Significance
Patients with Hutchinson–Gilford progeria syndrome (HGPS) almost always die of cardiovascular disease in their teens. Moreover, overlapping cardiovascular pathologies have been identified in HGPS and geriatric patients, including the loss of vascular smooth muscle cells in large vessels. However, how progerin leads to smooth muscle cell loss is still largely unknown. Here, using induced pluripotent stem cells as a model, we examined the role of progerin in smooth muscle cell differentiation and maintenance. Our study identified poly(ADP-ribose) polymerase 1 as a central regulator of smooth muscle cell survival in progeria and elucidated a molecular pathway underlying heart disease in progeria. This study may provide key insights into future medical interventions for the cardiovascular dysfunction in progeria and normal aging.
Abstract
Hutchinson–Gilford progeria syndrome (HGPS) is a severe human premature aging disorder caused by a lamin A mutant named progerin. Death occurs at a mean age of 13 y from cardiovascular problems. Previous studies revealed loss of vascular smooth muscle cells (SMCs) in the media of large arteries in a patient with HGPS and two mouse models, suggesting a causal connection between the SMC loss and cardiovascular malfunction. However, the mechanisms of how progerin leads to massive SMC loss are unknown. In this study, using SMCs differentiated from HGPS induced pluripotent stem cells, we show that HGPS SMCs exhibit a profound proliferative defect, which is primarily caused by caspase-independent cell death. Importantly, progerin accumulation stimulates a powerful suppression of PARP1 and consequently triggers an activation of the error-prone nonhomologous end joining response. As a result, most HGPS SMCs exhibit prolonged mitosis and die of mitotic catastrophe. This study demonstrates a critical role of PARP1 in mediating SMC loss in patients with HGPS and elucidates a molecular pathway underlying the progressive SMC loss in progeria.
DNA damage often arises as a result of normal cellular processes. Reactive oxygen species (ROS), the byproducts of cellular metabolism, can damage DNA bases and block the progression of replication, leading to replication fork collapse and double-strand breaks (DSBs). DSBs can also be induced by environmental factors including irradiation, chemical agents, or UV light (1). A gradual accumulation of DSBs and a decline in DNA repair capacity are suggested to play a causative role in normal physiological aging (2). Defects in DNA damage repair result in at least three premature aging diseases: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy (3). In addition, impaired DNA repair has also been implicated in the development of age-related neurodegenerative diseases such as Alzheimer's disease, Parkinson disease, and Huntington disease (4).
At the cellular level, DSBs are potent inducers of cell death. If left unrepaired, DSBs can trigger p53-mediated cell cycle arrest and programmed cell death; on the other hand, if repaired inaccurately, DSBs can cause small or large scale chromosome alterations, which can lead to premature entry into mitosis and mitotic cell death (mitotic catastrophe) (5). Two separate pathways control the repair of DBSs: homologous recombination (HR) and nonhomologous end joining (NHEJ). HR repairs DSBs using the undamaged sister chromosome as a template, which effectively protects genome integrity. In contrast, NHEJ repairs DSBs by connecting two free chromosome ends together with little requirement for sequence homology, which leads to a high frequency of chromosome misarrangements (1). Normally these two pathways antagonize each other, and the choice between these two is under precise control by a group of regulators including 53BP1, BRCA1/2, and poly(ADP-ribose) polymerase 1 (PARP1) (6, 7).
Among these regulators, PARP1 acts as an essential molecular switch controlling the activities of HR and NHEJ pathways. The classic function of PARP1 is involved in sensing and initiating DNA single-strand break (SSB) repair. A previous study demonstrated that treating an HR-deficient cell line with a PARP1 inhibitor led to abnormal chromosome karyotypes and significantly reduced cell survival, suggesting that PARP1 mediates the suppression of NHEJ upon DSBs (6). This sensitivity to a PARP1 inhibitor in the HR-deficient cells could be a combined effect of the PARP1’s dual roles in DNA damage repair. First, inhibition of PARP1 hinders SSB repair, and the unrepaired SSBs develop into DSBs. More importantly, inhibition of PARP1 removes the suppression of NHEJ, which results in chromosome aberrations and subsequent cell death in these HR-deficient cells.
Hutchinson–Gilford progeria syndrome (HGPS), the most drastic form of premature aging diseases, is characterized by multiple aging-related clinical features including growth retardation, lipodystrophy, alopecia, bone abnormalities, and severe cardiovascular defects (8, 9). Patients with HGPS typically start to display premature onset of aging-related pathologies at 12–24 mo of age and die in their early teens of heart attacks or strokes. Over 80% of HGPS cases are caused by a de novo mutation (1824 C→T) in exon 11 of the human LMNA gene (10). This mutation activates an alternative splice donor site, leading to a truncated lamin A mutant named “progerin,” which bears a 50 amino acid deletion near the C terminus. This internal deletion interferes with lamin A processing and causes a permanent farnesylation of progerin (11). As a dominant negative (12), the accumulation of progerin inside the nucleus causes various cell biological phenotypes, including a misshapen nuclear morphology, loss of peripheral heterochromatin, global changes in gene transcription, and disrupted mitotic progression (13–15). Interestingly defective DNA damage repair has been found in HGPS cells and various progeria animal models, suggesting a likely involvement of genome instability in the manifestation of this drastic premature aging disease (16–18).
Among all of the clinical features associated with HGPS, the cardiovascular disease is the cause of death in almost all patients. Despite its great importance, investigation of the HGPS arterial pathology has been extremely limited mainly due to the low incidence of HGPS. One postmortem study on an HGPS patient found a severe loss of vascular smooth muscle cells (SMCs) in the aorta and carotid arteries, which might induce aortic dilatation and plaque rupture (19). Moreover, two independent in vivo studies on HGPS transgenic mouse models (the G608G BAC and LmnaG609G models, respectively) confirmed the loss of SMCs in large vessels (20, 21). Significantly, this SMC loss could be largely rescued by treating the mice with farnesyltransferase inhibitors (FTIs) or progerin-suppressing morpholinos, indicating a causal connection between the presence of progerin and the SMC loss phenotype (22, 23). A previous study showed generation and recapitulation of premature aging phenotypes of HGPS SMCs from induced pluripotent stem (iPS) cells (24). However, it remains unclear how progerin accumulation results in SMC loss in large vessels.
In this study, we investigate how progerin induces cell death in SMCs. We present data that identify PARP1 as a central regulator of SMC survival in HGPS. We further demonstrate that the accumulation of progerin causes PARP1 down-regulation, which stimulates the error-prone NHEJ responses in S phase HGPS SMCs. Consequently, HGPS SMCs encounter cell division problems and die in mitosis through a caspase-independent mitotic catastrophe pathway.
Results
Normal and HGPS iPS Cells Show Comparable Differentiation Potency into SMC Lineage.
To study HGPS SMC defects, SMCs were generated from iPS cells derived from HGPS and normal skin fibroblasts. Analysis of HGPS iPS cells revealed the expression of Oct4, Nanog, SSEA4, and Tra1-60, indicating that these HGPS iPS cells contain essential pluripotency factors, as well as normal chromosome number and size (Fig. S1 A–C). Alkaline phosphatase (AP) staining and embryoid body (EB) formation assay further demonstrated the undifferentiated state and the differentiation potential of these iPS cells, respectively (Fig. S1 A and E). In accordance with previous reports, the expression of lamin A/C was repressed in both normal and HGPS iPS cells (Fig. S1D).
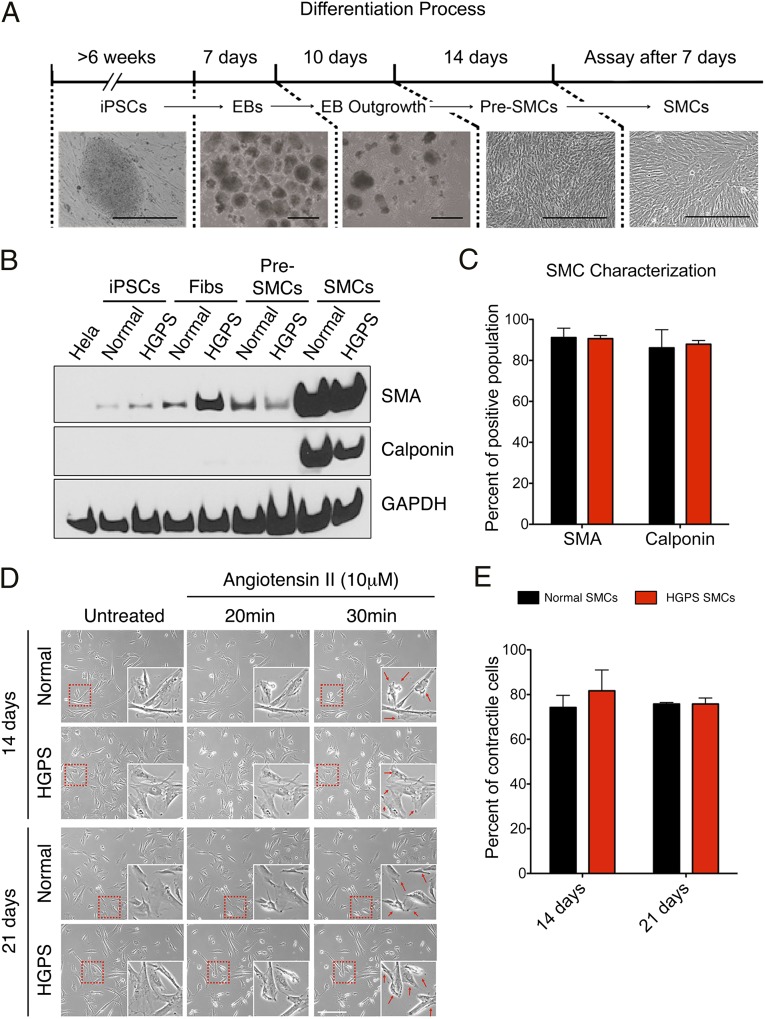
SMCs were induced from iPS cells using a previously established protocol (Fig. 1A, details in Methods) (25). After 7 d in the SMC differentiation medium, immunofluorescence revealed the expression of SMC-specific markers SMC–α-actin (SMA) and calponin in both normal and HGPS lines (Fig. S2A). In accordance with the immunofluorescence observation, Western blotting analysis and RT-quantitative PCR (qPCR) showed a significant up-regulation of SMA, calponin, or SM22-α in SMCs relative to iPS cells, fibroblasts, and pre-SMCs (Fig. 1B and Fig. S2 B and C). Notably, the expression of smooth muscle myosin heavy chain (smMHC), a marker for terminally differentiated SMCs, was not up-regulated in normal and HGPS SMCs, suggesting that the SMCs in our system were not terminally differentiated (Fig. S2B). FACS analysis using fluorescence-labeled SMA or calponin revealed that, after 14 d in SMC differentiation medium, >80% of the cells were induced to express SMA and calponin (Fig. 1C). Western blotting analysis suggested that the expression of these SMC markers was stable for at least 2 wk in SMC differentiation medium (from day 14 to day 28) (Fig. S2C). Next, we carried out contractility analysis on normal and HGPS SMCs. Cells at days 14 and 21 were treated with a vascular constrictor, angiotensin II. More than 70% of SMCs displayed various levels of contraction 30 min after stimulation (Fig. 1 D and E) and exhibited Ca2+ influx (Movies S1 and S2). Taken together, these results indicated that both normal and HGPS iPS cells were differentiated into SMCs with similar high efficiency.
Fig. 1.
Characterization of SMCs differentiated from normal and HGPS iPS cells. (A) Schematic diagram of SMC differentiation timeline and representative phase contrast images underneath each denoted stage. EB, embryoid body. (Scale bar, 400 μm.) (B) Western blot with anti-SMA, calponin, and GAPDH antibodies on the cell samples at various differentiation stages. (C) Percentages of SMA and calponin positive population in normal and HGPS SMCs at day 14. Results were generated from two biological replicates. (D) Representative phase control images showing the contraction of normal and HGPS SMCs at days 14 and 21 upon angiotensin II treatment. Cell contraction at 30 min after stimulation was indicated by red arrows in the enlarged images. (Scale bar, 400 μm.) (E) Quantification of D, showing the percentage of contracted SMCs upon stimulation. Results were generated from three independent experiments.
To measure progerin amounts during SMC differentiation, Western blotting analysis and immunofluorescence were conducted, which revealed a gradual accumulation of progerin in HGPS cells during SMC differentiation (Fig. S3A–C). Senescence-associated β-galactosidase (SA–β-gal) staining suggested that HGPS SMCs were more senescent than normal SMCs at similar stages (Fig. S4A). Interestingly, p16ink4a, a senescence marker and cyclin-dependent kinase inhibitor (26), was found to be up-regulated in HGPS SMCs relative to the normal controls (Fig. S4 B and C). Next, we examined whether the senescence in HGPS SMCs was related to shortened telomeres in HGPS, as suggested previously (24). RT-PCR using a pair of human telomerase reverse transcriptase (hTERT)-specific primers revealed that hTERT was robustly expressed in iPS cells but undetectable in both normal and HGPS SMCs (Fig. S4D), indicating that telomerase expression was significantly inhibited upon SMC differentiation. Using quantitative telomere PNA-FISH, as described in Cao et al. (27), we found no evidence of shortened telomeres in HGPS SMCs as well as in primary vascular SMCs isolated from G608G transgenic mice (20), in which the SMC loss phenotype was first discovered (Fig. S4 E and F).
Progerin Accumulation Leads to Caspase-Independent Cell Death in SMCs.
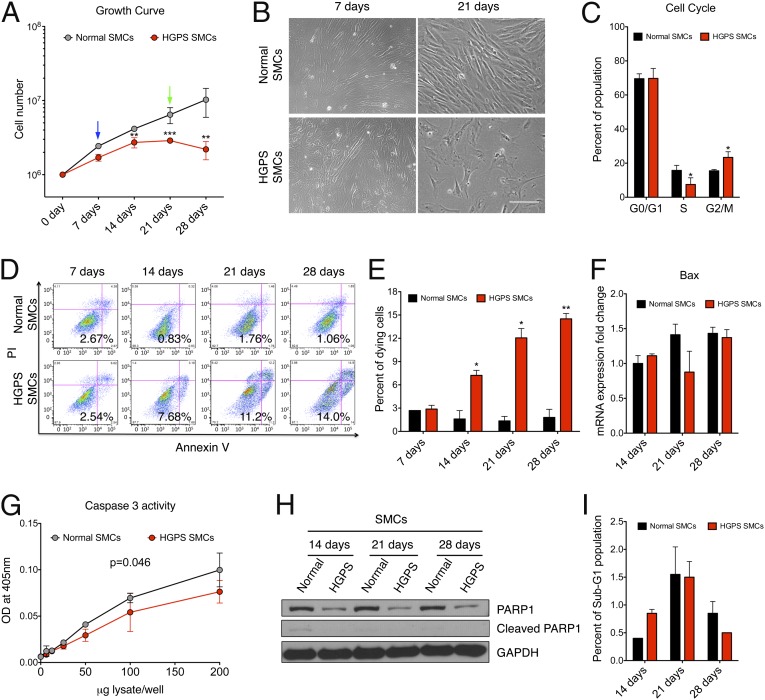
To evaluate cell proliferation potential, growth curves were plotted for HGPS and normal SMCs (Fig. 2 A and B). This experiment showed that, whereas normal SMCs kept a constant proliferation rate, the cell number of HGPS SMCs stopped increasing after day 14. We reasoned that this proliferative defect could be caused by either premature senescence or progerin-induced cell death. To distinguish these two possibilities, we conducted a cell cycle analysis at day 14. If premature senescence of HGPS SMCs was the dominant cause, the majority of HGPS SMCs should have been arrested in the G0/G1 phase of the cell cycle. To our surprise, cell cycle analysis revealed that the percentage of G0/G1 population of HGPS SMCs showed no significant changes compared with the control (∼70% in both), but there was an obvious increase of the mitotic population (G2/M) in HGPS SMCs (Fig. 2C). This result suggested that despite being more senescent than normal SMCs, HGPS SMCs did not lose their proliferative ability. Thus, we speculated that the proliferation defect in HGPS SMCs was likely caused by cell death occurring during cell division.
Fig. 2.
Increased caspase-independent cell death in HGPS SMCs. (A) Growth curve of normal and HGPS SMCs during differentiation. Results were generated from three biological replicates. *P < 0.05, **P < 0.01. Blue and green arrows referred to the images at days 7 and 21 in B, respectively. (B) Representative phase contrast images of normal and HGPS SMCs at days 7 and 21 during SMC differentiation. (Scale bar, 200 μm.) (C) Cell cycle analysis of normal and HGPS SMCs at day 14, illustrating the distribution of G0/G1, S, and G2/M phases. Results were generated from three biological replicates. *P < 0.05. (D) PI-annexin V flow cytometry analysis on normal and HGPS SMCs at various time points during SMC differentiation. The gates were set according to the positive and negative controls as suggested by the manufacturer. The cells in the Lower Right quadrant were scored as the dying cell population. (E) Quantification of D, showing the percentage of dying population based on the PI-annexin V assay. Results were generated from three biological replicates. *P < 0.05, **P < 0.01. (F) RT-qPCR analysis with Bax-specific primers on normal and HGPS SMCs at different time points during SMC differentiation. (G) Basal caspase 3 activity of normal and HGPS SMCs at day 14. Results were generated from two independent biological replicates. (H) Western blot with anti-PARP1 and GAPDH antibodies on normal and HGPS SMCs at indicated time points during differentiation. (I) Percentage of the sub-G1 population in normal and HGPS SMCs at days 14, 21, and 28.
To analyze cell death, next, we carried out propidium iodide (PI)-annexin V assay in both normal and HGPS SMCs at weekly time points during SMC differentiation. Indeed, HGPS-SMCs displayed an 8- to 10-fold increase in cell death compared with controls at days 14, 21, and 28 (Fig. 2 D and E). Notably, at the early time point day 7 when no obvious growth defect was detected (Fig. 2B), both normal and HGPS SMCs showed similar percentages of cell death, implying a relatively healthy state of HGPS SMCs at the beginning of SMC differentiation (Fig. 2 D and E). These results supported our speculation that the proliferation defect in HGPS SMCs was caused by cell death.
Because the PI-annexin V assay has frequently been applied in apoptosis studies, we measured the mRNA level of Bcl2-associated protein X (Bax), a major proapoptotic gene acting upstream of caspases (28). Interestingly, no significant change in the expression of Bax was observed in HGPS SMCs compared with the normal control (Fig. 2F), implying that caspases were not activated in the HGPS SMCs. To further investigate this, we checked the activity of caspase 3. Activity assay revealed that caspase 3 was not activated, but slightly suppressed in HGPS SMCs (Fig. 2G). Consistently, we did not detect any cleaved PARP1 protein, a well-known substrate of caspases, in either sample (Fig. 2H). Moreover, we did not find any obvious sub-G1 population in HGPS SMCs, indicating that there was no DNA fragmentation (Fig. 2I). Taken together, these data suggested that cell death in HGPS SMCs may be executed in a caspase-independent manner.
It has been shown that treatment with high dosages (micromolar) of camptothecin (CPT), an isotopomerase II inhibitor, can induce caspase-dependent apoptosis in normal cells (29). Thus, as a positive control to the above experiments, we treated both normal and HGPS SMCs with 20 μM CPT. As shown in Fig. S5, in control SMCs, we detected an up-regulation of Bax mRNA by 4-fold and caspase 3 activity by nearly 10-fold after 72 h in CPT treatment (Fig. S5 A and B). Consistently, cleaved PARP1 was clearly detected by Western blotting in these cells (Fig. S5C). However, HGPS SMCs appeared quite unresponsive to CPT induction, which failed to activate Bax expression, and exhibited a delay of over 48 h in caspase 3 activation and PARP1 cleavage (Fig. S5 A–C). PI-annexin V assay further confirmed no significant increase in cell death in HGPS SMCs after CPT treatment (Fig. S5D). These results suggest that, in the presence of progerin, caspases become less responsive to apoptotic stimuli like CPT. Collectively, we concluded that progerin accumulation in SMCs led to increased cell death, likely via caspase-independent pathways.
Progerin Accumulation Down-Regulates PARP1 in SMCs.
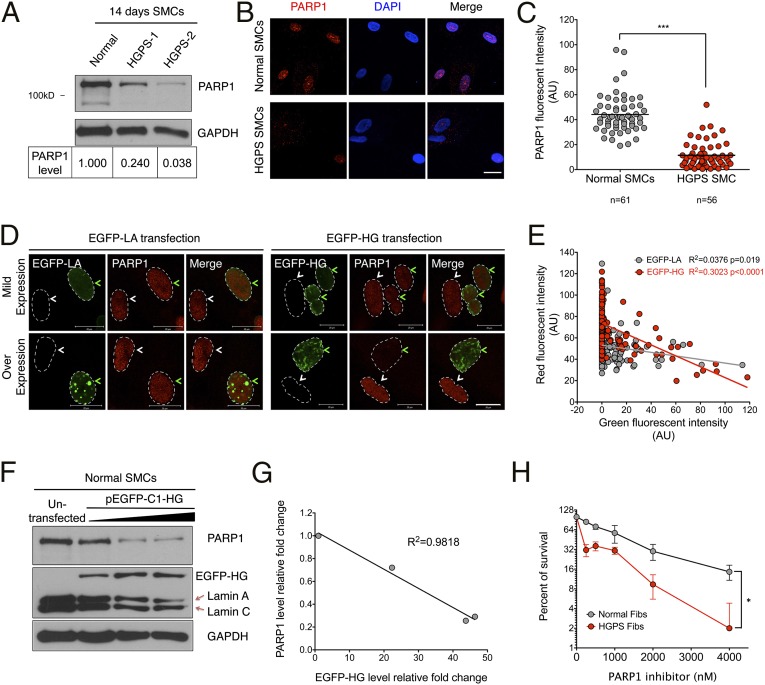
When probing PARP1 cleavage in the caspase studies, we observed that the levels of PARP1 protein were always lower in HGPS SMCs compared with the normal control at days 14, 21, and 28 (Fig. 2H and Fig. S5C), whereas no obvious change was detected at day 7 (Fig. S6A). To confirm this observation, we measured PARP1 levels in two different HGPS SMC lines at day 14 and found that the PARP1 levels in both were decreased by at least 75% relative to the control (Fig. 3A). Immunofluorescence showed a significant decrease in PARP1 nuclear staining in HGPS SMCs at day 14, compared with the control (Fig. 3 B and C). Moreover, in primary skin fibroblasts from HGPS patients and a transgenic animal model G608G (20), similar PARP1 reduction was found in late passage cells (Fig. S6 B–E). Resembling the cell cycle analysis in HGPS SMCs (Fig. 2C), a significant increase in the G2/M population was detected in late passage HGPS fibroblasts (Fig. S6 F and G). Notably, there was no significant change in PARP1 protein or cell cycle distribution in early passage HGPS fibroblasts, similar to the day 7 HGPS SMCs (Fig. S6A). This suggests that the PARP1 reduction phenotype is not exclusive for SMCs, but a consequence of progerin accumulation in each cell type. As an initial executor of SSB repair and a key mediator of DNA DSB repair, PARP1 plays a crucial role in maintaining genome stability. Thus, we next examined whether the PARP1 reduction was relevant to increased cell death in progerin-expressing SMCs.
Fig. 3.
Progerin down-regulates PARP1. (A, Upper) Western blot with anti-PARP1 and GAPDH antibodies on two HGPS SMC lines (HGADFN167 and HGADFN164) and a normal control SMC line at day 14. (Lower) Quantification of PARP1 in each sample. The intensity of PARP1 was first normalized to corresponding GAPDH and then normalized to the control SMC. (B) Representative images of immunofluorescence staining with anti-PAPR1 antibody in normal and HGPS SMCs at day 14. (Scale bar, 50 μm.) (C) Quantification of B, showing the PARP1 red fluorescence intensity. ***P < 0.001. (D) Representative images of immunofluorescence staining with anti-PARP1 in normal SMCs transfected with pEGFP-C1-LA or pEGFP-C1-HG plasmids. The transfected and untransfected cells are indicated by green and white arrows, respectively. The nuclei were outlined by white dashed lines. (Scale bar, 20 μm.) (E) Quantification of D, showing the correlation between EGFP-HG or EGFP-LA fluorescence intensity (x axis) and PARP1 fluorescent intensity (y axis). More than 100 cells were randomly picked for each group. (F) Western blot with anti-PARP1 and GAPDH antibodies on normal SMCs transfected with different amounts of pEGFP-C1-HG plasmid. (G) Quantification of F, showing a negative-linear relationship between PARP1 (y axis) and progerin (x axis) protein amounts. Signals were normalized to the untransfected control. (H) Survival curve of early passage normal and HGPS fibroblasts upon a PARP1 inhibitor (Olaparib) treatment. Results were generated from two independent experiments. *P < 0.05.
To determine the effects of progerin on PARP1, normal SMCs were transfected with EGFP-progerin (EGFP-HG) or EGFP-lamin A (EGFP-LA) and subsequently stained with an anti-PARP1 antibody. As shown in Fig. 3D, the PARP1 staining intensities were similar in the nontransfected cells (white arrows) as those in the cells expressing EGFP-LA or low amounts of EGFP-HG (green arrows). In contrast, in high expressers of EGFP-HG, PARP1 signals were significantly reduced (Fig. 3D). Quantification of fluorescence intensity further indicated that, as the amounts of EGFP-HG increased, PARP1 decreased at a significantly faster rate than when EGFP-LA amounts increased (Fig. 3E), supporting the belief that PARP1 reduction is associated with the amount of progerin.
Next, to directly investigate the causative relationship between progerin accumulation and PARP1 down-regulation, increasing amounts of EGFP-HG were transfected and expressed in normal SMCs. We found that the levels of PARP1 gradually decreased as the amounts of EGFP-HG increased (Fig. 3F). Quantification confirmed a negative linear relationship between the amounts of PARP1 and EGFP-HG (R2 = 0.9818, Fig. 3G). Notably, when progerin was ectopically expressed, a corresponding decrease of endogenous lamin A and C was observed, implying a potential feedback loop in SMCs to control the total amount of A-type lamins. Moreover, down-regulation of the progerin expression with a previously described progerin shRNA (24) led to a significant increase in PARP1 in HGPS SMCs (Fig. S7). Taken together, these results show that progerin resulted in PARP1 reduction in SMCs. In an attempt to address the functional importance of PARP1 in progerin-expressing cells, we applied a PARP1 inhibitor to the early passage HGPS fibroblasts, which expressed similar amounts of PARP1 as the control cells (Fig. S6B). As shown in Fig. 3H, HGPS fibroblasts showed much higher sensitivity to the PARP1 inhibitor than control cells (Fig. 3H), suggesting an indispensable role of PARP1 in progeria cell survival.
PARP1 Mislocalization Is Associated with Disrupted Ran Gradient in HGPS SMCs.
To investigate how progerin down-regulates PARP1, we compared PARP1 mRNA levels in normal and HGPS SMCs using RT-qPCR. Furthermore, we overexpressed progerin in normal SMCs and examined its effect on PARP1 transcription (Fig. S8). These data revealed no major changes in mRNA levels of PARP1 in those progerin-expressing SMCs, suggesting the observed down-regulation of PARP1 might result from compromised protein maintenance.
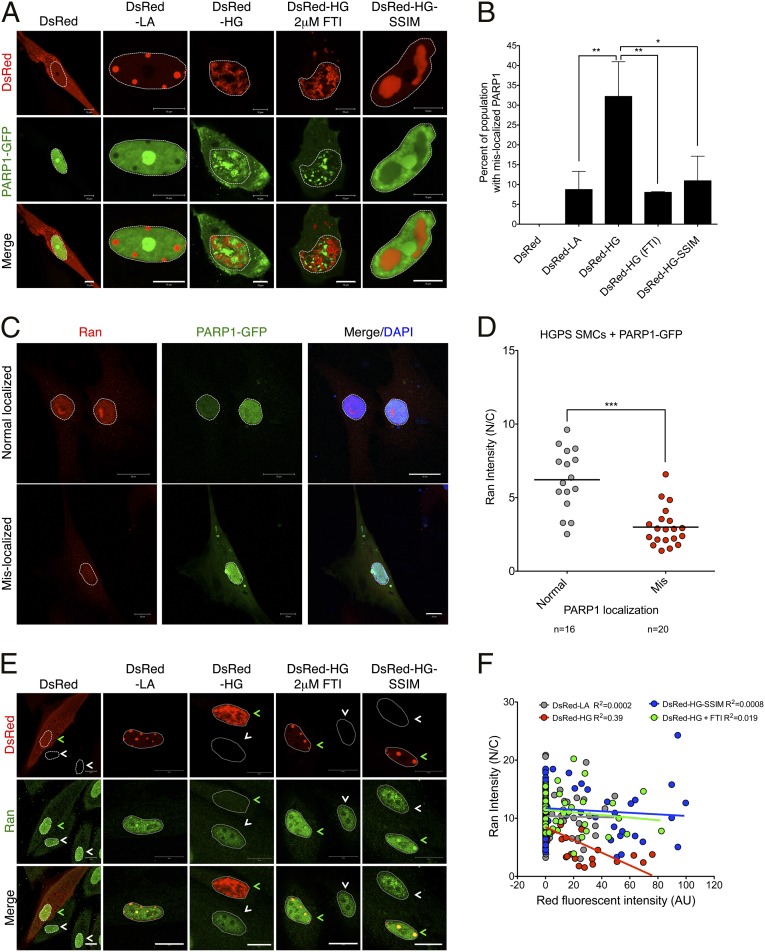
To examine how progerin may affect PARP1 protein stability, we overexpressed PARP1-GFP, as described previously (30), together with DsRed-LA or DsRed-HG in normal SMCs. As expected, PARP1-GFP was localized inside the nucleus when coexpressed with the vector control or DsRed-LA (Fig. 4A). In addition, consistent with a previous study (31), we observed that PARP1-GFP became concentrated in the nucleoli when overexpressed (Fig. 4A), reflecting its roles in ribosome biogenesis. However, in DsRed-HG transfected SMCs, more than 30% of cells showed cytoplasmic localization of PARP1-GFP (Fig. 4 A and B). These results implied that accumulated progerin excluded a fraction of PARP1 from the nucleus, and this fraction might be subjected to protease/proteasome-mediated degradation.
Fig. 4.
Progerin induces PARP1 mislocalization and Ran gradient disruption. (A) Representative images showing normal SMCs coexpressing PARP1-GFP with DsRed, DsRed-LA, DsRed-HG (with or without 2 μM FTI), or DsRed-HG-SSIM. Nuclei was outlined by white dashed lines. (Scale bar, 10 μm.) (B) Quantification of A, showing the percentage of PARP1-GFP–mislocalized cells under indicated conditions. More than 100 cells were counted for each group. Results were generated from three independent experiments. *P < 0.05, **P < 0.01. (C) Representative images of immunofluorescence staining with anti-Ran antibody on HGPS SMCs transfected with PARP1-GFP. Cells with both normally or mislocalized PARP1-GFP were shown. The nuclei were outlined by white dashed lines. (Scale bar, 20 μm.) (D) Quantification of C, showing the Ran gradient in HGPS SMCs transfected with PARP1-GFP. ***P < 0.001. (E) Representative images of immunofluorescence staining with anti-Ran antibody in normal SMCs transfected with DsRed, DsRed-LA, DsRed-HG (with or without 2 μM FTI) or DsRed-HG-SSIM. The transfected and untransfected cells were indicated by green and white arrows, respectively. The nuclei were outlined by white dashed lines. (Scale bar, 20 μm.) (F) Quantification of E, showing the correlation between DsRed fluorescent intensity and Ran gradient (Ran N/C) in N and C refer to the nucleus and cytoplasm, respectively. More than 50 cells were randomly picked for each group.
Next, we tested whether blocking the farnesylation of progerin could restore PARP1’s normal localization. To study this possibility, we used HG-SSIM, a nonfarnesylatable mutant of progerin that carries a cysteine to serine mutation on the CSIM motif at the C terminus (32), and coexpressed it with PARP1-GFP in normal SMCs. As reported previously (32), DsRed-HG-SSIM no longer located at the inner nuclear membrane but was sequestered in giant aggregates in nucleoplasm (Fig. 4A). Importantly, in the majority of the DsRed-HG-SSIM–transfected cells, we observed that the normal nuclear localization of PARP1-GFP was restored (Fig. 4A). Quantification revealed that the percentage of PARP1-mislocalized cells was significantly decreased to 10% in the DsRed-HG-SSIM–expressing cells, compared with roughly 30% in DsRed-HG–expressing cells (Fig. 4B). To further confirm this result, a FTI was used to treat DsRed-HG–expressing cells. As expected, the FTI treatment also rescued the mislocalization phenotype of PARP1-GFP and significantly reduced the mislocalized population (Fig. 4 A and B). These data suggested that the abnormal anchorage of progerin to the nuclear envelope was the main causes of PARP1 mislocalization.
We speculated that the PARP1 mislocalization in HGPS SMCs might be caused by a compromised nuclear import system mediated by a disrupted Ran gradient, which has been reported in HGPS fibroblasts (33, 34). To test this idea, we first examined the Ran gradient in normal and HGPS SMCs at days 7, 14, and 21 using immunofluorescence. As shown in Fig. S9A, HGPS SMCs displayed significantly decreased Ran signals in the nucleus, compared with the control cells, indicating that the Ran gradient [nuclear/cytoplasmic (N/C) ratio, Fig. S9B] was disrupted in HGPS SMCs. Moreover, HGPS SMCs at days 14 and 21 exhibited even lower N/C ratios than the same cells at day 7 (Fig. S9B), which is in agreement with the observation of a gradual reduction of PARP1 levels over time in HGPS SMCs. To further test the correlation between Ran gradient disruption and PARP1 mislocalization, HGPS SMCs at day 14 were transfected with PARP1-GFP and subsequently stained with Ran antibody. About 30% of the transfected HGPS SMCs displayed the PARP1 mislocalization phenotype (Fig. 4C). Importantly, these cells also showed significantly lower N/C ratios compared with the cells with normal nuclear PARP1-GFP (Fig. 4D).
Similar to Fig. 4A, we examined the Ran gradient in normal SMCs transfected with DsRed-HG or DsRed-LA. Unlike LA, which did not interfere with the Ran gradient (Fig. 4E), HG expression significantly weakened nuclear signals of Ran (Fig. 4E). Quantification revealed that the N/C ratios of Ran negatively correlated with progerin amounts (Fig. 4F). Furthermore, inhibition of progerin farnesylation through FTI or HG-SSIM mutant rescued the Ran gradient disruption (Fig. 4 E and F). Collectively, these data suggested that abnormal anchorage of progerin to the nuclear envelope disrupted Ran gradients, thereby affecting nuclear import of PARP1 in HGPS SMCs.
Compromised HR and Activated NHEJ in S/G2 Phase HGPS SMCs.
Previous studies reported that inhibition or silencing of PARP1 could lead to cell death in HR-deficient cells, due to NHEJ activation and consequent chromosome misarrangments (7). Given PARP1’s essential function in HR-deficient cells, we next examined the repair efficiencies of HR and NHEJ in HGPS SMCs.
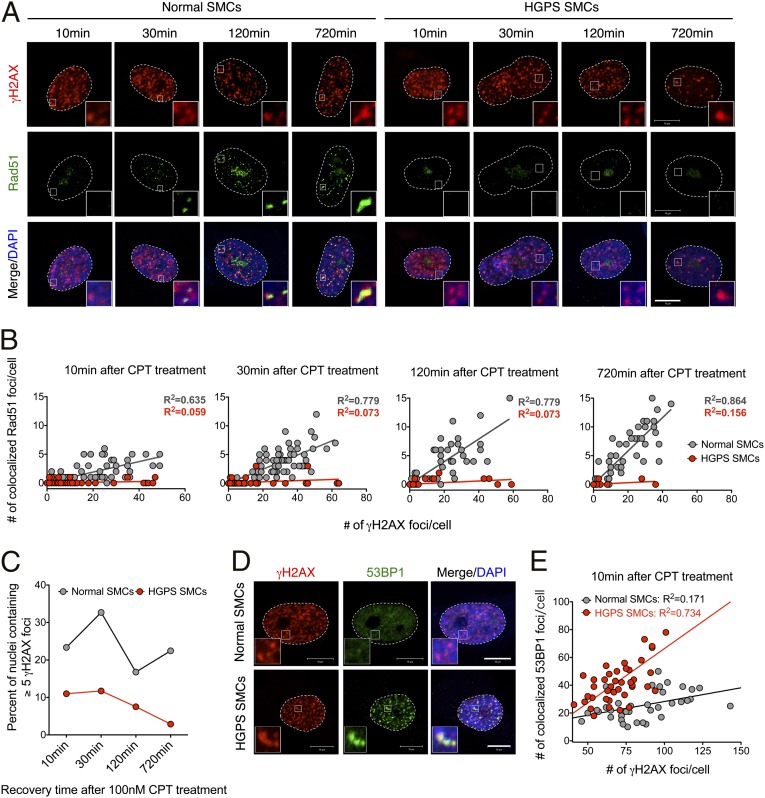
Because HR is the major mechanism for replication-associated DSB repair in S/G2 phase, we induced DNA damage with CPT again to mimic replication fork collapse (1). Different from the CPT experiment shown in Fig. S5, here, CPT was used at a very low concentration (100 nM), thus only S/G2 phase cells were affected and no global apoptosis was induced (35). At 10 min, 30 min, 120 min, and 720 min after CPT treatment, γH2AX and Rad51 signals were examined using immunofluorescence. After 10 min of CPT treatment, γH2AX foci were clearly identified in both normal and HPGS-SMCs, indicating that CPT had induced DSBs in both (Fig. 5A). Rad51 plays an essential role in HR-mediated DSB repair. In normal SMCs, the formation of Rad51 foci started to appear at the 30-min time point, and at 720 min, most of these foci became colocalized with γH2AX foci (Fig. 5A). Conversely, no distinct Rad51 foci were observed in HGPS SMCs throughout the whole time course (Fig. 5A). To elucidate this phenomenon, over 100 cells were randomly picked at each time point, and each cell was plotted according to the numbers of γH2AX and colocalized Rad51 foci (as x and y). In normal SMCs, Rad51 foci became increasingly colocalized with γH2AX foci as shown by an increase in the slope of the trend line during the time course. However, in HGPS SMCs, the slope of the trend line remained around 0 at all time points (Fig. 5B), revealing a completely defective HR in these HGPS SMCs. Notably, the percentage of γH2AX-positive nuclei in Fig. 5C is reduced in HGPS SMCs compared with normal controls under 100 nM CPT treatment, probably due to the reduced S phase cell population in HGPS SMCs (Fig. 2C and Fig. 6A).
Fig. 5.
HGPS SMCs show impaired HR and activated NHEJ. (A) Representative image showing immunofluorescence staining with anti-γH2AX and Rad51 antibodies on normal and HGPS SMCs recovered for various amounts of time after 100 nM CPT treatment. (Scale bar, 10 μm.) (B) Quantification of A, showing of the number of γH2AX (x axis) and colocalized Rad51 (y axis) foci in normal and HGPS SMCs at different time points. More than 100 cells were counted for each case. The trend lines of normal (in gray) and HGPS (in red) SMCs were also presented. (C) The percentage of nuclei containing more than five γH2AX foci in normal and HGPS SMCs at each indicated time point after 100 nM CPT treatment. (D) Representative images of immunofluorescence staining with anti-γH2AX and 53BP1 antibodies on normal and HGPS SMCs at 10 min after 100 nM CPT treatment. (Scale bar, 10 μm.) (E) Quantification of D, showing the number of γH2AX (x axis) and colocalized 53BP1 (y axis) foci in normal and HGPS SMCs at different time points. Over 50 cells were analyzed.
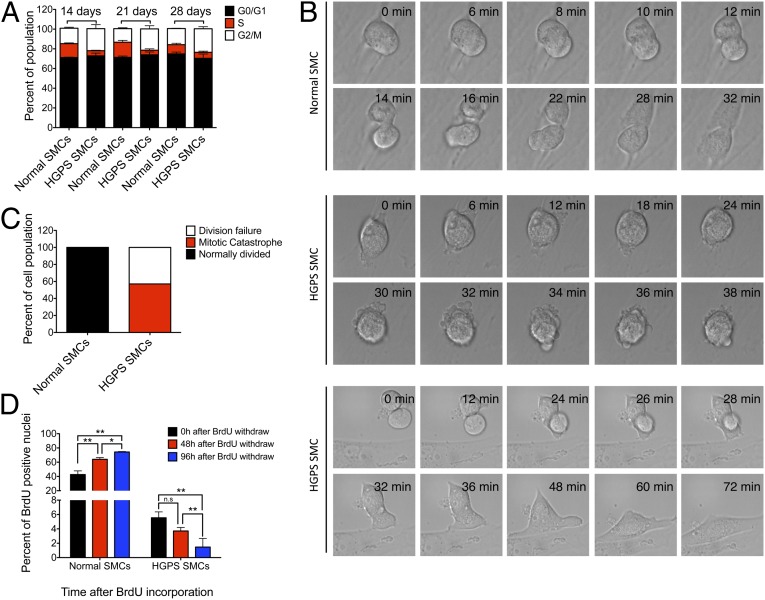
Fig. 6.
Mitotic catastrophe in HGPS SMCs. (A) Cell cycle analysis of the normal and HGPS SMCs at denoted time points. Results were generated from two biological replicates. (B) DIC live cell imaging of normal and HGPS SMCs at day 21. (Top) Normal mitotic division of a normal SMC; (Middle) mitotic catastrophe of an HGPS SMC; and (Bottom) failed mitosis of an HGPS SMC. (C) Percentage of normal and defective mitosis in normal and HGPS SMCs. n = 4 and 7 for normal and HGPS SMCs, respectively. (D) Quantification of the percentages of BrdU-positive nuclei at 48 h and 96 h after BrdU pulse in normal and HGPS SMCs at day 14. Results were generated from two independent experiments. *P < 0.05 and **P < 0.01.
With decreased PARP1 and a completely defective HR, DSBs will be fixed through the error-prone mechanism NHEJ. Indeed, in HGPS SMCs, the percentile of γH2AX-positive nuclei and the average number of γH2AX foci in each nucleus were gradually reduced after CPT treatment (Fig. 5 B and C), suggesting the NHEJ pathway was active in repairing these DSBs caused by CPT. 53BP1 can promote an NHEJ response to DNA damage (36). To assay the activity of NHEJ in S phase HGPS SMCs, we checked the formation of 53BP1 foci after 100 nM CPT treatment. We found that, whereas no apparent 53BP1 foci formed in normal SMCs, distinct 53BP1 foci started to colocalize with γH2AX at 10 min after treatment in HGPS SMCs (Fig. 5 D and E), suggesting that normal SMCs used HR to fix DSBs at S phase, whereas HGPS SMCs repair DSBs through NHEJ.
Mitotic Catastrophe in HGPS SMCs.
Based on the observations of increased caspase-independent cell death, down-regulated PARP1, defective HR, and activated NHEJ, we hypothesized that DSBs were repaired incorrectly by NHEJ, which resulted in problems during mitotic division, leading to mitotic catastrophe of HGPS SMCs. To test this hypothesis, cell cycle analysis was performed at days 14, 21, and 28 in SMC differentiation medium. Consistent with the data shown in Fig. 2, at each time point, we observed a significant increase in the G2/M population and a corresponding reduction in S phase population in HGPS SMCs compared with the normal control (Fig. 6A). This observation further supported the notion that mitosis is defective in HGPS. Taken together, this result indicated prolonged mitosis in HGPS SMCs, reflecting mitotic defects.
Next, to directly visualize the M phase of HGPS SMCs, we performed differential interference contrast (DIC) live cell imaging. Whereas all normal SMCs proceeded through mitosis smoothly (Fig. 6B, Top), HGPS SMCs either died and were destroyed during cell division (mitotic catastrophe) or exited mitosis without dividing but survived (Fig. 6B, Middle and Bottom, respectively). Statistic analysis confirmed that about 50% of the HGPS SMCs displayed mitotic catastrophe, and the rest failed to divide (Fig. 6C). In support of this observation, metaphase spectral karyotyping revealed ∼6.7% aneuploidy and 20% polyploidy in dividing HGPS SMCs (Fig. S10). Next, a BrdU pulse-chase experiment was conducted to follow the fate of S phase SMCs. Forty-eight and 96 h after the BrdU pulse, immunofluorescence staining of BrdU was carried out. In normal SMCs, the percentage of the BrdU positive nuclei gradually increased from 20% to 80%, reflecting normal cell division. In contrast, the BrdU-positive nuclei in HGPS SMCs gradually decreased from 6% at time 0 h to less than 2% at 96 h after pulse (Fig. 6D). This result indicated that over 60% of the S phase HGPS SMCs were not able to proceed through the cell division and died during cell division.
Discussion
Differentiated SMCs from iPS Cells.
Children with progeria die almost exclusively from cardiovascular disease. In 2006, Varga et al. reported the first evidence of progressive vascular SMC loss in large vessels of an HGPS transgenic mouse model (G608G BAC) (20). SMC loss might undermine vessel architecture and induce inflammation, vessel calcification, and excessive atherosclerosis (19). The mechanisms underlying progerin-induced SMC loss had not been explored, which was at least partially due to the technical difficulties in obtaining sufficient amounts of vascular SMCs from progeria animal models. Here, this problem was overcome by the iPS-mediated differentiation system that generated a large amount of patient SMCs. We found that these differentiated cells were induced to express a number of established SMC markers and exhibited functional characteristics of SMCs (Fig. 1, Fig. S2, and Movies S1 and S2). However, the expression of smMHC, a marker for fully differentiated SMCs (37), was not up-regulated (Fig. S2B), which suggested that the SMCs in our experiments were not terminally differentiated. Indeed, the proliferation assay (Fig. 2 A and B) showed that the normal SMCs could proliferate in SMC differentiation medium for at least 28 d. In vivo, SMC proliferation is a characteristic of synthetic SMCs, and contractile SMCs do not proliferate (37). Under pathological conditions, SMCs switch from the contractile to the synthetic phenotype (37, 38). Thus, we speculate that by using these iPS-generated SMCs, we may be studying how progerin affects SMCs responses to pathological conditions in vivo.
The Proliferative Defect in HGPS SMCs Is Primarily Caused by Caspase-Independent Cell Death, Not by Premature Senescence.
The HGPS SMCs expressed progerin, a hallmark of HGPS cells (Fig. S3), and stopped proliferating after 2 wk in SMC culture (Fig. 2 A and B). Initially, we had hypothesized that this severe growth defect was caused by premature senescence, as previously suggested by Liu and colleagues (24) and supported by our senescence analysis with SA–β-Gal assay and p16 expression (Fig. S4 A–C). However, to our surprise, cell cycle analysis indicated that the HGPS SMCs did not rest at G0, but actively reentered the cell cycle (Figs. 2C and 6A). Moreover, we found a significant delay in G2/M phase in the HGPS SMCs (Figs. 2C and 6A). Additional experiments showed that HGPS SMCs died in SMC differentiation medium in a caspase-independent manner (Fig. 2 D–I and Fig. S5). Based on these results, we concluded that this caspase-independent cell death accounted for the proliferation phenotypes in HGPS SMCs.
PARP1 Is a Key Regulator for SMC Survival in Progeria.
In addition to its well-recognized role in DNA SSB repair, several studies revealed a new role of PARP1 in protecting HR and suppressing NHEJ (6, 39, 40). Here, we found that the PARP1 protein levels were reduced in HGPS SMCs after day 14 in SMC culture as well as in late passage HGPS fibroblasts (Fig. 3 A and B and Fig. S6 A–E). Importantly, the progerin overexpression (Fig. 3 F and G) and shRNA experiments (Fig. S7) revealed a direct inverse relationship between the amounts of PARP1 and progerin (Fig. 3 F and G), indicating that progerin is a powerful suppressor of PARP1. The PARP1 inhibitor experiment in HGPS fibroblasts further revealed an essential role of PARP1 in cell survival (Fig. 3H). We speculate that the sensitivity of PARP1 to progerin may vary in different cell types. In fibroblasts, PARP1 may be less influenced by progerin accumulation than in SMCs, therefore PARP1 down-regulation was not obvious until late passage fibroblast cells. These data also suggest that PARP1 is indispensable in all progerin-expressing cell types, and the levels of PARP1 reduction may be positively correlated with the severity of phenotypes in HGPS patients.
Our next question was how does progerin inhibit PARP? Quantitative RT-PCR analysis showed a slight, insignificant reduction in PARP1 mRNA in the presence of progerin, suggesting that progerin likely affects PARP1 posttranslationally (Fig. S8). Microscopy analysis further revealed that a fraction of PARP1 was mislocalized to the cytosol in the presence of large amounts of progerin (Fig. 4 A and B), and this progerin-mediated PARP1 mislocalization was likely due to the abnormal anchorage of progerin to the nuclear envelope (Fig. 4 A and B). Previous studies demonstrated abnormal nuclear pore clustering and a disrupted Ran gradient across the nuclear membrane in HGPS fibroblasts (13, 33, 34). We speculated that this mislocalization of PARP1 was a novel effect of the disrupted RanGTP gradient. Indeed, a weakened Ran gradient (Fig. S9) and a strong correlation between these two (Fig. 4 C–E) were found in HGPS SMCs.
It has been shown that NHEJ is active in all cell cycle phases, whereas HR is particularly important in S/G2 (1). Moreover, DSBs produced by replication fork inhibitors are primarily repaired by HR (1). Using an isotopomerase II inhibitor (CPT) at a nanomolar concentration that only affects S/G2 phase cells, our analysis revealed a completely deficient HR and an elevated NHEJ response in S phase HGPS SMCs (Fig. 5). When the error-prone NHEJ pathway is used in DNA repair, irreversible chromosome aberrations can occur, resulting in complicated consequences (6, 7). Most commonly, cells will spend more time in M phase, attempting to resolve the problems (41). If entangled chromosomes are unresolvable, cells will die through mitotic catastrophe (41). Indeed, our cell cycle analysis revealed a palpable G2/M delay in HGPS SMCs (Fig. 6A) and late passage fibroblasts (Fig. S6 F and G), suggesting that these cells were encountering problems during cell division. Live cell imaging and spectral karyotyping experiments further demonstrated that HGPS SMCs either died in mitosis or failed to divide, forming binucleated cells (Fig. 6 B and C and Fig. S10). An additional line of supportive evidence arose from a BrdU pulse-chase assay, which indicated that most of the labeled S phase HGPS SMCs could not proceed through the M phase (Fig. 6D). Previous studies also have shown mitotic delay and binucleation in HGPS fibroblast cells (14). Again, the mitotic delay and cell death in HGPS SMCs were more prominent than those observed in HGPS fibroblasts, possibly due to a greater PARP1 reduction in HGPS SMCs.
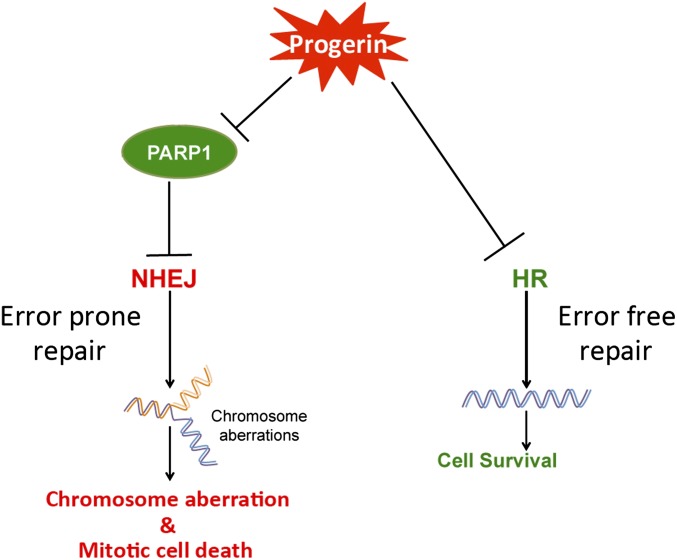
DNA damage has long been implicated in HGPS pathogenesis. In 2005, the Zhou group published a study elucidating a deficient HR, an overactive NHEJ, and defective DNA damage repair in primary HGPS fibroblasts and mouse embryonic fibroblasts (MEFs) from a progeria model (16). In 2011, Zhang and colleagues generated the first iPS cell model for HGPS and reported increased DNA damage in differentiated HGPS SMCs (42). In addition, Liu et al. reported a decrease of DNA PKCs in differentiated HGPS SMCs, an enzyme with an established role in the NHEJ response (24). In line with these previous reports, we reported a previously unidentified and essential role of PARP1 in mediating DNA damage repair and SMC survival in HGPS. Our data can be summarized in the model presented in Fig. 7: the accumulation of progerin causes PARP1 down-regulation, which results in the activation of the NHEJ pathway in the HR-deficient HGPS SMCs. The error-prone NHEJ further leads to chromosome aberrations and mitotic catastrophe in HGPS SMCs.
Fig. 7.
Working model. The accumulation of membrane-associated progerin not only causes HR deficiency via an unknown mechanism, but also causes PARP1 down-regulation, likely through interfering with PARP1 nuclear import. This PARP1 reduction results in the activation of the NHEJ pathway in the HR-deficient HGPS SMCs, leading to error-prone DNA repair, chromosome aberration, and mitotic catastrophe in HGPS SMCs.
Insights into Future Therapeutics of HGPS and Normal Vascular Aging.
Based on our model, to rescue the SMC loss in HGPS, one solution is to restore normal HR function. Whereas the precise pathway for progerin to interfere with HR remains to be understood, previous studies have provided some valuable insights. One emerging hypothesis is that progerin disrupts chromatin remodeling after DSBs, hence blocking the access of DNA damage repair factors to damaged regions. This idea is supported by a number of studies demonstrating global changes in genome organization and the epigenome in HGPS cells (43, 44). Importantly, recent studies have suggested that changes in H3K9me3 levels in HGPS cells can affect genome maintenance and the compromised ATM-mediated phosphorylation of KAP-1 impairs DNA repair in Zmpste24-deficient MEFs (17, 18), providing a possible molecular connection between chromatin modifications and the DNA repair machinery. Other lines of studies have suggested a role of Xeroderma pigmentosum group A (XPA) protein in excluding normal access of DNA repair machinery to DSBs in HGPS cells (45, 46). Whereas the detailed mechanisms underlying HR deficiency in HGPS are still being elucidated, we predict that specific treatments that promote targeting of HR repair proteins to DSB termini are expected to reduce or diminish the SMC loss phenotype associated with progerin accumulation.
Increasing evidence suggests the involvement of progerin in the normal aging process (27, 47, 48). Significantly, overlapping cardiovascular pathology was identified in HGPS and geriatric patients (49). Additional studies demonstrated an accumulation of unprocessed farnesylated prelamin A (a longer version of progerin) in vasculature (50, 51). It will be of great interest to determine whether HR becomes partially impaired in aged vasculature and whether a similar PARP1-mediated mitotic cell death mechanism also contributes to normal vascular aging. In summary, the present study uncovers what we believe is the first study to elucidate the DNA damage response pathway underlying the vascular disease in HGPS.
Methods
Cell Lines.
Primary skin fibroblasts carrying the classic G608G mutation (HGADFN167, HGADFN164) and a control fibroblast line (HGFDFN168) were obtained from the Progeria Research Foundation and grown under the conditions as previously described (14). These fibroblasts were transfected with retroviral mixtures of KLF4, SOX2, OCT4, and C-MYC for iPS cell induction, as reported previously (52). Two independently isolated iPS colonies from HGPS HGADFN167 and normal HGFDFN168 (father of HGADFN167), as well as another previously characterized HGPS iPS colony (HGADFN164) (52), were used in this study. In addition, we also tested two iPS lines isolated and characterized by the Progeria Research Foundation (HGADFN167 iPS 1J and HGFDFN168 iPS 1D2). All HGPS iPS colonies revealed very similar results. Only representative experiments are shown.
Antibodies.
Rabbit anti-progerin was previously described (53). Other antibodies used in immunoblotting and immunofluorescence studies were obtained from the following sources: mouse anti-lamin A/C (Mab3211; Millipore), mouse Cy3-anti-SMA (C6198; Sigma), rabbit anti-calponin (04–589; Millipore), mouse anti–β3-tubulin (sc-58888; Santa Cruz), mouse anti-vimentin (sc-6260; Santa Cruz), mouse anti–α-fetoprotein (sc-166325; Santa Cruz), rabbit anti-p16ink4a (sc-468; Santa Cruz), rabbit anti-PARP1 (9542; Cell Signaling), mouse anti-PARP1 (F-2; Santa Cruz), rabbit anti-Ran (Abcam), mouse anti-γH2AX (05–636; Millipore), rabbit anti-Rad51 (ab63801; Abcam), mouse anti-BrdU (555627; BD) and mouse anti-GADPH (ab8245; Abcam). Antibodies used for iPS cell characterization were previously described (52).
Statistical Analysis.
To compare two groups (normal and HGPS), a two-tailed Student t test was used. Results are presented as mean ± SD, and a P value of <0.05 was considered significant. Statistics were carried out by GraphPad Prism 5.0.
For full details of other methods, see SI Methods.
Supplementary Material
Acknowledgments
We thank members in the K.C. laboratory, Dr. Francis Collins [National Institutes of Health (NIH)] and Dr. Steven Fisher (University of Maryland School of Medicine) for helpful discussions; Ken Class and Amy Beaven (flow cytometry and imaging cores at the University of Maryland College Park) for technical support; Drs. Norma Andrews and Andrew Flannery (University of Maryland) for help on the live cell imaging; Dr. George Daley (Harvard University) for sharing the pMIG retroviral vectors; Dr. Amalia Dutra (National Human Genome Research Institute/NIH) for her help on SKY analysis; and Dr. Guy Poirier (Université Laval) for the generous gift of PARP1 plasmids. This work was supported by NIH/National Institute on Aging Grant R00AG029761 (to K.C.) and by Maryland Stem Cell Fund exploratory Grant 2011-MSCRFE-0124-00 (to K.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.-M.F. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320843111/-/DCSupplemental.
References
- 1.Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23(16):5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: Causal or correlative? Nucleic Acids Res. 2007;35(22):7417–7428. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraemer KH, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145(4):1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 5.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: Mitotic catastrophe. Cell Death Differ. 2008;15(7):1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 6.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci USA. 2011;108(8):3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon LB, et al. Disease progression in Hutchinson-Gilford progeria syndrome: Impact on growth and development. Pediatrics. 2007;120(4):824–833. doi: 10.1542/peds.2007-1357. [DOI] [PubMed] [Google Scholar]
- 9.Merideth MA, et al. Phenotype and course of Hutchinson-Gilford progeria syndrome. N Engl J Med. 2008;358(6):592–604. doi: 10.1056/NEJMoa0706898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423(6937):293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capell BC, Collins FS. Human laminopathies: Nuclei gone genetically awry. Nat Rev Genet. 2006;7(12):940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 12.Fong LG, et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J Clin Invest. 2006;116(3):743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101(24):8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao K, Capell BC, Erdos MR, Djabali K, Collins FS. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc Natl Acad Sci USA. 2007;104(12):4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dechat T, et al. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc Natl Acad Sci USA. 2007;104(12):4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, et al. Genomic instability in laminopathy-based premature aging. Nat Med. 2005;11(7):780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- 17.Liu B, Wang Z, Ghosh S, Zhou Z. Defective ATM-Kap-1-mediated chromatin remodeling impairs DNA repair and accelerates senescence in progeria mouse model. Aging Cell. 2013;12(2):316–318. doi: 10.1111/acel.12035. [DOI] [PubMed] [Google Scholar]
- 18.Liu B, et al. Depleting the methyltransferase Suv39h1 improves DNA repair and extends lifespan in a progeria mouse model. Nat Commun. 2013;4:1868. doi: 10.1038/ncomms2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López-Candales A, et al. Decreased vascular smooth muscle cell density in medial degeneration of human abdominal aortic aneurysms. Am J Pathol. 1997;150(3):993–1007. [PMC free article] [PubMed] [Google Scholar]
- 20.Varga R, et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2006;103(9):3250–3255. doi: 10.1073/pnas.0600012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villa-Bellosta R, et al. Defective extracellular pyrophosphate metabolism promotes vascular calcification in a mouse model of Hutchinson-Gilford progeria syndrome that is ameliorated on pyrophosphate treatment. Circulation. 2013;127(24):2442–2451. doi: 10.1161/CIRCULATIONAHA.112.000571. [DOI] [PubMed] [Google Scholar]
- 22.Capell BC, et al. A farnesyltransferase inhibitor prevents both the onset and late progression of cardiovascular disease in a progeria mouse model. Proc Natl Acad Sci USA. 2008;105(41):15902–15907. doi: 10.1073/pnas.0807840105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osorio FG, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3(106):ra107. doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 24.Liu GH, et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature. 2011;472(7342):221–225. doi: 10.1038/nature09879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie CQ, et al. A highly efficient method to differentiate smooth muscle cells from human embryonic stem cells. Arterioscler Thromb Vasc Biol. 2007;27(12):e311–e312. doi: 10.1161/ATVBAHA.107.154260. [DOI] [PubMed] [Google Scholar]
- 26.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao K, et al. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest. 2011;121(7):2833–2844. doi: 10.1172/JCI43578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74(4):609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 29.Ulukan H, Swaan PW. Camptothecins: A review of their chemotherapeutic potential. Drugs. 2002;62(14):2039–2057. doi: 10.2165/00003495-200262140-00004. [DOI] [PubMed] [Google Scholar]
- 30.Haince JF, et al. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283(2):1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 31.Boamah EK, Kotova E, Garabedian M, Jarnik M, Tulin AV. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012;8(1):e1002442. doi: 10.1371/journal.pgen.1002442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capell BC, et al. Inhibiting farnesylation of progerin prevents the characteristic nuclear blebbing of Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2005;102(36):12879–12884. doi: 10.1073/pnas.0506001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley JB, et al. The defective nuclear lamina in Hutchinson-Gilford progeria syndrome disrupts the nucleocytoplasmic Ran gradient and inhibits nuclear localization of Ubc9. Mol Cell Biol. 2011;31(16):3378–3395. doi: 10.1128/MCB.05087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Snow CJ, Dar A, Dutta A, Kehlenbach RH, Paschal BM. Defective nuclear import of Tpr in Progeria reflects the Ran sensitivity of large cargo transport. J Cell Biol. 2013;201(4):541–557. doi: 10.1083/jcb.201212117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray Chaudhuri A, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19(4):417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 36.Callen E, et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell. 2013;153(6):1266–1280. doi: 10.1016/j.cell.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rensen SS, Doevendans PA, van Eys GJ. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth Heart J. 2007;15(3):100–108. doi: 10.1007/BF03085963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rzucidlo EM, Martin KA, Powell RJ. Regulation of vascular smooth muscle cell differentiation. J Vasc Surg. 2007;45(Suppl A):A25–A32. doi: 10.1016/j.jvs.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Hochegger H, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25(6):1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu H, et al. Identification of poly (ADP-ribose) polymerase-1 (PARP-1) as a novel Kruppel-like factor 8-interacting and -regulating protein. J Biol Chem. 2011;286(23):20335–20344. doi: 10.1074/jbc.M110.215632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12(6):385–392. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J, et al. A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell. 2011;8(1):31–45. doi: 10.1016/j.stem.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103(23):8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCord RP, et al. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23(2):260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Musich PR, Zou Y. DNA-damage accumulation and replicative arrest in Hutchinson-Gilford progeria syndrome. Biochem Soc Trans. 2011;39(6):1764–1769. doi: 10.1042/BST20110687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musich PR, Zou Y. Genomic instability and DNA damage responses in progeria arising from defective maturation of prelamin A. Aging (Albany, NY Online) 2009;1(1):28–37. doi: 10.18632/aging.100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312(5776):1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McClintock D, et al. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS ONE. 2007;2(12):e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olive M, et al. Cardiovascular pathology in Hutchinson-Gilford progeria: Correlation with the vascular pathology of aging. Arterioscler Thromb Vasc Biol. 2010;30(11):2301–2309. doi: 10.1161/ATVBAHA.110.209460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Drozdov I, Shroff R, Beltran LE, Shanahan CM. Prelamin A accelerates vascular calcification via activation of the DNA damage response and senescence-associated secretory phenotype in vascular smooth muscle cells. Circ Res. 2013;112(10):e99–e109. doi: 10.1161/CIRCRESAHA.111.300543. [DOI] [PubMed] [Google Scholar]
- 51.Ragnauth CD, et al. Prelamin A acts to accelerate smooth muscle cell senescence and is a novel biomarker of human vascular aging. Circulation. 2010;121(20):2200–2210. doi: 10.1161/CIRCULATIONAHA.109.902056. [DOI] [PubMed] [Google Scholar]
- 52.Xiong ZM, LaDana C, Wu D, Cao K. An inhibitory role of progerin in the gene induction network of adipocyte differentiation from iPS cells. Aging (Albany, NY Online) 2013;5(4):288–303. doi: 10.18632/aging.100550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao K, et al. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson-Gilford progeria syndrome cells. Sci Transl Med. 2011;3(89):89ra58. doi: 10.1126/scitranslmed.3002346. [DOI] [PubMed] [Google Scholar]