen conservation of the unique mode of IscR promoter sequence recognition and discrimination. Despite extensive conservation of the DNA-binding surface, apo-IscRTp was unable to recognize the heterologous hya promoter from E. coli. Residue at position 40 played a pivotal role in this process because relief of steric hindrance (P40S mutant) was sufficient to promote binding. These subtle differences in specificity highlight the precise tailoring of each IscR molecule to its cognate partners, despite overall conservation of the recognition mechanism.
Similar to the E. coli molecule (12), the clusterless form of the protein binds to the here-identified T. potens suf (type-2) promoter whereas the previously unidentified type-1 promoter (isc) is recognized by holo-IscR. In the absence of the Fe/S cluster, the strictly conserved E43 residue is pivotal for discriminating between type-1 and type-2 promoters by establishing specific interactions with an invariant CC dinucleotide in type-2 sequences. In fact, mutation of this residue to an uncharged alanine seems to mimic the cluster-induced specificity switch of IscR, allowing the clusterless regulator to recognize and to bind to type-1 promoter sequences (13). In apo-IscRTp, the E43A mutation promotes binding to two sequences upstream of the T. potens iscRSU operon. These regions are highly homologous to the E. coli isc sequences recognized by both holo- and mutant apo-IscR E43A. Given that one of these sequences (iscTp_4) contains a −35-element sequence, we propose that T. potens holo-IscR may act as a repressor of Fe/S biogenesis by hindering RNA polymerase binding. As a whole, our results suggest a conserved regulation mechanism by IscR, where Fe/S cluster binding to this transcription regulator enables recognition of type-1 promoters, a process that is mimicked by the E43A mutation.
By using structurally relevant mutants, where serine replaces all putative cluster-coordinating cysteine residues, a network of polar interactions could be identified, connecting the cluster-binding region of one subunit to the DNA-contacting interface of its neighbor in the functional IscR dimer. Any perturbation resulting from cluster ligation could therefore be allosterically transmitted to the nucleic acid-binding region that comprises the conserved E43. Regulation through Fe/S cluster ligation allows T. potens IscR to act as a sensor and to be a central player of an auto-regulatory loop responsible for keeping the appropriate levels of cellular Fe/S cluster formation and delivery.
Methods
Protein Expression and Purification.
A synthetic iscr gene, encoding the same amino acid sequence as TherJR_1914 from the T. potens genome, except for a Gly-Leu insertion immediately downstream from the N-terminal methionine, was ordered from Eurofins MWG Operon. The E. coli iscr gene (b2531) fragment spanning nucleotides +4 to +489 of the IscR ORF was amplified from an E. coli K12 colony using specific primers. Both ORFs were cloned into the NdeI and XhoI sites of the expression vector pET30a (IscRTp-wt) or into the Acc65I and NcoI sites of the expression vector pETZ2_1a (43). The latter constructs were used to obtain the triple mutants (C92/101/107S for T. potens or C92/98/104S for E. coli) corresponding to the clusterless forms of the proteins (apo-IscRTp and apo-IscREc) by site-directed mutagenesis.
The N-terminal His6-tagged apo-IscRTp was overexpressed in E. coli BL21 (DE3) cells and the E. coli protein in E. coli BL21 Star (DE3) (Life Technologies). Briefly, cells were grown in LB medium at 37 °C until OD600 = 0.7. At this point, the temperature was decreased to either 25 °C (apo-IscREc) or 30 °C (apo-IscRTp), and the expression was induced with the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation after 4 h and lysed by incubation (60 min on ice with shaking) with 25 μg/mL chicken egg white lysozyme (Sigma). Clarified protein extracts in 20 mM sodium phosphate (pH 7.5), 0.5 M NaCl, 10 mM imidazole, 5% (vol/vol) glycerol, 150 mM arginine, and 2.5 mM β-mercaptoethanol (buffer A) were loaded onto a HisTrap HP column (GE Healthcare) preequilibrated in the same buffer, and bound proteins were eluted with buffer A containing 125 mM imidazole. The IscR-containing fractions were pooled, and the His6 and the solubility tags were removed by incubation with tobacco etch virus (TEV) protease at 4 °C concomitantly to an overnight dialysis against 20 mM sodium phosphate (pH 7.5), 0.2 M NaCl, 10 mM imidazole, 5% (vol/vol) glycerol, 150 mM arginine, and 2.5 mM β-mercaptoethanol. Pure recombinant IscR was separated from the expression tag and noncleaved material by a second immobilized-metal affinity chromatography (IMAC) step, in the same conditions as described above. The buffer was further exchanged for 10 mM Hepes (pH 7.5), 800 mM KCl, and 5% (vol/vol) glycerol using a HiPrep 26/10 (GE Healthcare) desalting column. The protein was either used immediately or flash-frozen in liquid nitrogen and stored at −80 °C until needed. Final protein concentrations were estimated by measuring the absorbance of the samples at 280 nm.
Point mutants apo-IscRTp E43A, apo-IscREc E43A, and apo-IscRTp P40S were generated by site-directed mutagenesis of the pETZ2_1a constructs. All IscR protein variants used for biochemical and crystallization experiments were expressed and purified as described for apo-IscRTp, except for apo-IscRTp-wt, which was purified by a single IMAC step followed by desalting on a HiPrep 26/10 column (GE Healthcare). The E. coli cysteine desulfurase IscS used in reconstitution assays was expressed and purified as described previously (42).
Fe/S Cluster Reconstitution.
Reconstitution of the Fe/S cluster of apo-IscRTp-wt was performed under oxygen-depleted atmosphere (<3 ppm O2) in an anaerobic chamber (Belle Technology). All buffers used were sparged with nitrogen gas for 20 min and kept in the anaerobic chamber for at least 12 h before use. Small volumes of aerobically purified proteins (apo-IscRTp-wt and E. coli IscS), as well as sodium dithionite, cysteine, and iron (II) sources were equilibrated in the same low-oxygen conditions for 1 h.
An apo-IscRTp-wt solution (25 μM) was mixed with ammonium iron (II) sulfate (Sigma) and l-cysteine (Sigma) in 20-fold and eightfold molar excess, respectively. The reaction [in 50 mM Tris (pH 8), 150 mM NaCl, 5 mM DTT] was started after 10 min by addition of 2 μM E. coli IscS, and Fe/S cluster formation was followed by monitoring absorbance at 420 nm. Upon reaction completion, R-IscRTp-wt was purified by IMAC (His-Buster Nickel spin columns; Amocol). Absorption spectra (250–750 nm) were recorded immediately after purification and upon sample reduction with 2 mM sodium dithionite. Control reactions were performed omitting l-cysteine (nonreconstituted IscRTp-wt; NR-IscRTp-wt).
Electrophoretic Mobility-Shift Assay.
Complementary oligonucleotides (Sigma) containing the sequence of the E. coli hya or of the T. potens isc promoter (Table S1) were annealed into double-stranded DNA by heating a 50-μM solution to 95 °C for 5 min in a water bath, followed by slowly (overnight) cooling to room temperature. For electrophoretic mobility-shift assay (EMSA) analysis using the complete sequence of the T. potens suf promoter region (Table S1), the sequence upstream of the sufC gene (TherJR_0923) was amplified by PCR using a synthetic template (Eurofins). DNA solutions (1 μM) were incubated with 7.5–10 μM purified protein at room temperature for 20 min in binding buffer [40 mM Tris⋅HCl (pH 8.0), 100–150 mM KCl, 5% (vol/vol) glycerol, and 1 mM DTT], and the resulting complexes were resolved on 8% (wt/vol) nondenaturing polyacrylamide gels using 1× TAE (40 mM Tris⋅HCl, 20 mM acetic acid, and 1 mM EDTA) as running buffer. DNA was detected by either ethidium bromide staining or chemiluminescent detection. For chemiluminescent detection, annealed DNA probes were end-labeled with digoxigenin using recombinant terminal transferase (Roche). The labeled probes (1.5 μmol) were mixed with 11.25 μmol purified IscR in 15 µL of binding buffer [40 mM Tris⋅HCl (pH 8.0), 10% (vol/vol) glycerol, 1 mM DTT, and 50 mM KCl]. For competition reactions, labeled probe was added after incubating the protein for 10 min with 100-fold molar excess competitor DNA. The samples were separated in 8% (wt/vol) nondenaturing polyacrylamide gels and electroblotted onto positively charged nylon membrane (GE Healthcare), and the digoxigenin-labeled probes were detected with anti-digoxigenin-AP antibody and the chemiluminescent substrate disodium 2-chloro-5-(4-methoxyspiro [1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.1(3,7)]decan]-4-yl)-1-phenyl phosphate (CDP-Star; Roche).
Circular Dichroism Measurements.
Binding of wild-type IscRTp to DNA was monitored by circular dichroism (CD) spectroscopy (44). CD spectra were recorded at 20 °C in 1-nm steps on a temperature-controlled Jasco J-815 spectropolarimeter, continuously purged with nitrogen gas. For each sample, the smoothed average of four spectra was considered. Briefly, either anaerobically reconstituted (R-IscRTp-wt) or nonreconstituted (NR-IscRTp-wt) and purified apo-IscRTp-wt was added incrementally to a sealed 10-mm-path quartz cuvette containing the iscTp_1 sequence (Table S1) in binding buffer, and CD spectra (260–320 nm) were recorded.
Microscale Thermophoresis Assays.
Interactions between IscR variants (apo-IscRTp, apo-IscRTp E43A, apo-IscRTp P40S, and apo-IscREc E43A) and the different DNA sequences (iscTp_3, iscTp_4, iscTp_5, hyaEc, and iscbEc) (Fig. 5A and Table S1) were assessed using microscale thermophoresis (45) with a Monolith NT.115 instrument (NanoTemper Technologies). Purifed proteins (20 μM) were labeled using the Monolith NT.115 Protein Labeling Kit RED-NHS (NanoTemper Technologies) according to the manufacturer’s instructions. Labeled proteins were diluted to 50 nM in assay buffer [40 mM Tris⋅HCl (pH 7.9), 150 mM KCl, 5% (vol/vol) glycerol, 1 mM DTT, 0.1% (vol/vol) Tween 20]. Ligand dilutions were prepared in assay buffer without Tween 20 and mixed with each protein sample at a volume ratio of 1:1. Measurements with apo-IscRTp, apo-IscRTp P40S, and apo-IscRTp E43A were performed in standard capillaries whereas hydrophilic capillaries were used for measurements with apo-IscREc E43A. For each interaction, data from at least two independent runs were averaged, and the average curve was fitted with NTAnalysis software (NanoTemper Technologies).
Crystallization of apo-IscRTp and apo-IscREc:hya Complex.
Initial crystallization conditions for apo-IscRTp were screened at 20 °C using the sitting-drop method with commercial sparse-matrix crystallization screens. Drops consisting of equal volumes (1 µL) of protein (at 20 mg/mL) and precipitant solution were equilibrated against a 300-µL reservoir. Crystals were obtained after 2 d using 0.1 M Bis-Tris (pH 6.5) and 3 M NaCl as precipitant. Before data collection, crystals were cryoprotected by immersing them briefly in a 1:1 mixture of precipitant solution and 40% (vol/vol) of 2 mg/mL NDSB-201 (3-(1-pyridino)-1-propane sulfonate) solution in ethylene glycol and flash-cooled in liquid nitrogen (46). Selenomethionyl apo-IscRTp crystallized in the same conditions and was cryoprotected following the procedure described above.
A 3.8-fold molar excess of apo-IscREc was mixed with double-stranded oligonucleotide (prepared as described in Electrophoretic Mobility-Shift Assay) comprising region −30 to −55 of the E. coli hya promoter sequence with a single-base 5′ overhang (hya_26_OH) (Table S1) and incubated at room temperature for 30 min. The complex was either used immediately or flash frozen in liquid nitrogen and stored at −80 °C. Initial crystallization conditions were established at the High Throughput Crystallization Laboratory of the European Molecular Biology Laboratory, using the sitting-drop method. Crystals were obtained at 20 °C, from 0.2-µL drops composed of identical volumes of complex solution [350 μM protein and 92 μM oligonucleotide in 40 mM Tris⋅HCl (pH 8.0), 150 mM KCl, 10% (vol/vol) glycerol, 1 mM DTT] and of precipitant [0.1 M citric acid (pH 4.0 or 6.0), 1 M lithium chloride, 20% (wt/vol) PEG 6000]. Better and larger crystals could be obtained from the condition at pH 4.0 using the hanging-drop vapor diffusion method. The optimized crystals were cryoprotected in the same conditions as the apo-IscRTp crystals.
Data Collection and Processing.
X-ray diffraction data were collected from cooled (100 K) single crystals at synchrotron beam lines ID29 (apo-IscRTp and Se-Met apo-IscRTp) (47) and ID23-EH2 (apo-IscREc:hya complex) (48) of the European Synchrotron Radiation Facility. The apo-IscRTp data were recorded on a Pilatus 6M detector (Dectris) using a wavelength of 0.9763 Å (native dataset) or 0.9792 Å (Se-Met dataset). For the native data, 1,200 images were collected in 0.1° oscillation steps with 0.1-s exposure per frame whereas, for the Se-Met data, 3,600 images were recorded in 0.1° oscillation steps with 0.037-s exposure per frame. The apo-IscREc:hya complex data were recorded on a MX-225 detector (Marresearch) using a wavelength of 0.8726 Å. One hundred images were collected in 0.95° oscillation steps with 5.43-s exposure per frame. Diffraction data were integrated with XDS (49), scaled with XSCALE (50), and reduced with utilities from the CCP4 program suite (51). Data collection statistics are summarized in Table 2.
Table 2.
Statistics of data collection, processing, and refinement
| Dataset | T. potens IscR* (native) | T. potens IscR* (Se-Met) | E. coli IscR-DNA complex* |
| Crystallographic analysis | |||
| Wavelength, Å | 0.9763 | 0.9792 | 0.8726 |
| Space group | P41 | P41 | P212121 |
| Unit cell dimensions, Å | a = b = 53.6; c = 118.4 | a = b = 53.4; c = 118.7 | a = 49.0; b = 75.8; c = 173.4 |
| Resolution range, Å | 53.6–1.60 (1.69–1.60) | 48.7–2.47 (2.61–2.47) | 46.0–2.49 (2.62–2.49) |
| Reflections (measured/unique) | 196,159/43,750 (28,394/6,325) | 111,297/11,861 (13,614/1,647) | 87,182/23,387 (12,463/3,244) |
| Completeness, % | 99.7 (98.7) | 99.1 (94.1) | 99.3 (96.3) |
| Multiplicity | 4.5 (4.5) | 9.4 (8.3) | 3.7 (3.8) |
| Rmerge† | 0.046 (1.299) | 0.190 (1.573) | 0.103 (0.911) |
| Rpim‡ | 0.024 (0.688) | 0.064 (0.555) | 0.061 (0.531) |
| 〈I/σ(I)〉 | 14.3 (1.6) | 7.3 (1.4) | 8.6 (1.5) |
| Monomers per asymmetric unit | 2 | 2 | 2 |
| Mathews coefficient, Å3⋅Da−1 | 2.53 | 2.52 | 3.13 |
| Solvent content, % | 51.4 | 51.2 | 60.7 |
| Structure refinement | |||
| Resolution range, Å | 48.8–1.60 | — | 46.0–2.49 |
| Rfactor§/Free Rfactor¶ | 0.202/0.220 | — | 0.207/0.251 |
| Unique reflections (work/test set) | 41,642/1,973 | — | 22,057/1,193 |
| Water molecules | 156 | — | 15 |
| Total no. of atoms | 2,363 | — | 2,999 |
| No. of macromolecule atoms | 2,205 | — | 2,984 |
| rmsd bond lengths, Å | 0.011 | — | 0.008 |
| rmsd bond angles, ° | 1.09 | — | 1.38 |
| Average overall B factor, Å2 | 38.1 | — | 77.7 |
| Ramachandran favored, % | 97.5 | — | 96.0 |
| Ramachandran outliers, % | 0.0 | — | 0.4 |
| PDB entry | 4cic | — | 4chu |
Values in parentheses correspond to the outermost resolution shell. Each dataset was recorded from a single crystal.
Rmerge = ∑hkl∑i|Ii(hkl) – 〈I(hkl)〉|/∑hkl∑iIi(hkl), where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections.
Rpim = ∑hkl[1/(N – 1)]1/2 ∑i |Ii(hkl) – 〈I(hkl)〉|/∑hkl ∑iIi(hkl), where Ii(hkl) is the observed intensity and 〈I(hkl)〉 is the average intensity of multiple observations of symmetry-related reflections.
Rfactor = ∑||Fo| − |Fc||/∑|Fo|, where |Fo| and |Fc| are observed and calculated structure factor amplitudes, respectively.
Free Rfactor is the cross-validation Rfactor computed for a randomly chosen subset of 5% of the total number of reflections, which were not used during refinement.
Structure Solution and Refinement.
The structure of apo-IscRTp was solved by single-wavelength anomalous diffraction using the anomalous signal of selenium-substituted crystals with the SHELXC/SHELXD/SHELXE pipeline (52) and the HKL2MAP GUI (53). The resulting electron density maps were readily interpretable. The structure of the apo-IscREc:hya complex was solved by molecular replacement with PHASER (54) using a truncated version of the refined apo-IscRTp structure as search model. For both structures, alternating cycles of model building with COOT (55) and of refinement with PHENIX (56) were performed until model completion. For the apo-IscRTp structure, the final model comprises residues Gly-3 to Gly85 and Ser101 to Ile149 for subunit A and Gly-3 to Gly85 and Ser101 to Tyr148 for subunit B whereas the apo-IscREc:hya complex comprises residues Met0 to Asp88 and Lys103 to Ser139 for subunit A, and Gly1 to Asp84 and Gln93 to Val135 for subunit B. Model refinement statistics are summarized in Table 2.
Supplementary Material
Acknowledgments
We thank Jorge Vieira for help with Automatic Detection of Positively Selected Sites. We acknowledge the European Synchrotron Radiation Facility (ESRF) for provision of synchrotron radiation facilities and thank the ESRF staff for help with data collection. Microscale thermophoresis data collection was carried out at the Campus Science Support Facilities Protein Technologies Facility (www.csf.ac.at). This work was funded by Fundo Europeu de Desenvolvimento Regional through the Operational Competitiveness Programme-COMPETE and by national funds through Fundação para a Ciência e a Tecnologia under project FCOMP-01-0124-FEDER-028116 (PTDC/BBB ‐ BEP/2127/2012) and PhD Fellowship SFRH/BD/66461/2009 (to J.A.S.). The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (Grant Agreement 283570).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4CHU and 4CIC).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1322728111/-/DCSupplemental.
References
- 1.Beinert H, Holm RH, Münck E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science. 1997;277(5326):653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Ayala-Castro C, Saini A, Outten FW. Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev. 2008;72(1):110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fontecave M, Choudens SO, Py B, Barras F. Mechanisms of iron-sulfur cluster assembly: The SUF machinery. J Biol Inorg Chem. 2005;10(7):713–721. doi: 10.1007/s00775-005-0025-1. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi Y, Nakamura M. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J Biochem. 1999;126(5):917–926. doi: 10.1093/oxfordjournals.jbchem.a022535. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277(32):28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 6.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52(3):861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz CJ, et al. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98(26):14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60(4):1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 9.Lee KC, Yeo WS, Roe JH. Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. J Bacteriol. 2008;190(24):8244–8247. doi: 10.1128/JB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo WS, Lee JH, Lee KC, Roe JH. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol. 2006;61(1):206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 11.Giel JL, et al. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol Microbiol. 2013;87(3):478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesbit AD, Giel JL, Rose JC, Kiley PJ. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J Mol Biol. 2009;387(1):28–41. doi: 10.1016/j.jmb.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajagopalan S, et al. Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat Struct Mol Biol. 2013;20(6):740–747. doi: 10.1038/nsmb.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riboldi GP, Verli H, Frazzon J. Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein. BMC Biochem. 2009;10:3. doi: 10.1186/1471-2091-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riboldi GP, de Oliveira JS, Frazzon J. Enterococcus faecalis SufU scaffold protein enhances SufS desulfurase activity by acquiring sulfur from its cysteine-153. Biochim Biophys Acta. 2011;1814(12):1910–1918. doi: 10.1016/j.bbapap.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Albrecht AG, et al. SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J Bacteriol. 2010;192(6):1643–1651. doi: 10.1128/JB.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrne-Bailey KG, et al. Complete genome sequence of the electricity-producing “Thermincola potens” strain JR. J Bacteriol. 2010;192(15):4078–4079. doi: 10.1128/JB.00044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson HK, et al. Surface multiheme c-type cytochromes from Thermincola potens and implications for respiratory metal reduction by Gram-positive bacteria. Proc Natl Acad Sci USA. 2012;109(5):1702–1707. doi: 10.1073/pnas.1112905109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riboldi GP, de Mattos EP, Frazzon J. Biogenesis of [Fe-S] cluster in Firmicutes: An unexploited field of investigation. Antonie van Leeuwenhoek. 2013;104(3):283–300. doi: 10.1007/s10482-013-9966-5. [DOI] [PubMed] [Google Scholar]
- 20.Zheng L, Cash VL, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273(21):13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
- 21.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci USA. 2000;97(2):599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangachari K, et al. SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 2002;514(2-3):225–228. doi: 10.1016/s0014-5793(02)02369-4. [DOI] [PubMed] [Google Scholar]
- 23.Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW. SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry. 2010;49(43):9402–9412. doi: 10.1021/bi1011546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silberg JJ, Tapley TL, Hoff KG, Vickery LE. Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron-sulfur cluster assembly protein IscU. J Biol Chem. 2004;279(52):53924–53931. doi: 10.1074/jbc.M410117200. [DOI] [PubMed] [Google Scholar]
- 25.Trotter V, et al. The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Mol Microbiol. 2009;74(6):1527–1542. doi: 10.1111/j.1365-2958.2009.06954.x. [DOI] [PubMed] [Google Scholar]
- 26.Selbach BP, et al. Fe-S cluster biogenesis in Gram-positive bacteria: SufU is a zinc-dependent sulfur transfer protein. Biochemistry. 2014;53(1):152–160. doi: 10.1021/bi4011978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry. 2009;48(44):10644–10653. doi: 10.1021/bi901518y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian T, He H, Liu XQ. The SufBCD protein complex is the scaffold for iron-sulfur cluster assembly in Thermus thermophiles HB8. Biochem Biophys Res Commun. 2014;443(2):376–381. doi: 10.1016/j.bbrc.2013.11.131. [DOI] [PubMed] [Google Scholar]
- 29.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277(30):27353–27359. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 30.André G, et al. Global regulation of gene expression in response to cysteine availability in Clostridium perfringens. BMC Microbiol. 2010;10:234. doi: 10.1186/1471-2180-10-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen G, et al. SufR coordinates two [4Fe-4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J Biol Chem. 2007;282(44):31909–31919. doi: 10.1074/jbc.M705554200. [DOI] [PubMed] [Google Scholar]
- 32.Wang T, et al. The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J Bacteriol. 2004;186(4):956–967. doi: 10.1128/JB.186.4.956-967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi YS, et al. Identification of Pseudomonas aeruginosa genes crucial for hydrogen peroxide resistance. J Microbiol Biotechnol. 2007;17(8):1344–1352. [PubMed] [Google Scholar]
- 34.Runyen-Janecky L, et al. Role and regulation of iron-sulfur cluster biosynthesis genes in Shigella flexneri virulence. Infect Immun. 2008;76(3):1083–1092. doi: 10.1128/IAI.01211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng J, Zhang K, Liu J, Qiu G. Expression, purification, and characterization of iron-sulfur cluster assembly regulator IscR from Acidithiobacillus ferrooxidans. J Microbiol Biotechnol. 2008;18(10):1672–1677. [PubMed] [Google Scholar]
- 36.Shepard W, et al. Insights into the Rrf2 repressor family—the structure of CymR, the global cysteine regulator of Bacillus subtilis. FEBS J. 2011;278(15):2689–2701. doi: 10.1111/j.1742-4658.2011.08195.x. [DOI] [PubMed] [Google Scholar]
- 37.Ji Q, et al. Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem. 2012;287(25):21102–21109. doi: 10.1074/jbc.M112.359737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregoret LM, Rader SD, Fletterick RJ, Cohen FE. Hydrogen bonds involving sulfur atoms in proteins. Proteins. 1991;9(2):99–107. doi: 10.1002/prot.340090204. [DOI] [PubMed] [Google Scholar]
- 39.Zhou P, Tian F, Lv F, Shang Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009;76(1):151–163. doi: 10.1002/prot.22327. [DOI] [PubMed] [Google Scholar]
- 40.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384(Pt 2):201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turk V, et al. Cysteine cathepsins: From structure, function and regulation to new frontiers. Biochim Biophys Acta. 2012;1824(1):68–88. doi: 10.1016/j.bbapap.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prischi F, et al. Of the vulnerability of orphan complex proteins: The case study of the E. coli IscU and IscS proteins. Protein Expr Purif. 2010;73(2):161–166. doi: 10.1016/j.pep.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Olichon A, Surrey T. Selection of genetically encoded fluorescent single domain antibodies engineered for efficient expression in Escherichia coli. J Biol Chem. 2007;282(50):36314–36320. doi: 10.1074/jbc.M704908200. [DOI] [PubMed] [Google Scholar]
- 44.Carpenter ML, Kneale GG. Circular dichroism for the analysis of protein-DNA interactions. Methods Mol Biol. 1994;30:339–345. doi: 10.1385/0-89603-256-6:339. [DOI] [PubMed] [Google Scholar]
- 45.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9(4):342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira PJ, et al. Mycobacterium tuberculosis glucosyl-3-phosphoglycerate synthase: Structure of a key enzyme in methylglucose lipopolysaccharide biosynthesis. PLoS ONE. 2008;3(11):e3748. doi: 10.1371/journal.pone.0003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Sanctis D, et al. ID29: A high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. J Synchrotron Radiat. 2012;19(Pt 3):455–461. doi: 10.1107/S0909049512009715. [DOI] [PubMed] [Google Scholar]
- 48.Flot D, et al. The ID23-2 structural biology microfocus beamline at the ESRF. J Synchrotron Radiat. 2010;17(1):107–118. doi: 10.1107/S0909049509041168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pape T, Schneider TR. HKL2MAP: A graphical user interface for macromolecular phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 54.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reboiro-Jato D, et al. ADOPS—Automatic Detection Of Positively Selected Sites. J Integr Bioinform. 2012;9(3):200. doi: 10.2390/biecoll-jib-2012-200. [DOI] [PubMed] [Google Scholar]
- 58.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2 3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 59.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 5):510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 60.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.