Significance
The CMV glycoprotein (g) entry complex gH/gL is a significant target of the human immune system, eliciting production of highly neutralizing antibodies. MSL-109, a monoclonal antibody that binds to gH, potently neutralizes CMV entry. Here, viral mutants resistant to MSL-109 were generated and the epitope mapped on the model of the structure of CMV gH. We have used a cell-surface fluorescence assay to reveal novel interactions among CMV glycoproteins and probe the effect of MSL-109 on glycoprotein interactions. Our data confirm cross-talk between gH/gL and gB, the fusion protein. Moreover, we identify a homophilic interaction between gH/gL heterodimers that is perturbed by MSL-109. This study suggests a novel mechanism of neutralization, and highlights potential strategies for treatment of CMV disease.
Abstract
Cytomegalovirus (CMV) is a widespread opportunistic pathogen that causes birth defects when transmitted transplacentally and severe systemic illness in immunocompromised individuals. MSL-109, a human monoclonal IgG isolated from a CMV seropositive individual, binds to the essential CMV entry glycoprotein H (gH) and prevents infection of cells. Here, we suggest a mechanism for neutralization activity by MSL-109. We define a genetic basis for resistance to MSL-109 and have generated a structural model of gH that reveals the epitope of this neutralizing antibody. Using surface-based, time-resolved FRET, we demonstrate that gH/gL interacts with glycoprotein B (gB). Additionally, we detect homodimers of soluble gH/gL heterodimers and confirm this novel oligomeric assembly on full-length gH/gL expressed on the cell surface. We show that MSL-109 perturbs the dimerization of gH/gL:gH/gL, suggesting that dimerization of gH/gL may be required for infectivity. gH/gL homodimerization may be conserved between alpha- and betaherpesviruses, because both CMV and HSV gH/gL demonstrate self-association in the FRET system. This study provides evidence for a novel mechanism of action for MSL-109 and reveals a previously undescribed aspect of viral entry that may be susceptible to therapeutic intervention.
Human CMV is a β-group herpesvirus that causes severe complications in immunocompromised individuals. CMV infects between 60% and 80% of the adult population worldwide (1). As with other herpesviruses, CMV establishes a lifelong latency in the host but is largely asymptomatic among infected immunocompetent individuals (2). However, during severe immunosuppression (e.g., in the setting of hematopoietic stem cell transplantation and solid organ transplantation, or advanced HIV/AIDS), CMV reactivation or primary infection can result in life-threatening disease. In addition, the acquisition of primary CMV infection during pregnancy, although of little consequence to the mother, can have severe clinical consequences in the developing fetus (3, 4). The current therapy for CMV disease is treatment with either ganciclovir or valganciclovir, which are associated with significant toxicity and not approved for use in pregnant women or for congenitally damaged infants (5). CMV hyperimmunoglobulin (CMV-HIG; pooled human IgG from CMV-positive individuals) has demonstrated efficacy in certain solid organ transplant recipients and more recently found to show limited success in protecting infants from congenital CMV disease (1, 6, 7). These findings suggest that more potent or differently targeted antibody therapy may prove to be an effective and safe alternative to the current forms of CMV therapy.
Like other herpesviruses, CMV uses multiprotein entry complexes to initiate infection of host cells. Three glycoproteins, gB, gH, and gL, known as the “core fusion machinery,” are conserved in all herpesviruses and are required for entry (8, 9). gB, the most conserved of these glycoproteins, exists as a homotrimer and catalyzes membrane fusion during viral entry (9, 10). gH and gL form a heterodimer, and aid in conferring cell-type specificity to different herpesviruses, although their precise role has not been demonstrated. Recent work in herpes simplex virus-1 (HSV-1) indicates that when glycoprotein gD binds to its cellular receptor, it associates with gH/gL, and in turn gH/gL binds gB and subsequently triggers fusion as a result of a direct interaction (11, 12). These data suggest that, at least in HSV-1, gH/gL facilitates the activation and not the repression of gB. Complexes of gB:gH/gL have been detected from both HSV and CMV (13, 14). CMV lacks gD and requires the minimal complex of gB:gH/gL for entry into fibroblasts. For entry into monocytes, macrophages, epithelial cells, and endothelial cells, CMV requires the pentameric complex gH/gL/UL128/UL130/UL131 in addition to gB (10, 15–17). Although the mechanisms for formation and regulation of these multiprotein complexes have been elucidated in HSV, they are less well understood in CMV. Specifically, there remains uncertainty about the nature of these complexes, their role in infection, and how these complexes are targets of neutralization by antibodies.
Recently, the crystal structure of gH was determined from HSV-2, Epstein–Barr virus (EBV), and pseudorabies virus (PRV), and although the overall protein sequence conservation is low, the core structure of gH is conserved (12, 18, 19). gH has three distinct domains: the N-terminal domain that binds gL (domain H1), the central helical domain (domain H2), and the C-terminal β-sandwich domain (domain H3) (9, 12, 20). Domain H1 is the most divergent, whereas domains H2 and H3 are more conserved and present the same fold in gH from HSV-2, EBV, and PRV (9, 12, 20). The HSV-neutralizing antibody LP11 binds H1 and prohibits the interaction of gB with gH/gL, as shown by bimolecular complementation, revealing a face of gH involved in gB binding (12, 21, 22). Interestingly, the same face of gH/gL is implicated for gB binding in EBV, albeit more N-terminal on the heterodimer, within gL (23, 24). Another anti-HSV neutralizing antibody, 52S, inhibits cell–cell fusion and binds to gH at H2/H3 border (21). Compared with the position of LP11 epitope, 52S binds to the opposite face of gH/gL, consistent with the finding that 52S does not block the association of gH/gL with gB (21). It is unclear how this antibody blocks viral entry. Domain H3 of gH is the most highly conserved and several nonfunctional mutations map to H3 in both HSV-1 and EBV (22, 25). Moreover, a potent neutralizing mAb (CL59) for EBV is directed at this domain (25). Because this is the most conserved domain of gH, one can speculate that domain H3 serves a similar function in other herpesviruses, such as CMV. Of the prevalent herpesviruses, the least is known about CMV gH/gL, which lacks a solved structure and mechanistic studies involving neutralizing antibodies.
MSL-109 is a human monoclonal IgG originally isolated from spleen cells of a CMV seropositive individual. MSL-109 recognizes CMV gH complexes, and in vitro, blocks the infection of fibroblasts by laboratory and clinical strains of CMV (26). Both the mechanism of neutralization and how MSL-109 engages gH is unknown. MSL-109 has been evaluated in the clinic for the prevention of CMV infection following allogeneic hematopoietic stem cell transplantation and as adjuvant therapy for CMV retinitis in HIV-infected individuals (27, 28). Although MSL-109 did not demonstrate benefit in all-comers, analysis of a subset of the transplantation patients who were at high risk for primary CMV infection (donor-positive/recipient-negative patients) demonstrated that MSL-109 could confer protection (27). In addition, MSL-109 failed to demonstrate efficacy in HIV patients with CMV retinitis, possibly because of the immune-privileged nature of the eye (29). Ultimately, MSL-109 was not developed further. A recent study demonstrated a nongenetic viral resistance to MSL-109 in vitro and suggested that the clinical failure of MSL-109 was because of this novel mechanism of resistance (30). However, this study did not reveal the binding site or mechanism of action of MSL-109, because the mode of resistance was nongenetic.
In this study, MSL-109–resistant CMV was generated primarily through passage on epithelial cells and was found to have a genetic basis. Resistance mutations were mapped onto a structural model of gH, revealing a putative epitope for MSL-109 in the H2 domain of gH. Using a surface-based, time-resolved (TR) FRET method, we demonstrate interactions between viral glycoproteins in real-time, namely that gH/gL binds to gB and gH/gL binds to gH/gL, in the form of gH/gL:gH/gL homodimers. MSL-109 binding perturbs the gH/gL:gH/gL interaction but not the gB:gH/gL interaction, suggesting that MSL-109 engages gH at the gH/gL:gH/gL interface. Moreover, introduction of an epitope tag into the MSL-109 interface allows anti-tag antibody to prevent viral infection. This study suggests a mechanism of neutralization for MSL-109 in addition to revealing a previously undescribed homophilic interaction in CMV envelope glycoproteins required for viral entry.
Results
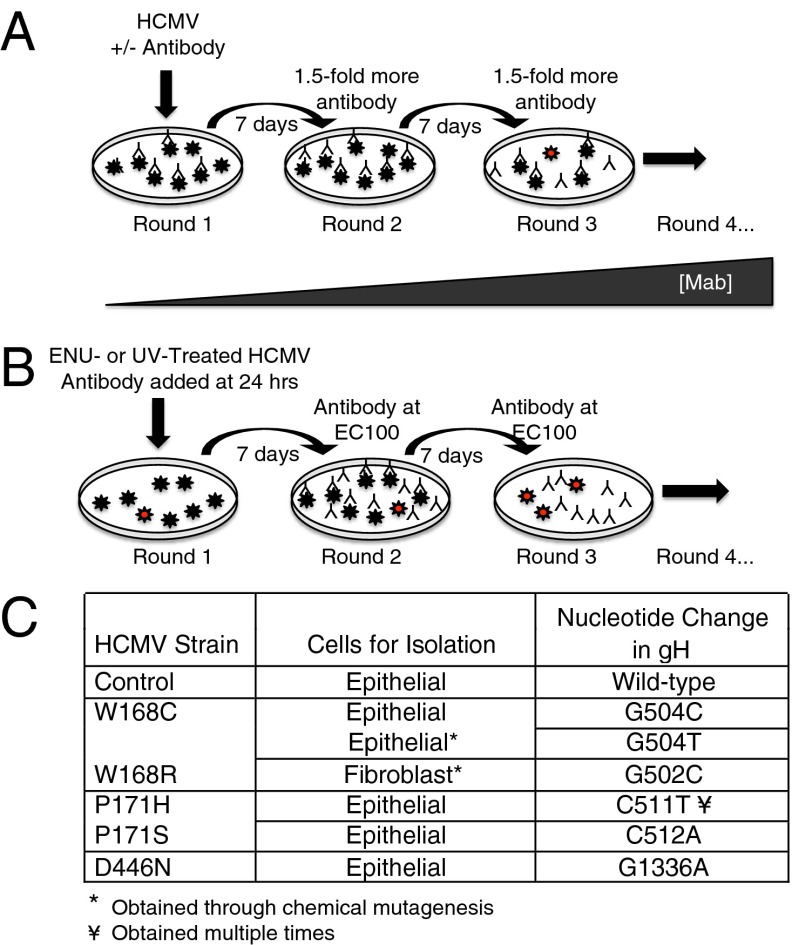
MSL-109 is a highly potent monoclonal antibody against gH that neutralizes CMV entry on a variety of cell types (Fig. S1). To understand the mechanism by which MSL-109 neutralizes viral entry, we passaged CMV VR1814 virus in epithelial cells (ARPE-19) or fibroblasts (MRC-5) in the presence of suboptimal concentrations of MSL-109 antibody and selected for genetic mutants resistant to neutralization (Fig. 1 A and B). The generation of resistance mutants on fibroblasts required mutagenesis of the virus before passaging. All strains resistant to MSL-109 possessed a single nonconservative amino acid mutation in gH, W168C/R, P171H/S, or D446N, compared with the wild-type control (Fig. 1C). Eleven independently selected strains resistant to MSL-109 were generated, encompassing five distinct nucleotide mutations in only three amino acids and no other mutations were found in the other entry glycoproteins (gB, gL, UL128, UL130, and UL131). Epithelial cells yielded the majority of the resistance mutations, with fibroblasts yielding a single mutation in the gH protein at residue position 168 only after prior mutagenesis.
Fig. 1.
Generation of MSL-109–resistant virus. (A) Virus at multiplicity of infection (MOI) of 1 was mixed with a suboptimal concentration MSL-109 and added to epithelial (ARPE-19) or fibroblast (MRC-5) cells. Each week, half of the supernatant was passaged onto new cells and antibody concentration was increased 1.5-fold to a final concentration of 10 × EC90. Virus is represented by stars, with resistant virus indicated by red stars. (B) Virus was exposed to mutagen (ENU or UV) and added to cells. Fully neutralizing amounts of MSL-109 was added to cells before round two of infection. (C) Table of mutations in gH protein isolated from MSL-109–resistant virus. The method and cell type used for mutant generation are included in the table. Control was propagated in ARPE-19 cells in parallel. Mutations in amino acids 168 and 171 were obtained independently multiple times, whereas D446N was obtained only once.
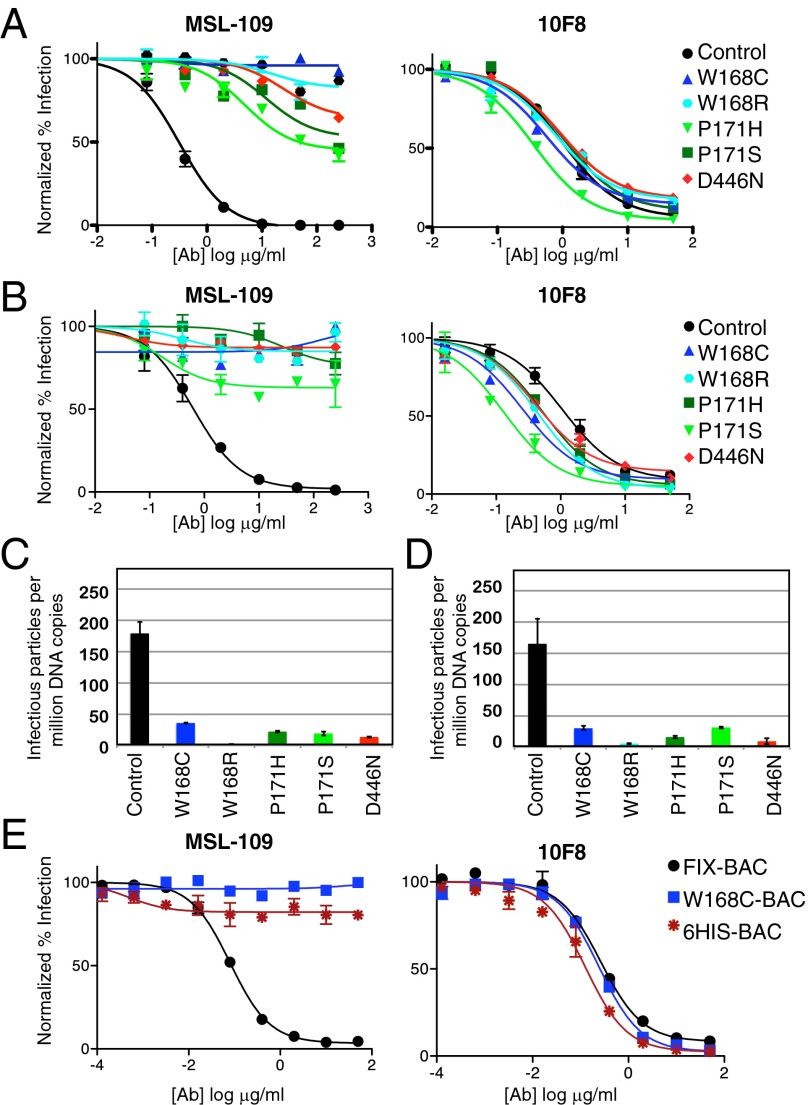
The resistant mutants demonstrated a range of resistance from 150-fold to >1,000-fold, with the W168C/R mutation resulting in complete loss of neutralization at 250 μg/mL on both fibroblasts and epithelial cells (Fig. 2 A and B). Of note, all mutations demonstrated cross-resistance, meaning that virus-harboring mutations resistant to MSL-109 generated on epithelial cells also showed resistance to MSL-109 on fibroblasts, suggesting that the mechanism of MSL-109 is independent of cell type. All resistant strains were sensitive to neutralization by a different anti-gH neutralizing monoclonal antibody, 10F8, indicating that gH/gL was still expressed on the surface of the virus (Fig. 2 A and B). When comparing the ability of MSL-109–resistant strains with that of the wild-type control strain (passaged in parallel without antibody pressure) to infect epithelial cells or fibroblasts, these strains were found to have an in vitro entry defect (5- to 20-times less efficient), suggesting reduced fitness (Fig. 2 C and D). Moreover, mutations that were generated in one cell type also showed entry defects in the other cell type, suggesting that gH may play a common role in cell entry regardless of cell type. To demonstrate that such a mutation is necessary and sufficient to generate MSL-109 resistance, we took advantage of a bacterial artificial chromosome (BAC) that contains the entire CMV genome and can produce infectious virus. Recapitulation of the W168C mutation in FIX-BAC (31) produced a virus that was insensitive to MSL-109, but still sensitive to 10F8, consistent with our initial resistant strains (Fig. 2E).
Fig. 2.
Characterization of strains resistant to MSL-109. (A and B) Neutralization assays with MSL-109 (Left) and 10F8 (Right) antibodies on MSL-109–resistant strains in (A) epithelial cells (ARPE-19) or (B) fibroblast cells (MRC-5). Antibody was serially diluted and mixed with a fixed amount of virus, such that the final virion concentration was a MOI of 1. Cells were infected for 18 h and quantitated using immunofluorescence with an anti–CMV-IE antibody. Data were log-transformed, normalized to infection without antibody, and graphed using Prism (GraphPad Software). (C and D) Cell entry assays with resistant strains on (C) epithelial cells or (D) fibroblasts. DNA copy number was determined using quantitative PCR of viral gene pp65 and was used to infect cells. Infected cells were quantitated and data graphed as infectious virus per million copies of DNA. Error bars represent the SD from six infections. (E) Neutralization assay with MSL-109 (Left) and 10F8 (Right) antibodies on FIX-BAC–derived strains.
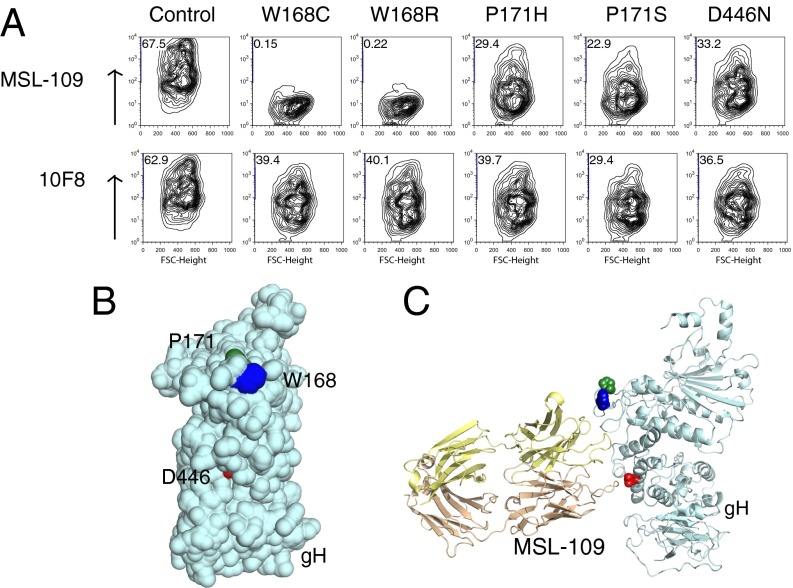
To examine how each mutation affected the ability of MSL-109 to bind gH, FACS was performed with MSL-109 on COS-7 cells transiently expressing gH/gL complexes at their surface (Fig. 3A). The gH/gL complexes harboring resistance mutations still expressed, as demonstrated by binding to 10F8. The W168C/R mutations that displayed resistance to MSL-109 also completely abolished MSL-109 binding to gH/gL (Fig. 3A). As expected, the mutations in P171 (P171H/S) and D446, which demonstrate only reduced resistance, were still able to bind to MSL-109.
Fig. 3.
MSL-109 interactions with wild-type and resistant gH/gL. (A) FACS profiles of MSL-109 or 10F8 antibody binding to cell-surface expressed gH/gL. COS-7 cells were transfected with plasmids bearing wild-type or mutated gH/gL/eGFP. Only GFP-expressing cells are shown. The y axis represents antibody binding (i.e., APC intensity), with positive cells given as a percent of GFP+ cells. (B) Theoretical model of CMV gH with resistance mutations highlighted in blue, green, and red. (C) Theoretical model of CMV gH with crystal structure of MSL-109 FAb (gold) docked onto gH using the resistance mutations (colored blue, green, and red) as boundaries.
Recently, the structure of gH was solved from three distantly related herpesviruses (12, 18, 19). We were able to model the protein sequence of CMV gH (except for the first part of domain H1) using these solved structures because of significant similarities in the structures of each domain. When the three residues that led to resistance were mapped onto the gH structural model, they localized to domain H2 and were all on the same face of the gH (Fig. 3B). We used the crystal structure of the MSL-109 Fab and the site of the resistance mutations on gH to model how the MSL-109 Fab may engage gH (Fig. 3C). Comparison of the predicted MSL-109 Fab “footprint” with the neutralizing antibody-binding sites of other herpesviruses revealed an overlying binding site with d6.3 on PRV gH, suggesting a common mechanism of neutralization between the alpha- and betaherpesviruses (Fig. S2).
To further validate the binding site of MSL-109 and demonstrate the importance of this site on gH for infection, we engineered a 6xHis epitope tag in the CMV FIX-BAC between W168 and P171, two residues critical for MSL-109 binding. The subsequent virus was infectious (6-His-BAC) and could be evaluated in neutralization assays (Fig. 2E). We found that the 6-His-BAC virus was resistant to MSL-109, but still sensitive to 10F8 (Fig. 2E). These data are consistent with the fact that MSL-109 no longer binds to cell surface-expressed gH/gL that harbors this tag (Fig. 4A). Interestingly, virus bearing this His-tag is now sensitive to neutralization by an anti-His antibody, confirming that this site on gH is important for infection (Fig. 4B).
Fig. 4.

Characterization of 6xHis tag insertion within the MSL-109 epitope. (A) FACS profiles of MSL-109 or 10F8 antibody binding to cell surface-expressed HA-gH/gL or HA-gH (6xHis)/gL. COS-7 cells were transfected with plasmids bearing HA-gH/gL or HA-gH (6xHis)/gL. The y axis represents antibody binding (i.e., APC intensity) with positive cells given as percent of GFP+ cells. (B) Neutralization assay with anti-His antibody on wild-type (black circles), W168C (blue squares), or 6xHis (red asterisks). Antibody was serially diluted and mixed with a fixed amount of virus, such that the final virion concentration was a MOI of 1. Cells were infected for 18 h and quantitated using immunofluorescence with an anti–CMV-IE antibody. Data were log-transformed, normalized to infection without antibody, and graphed using Prism (GraphPad Software).
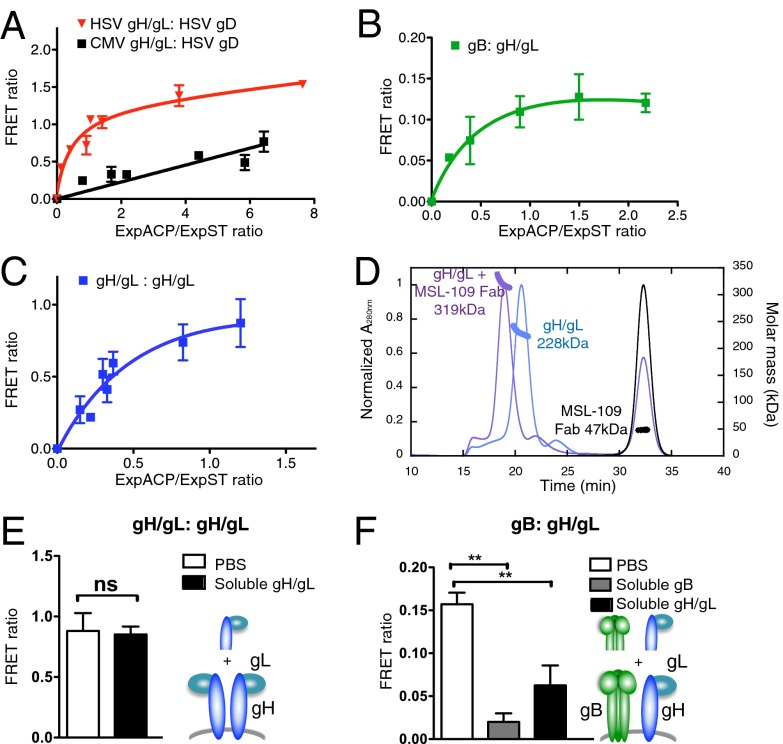
We sought to understand the mechanism by which MSL-109 neutralizes viral entry. Domain H2 of gH is conserved among herpesviruses, and is a target for neutralizing antibodies like LP11 that blocks gH-gB interaction in HSV-2 and d6.3 with unknown function in PRV. To query whether MSL-109 might inhibit gH–gB interaction, we took advantage of two cell-surface protein-labeling technologies: the SNAP- and acyl-carrier protein (ACP)-technologies combined with TR-FRET (32, 33). In our assay, glycoproteins bearing N-terminal fusion tags with either SNAP or ACP were covalently labeled with a donor (Lumi4-Tb)-conjugated benzyl-guanine or an acceptor (A647)-conjugated CoA, respectively. This technology allows for the detection of an energy transfer between the two fluorophores when in close proximity. The intensity of the signal is proportional to the distance between the donor and acceptor as well as to the number of proteins in a given interaction. This TR-FRET approach was originally implemented to study G protein-coupled receptor oligomerization (32) and was recently successfully adapted to immunoreceptors (34). To validate this method on viral glycoproteins, we examined the well-documented interaction of HSV-1 gH and HSV-1 gD. When a TR-FRET interaction is specific, the donor will be associated with increasing concentrations of acceptor until the maximal TR-FRET efficiency is reached, resulting in a saturation curve. In contrast, a TR-FRET signal resulting from a random collision will be fitted with a linear regression. A saturation curve was obtained for HSV-1 gH/gL and HSV-1-gD, indicative of a specific interaction between these two glycoproteins (Fig. 5A). As expected, HSV-1 gD did not associate with CMV gH/gL, further validating the method (Fig. 5A).
Fig. 5.
Glycoprotein interactions as monitored by TR-FRET. (A) TR-FRET saturation curve of HSV-1 ST-gH/gL with HSV-1 ACP-gD, or CMV ST-gH/gL with HSV-1 ACP-gD. Cells were cotransfected with a constant amount of HSV-1 ST-gH/gL or CMV ST-gH/gL and increasing amounts of HSV-1 ACP-gD. FRET and expression of the ACP and ST-proteins (ExpACP and ExpST) were measured. FRET ratio is shown as a function of the ExpACP/ExpST ratio. (B and C) TR-FRET saturation curves of CMV ST-gB with CMV ACP-gH/gL (B) or CMV ST-gH/gL with CMV ACP-gH/gL (C), as described in A. (D) SEC-MALS of soluble gH/gL (blue) and gH/gL in complex with MSL-109 Fab (purple) shown by UV absorbance (280 nM; Left axis). Molecular weight calculated from peak elutions and mass graphed as a thick line on the corresponding peak (Right axis). The second peak at ∼32.5 mL measures 47 kDa, consistent with excess Fab. (E) FRET ratio between ST-gH/gL and ACP-gH/gL after addition of PBS (white bar) or soluble gH/gL (black bar). (F) FRET ratio between ST-gB and ACP gH/gL recorded after addition of PBS (white bar), soluble gB (gray bar), or soluble gH/gL (black bar). Data in E are means ± SEM of triplicates from three independent experiments. Data in (A–C, and F) are means ± SEM of triplicates from a representative experiment.
Next, we examined interactions between CMV gB and gH/gL, and in addition, gH/gL and itself. The tag on the N terminus of the CMV glycoproteins still allowed for virus infectivity and neutralization by MSL-109 and 10F8 (Fig. S3). Using the TR-FRET assay, we were able to detect an interaction between CMV gB and CMV gH/gL (Fig. 5B). Unexpectedly, a homophilic interaction between the CMV gH/gL heterodimers was also observed (Fig. 5C). Although it is well-established that gH forms a heterodimer with gL, homodimers of gH/gL heterodimers (i.e., gH/gL:gH/gL) have not been previously reported. Moreover, we found that HSV-1 gH/gL was able to self-associate, suggesting conservation of homodimer formation (Fig. S4). As expected, we were also able to detect a CMV gB–gB interaction (Fig. S5A).
To confirm the homodimerization interaction of CMV gH/gL, we produced soluble recombinant gH/gL complex in baculovirus and using size-exclusion chromatography (SEC), we noted that the complex eluted in a single peak indicating a single species. We next submitted these fractions to multiangle light scattering (MALS) to determine absolute molar mass. Calculated by MALS, the molecular weight of soluble gH/gL is 228 kD, consistent with a 2:2 gH/gL:gH/gL complex (theoretical mass of 215 kD) (Fig. 5D). Binding of the MSL-109 Fab fragment to gH/gL shifts the SEC elution profile and the calculated molecular weight becomes 319 kD, predictive of a 2:2:2 complex (theoretical mass of 312 kD), in which each of the two gH/gL heterodimers is bound by one Fab fragment (Fig. 5D).
To evaluate the stability of the CMV glycoprotein interactions, we examined whether the addition of soluble CMV gB or gH/gL protein (i.e., extracellular domains) could disrupt the TR-FRET signal. We found that gH/gL or gB extracellular domains were unable to disrupt the gH-gH interaction (Fig. 5E) or the gB–gB interaction (Fig. S5A), respectively, but did disrupt the gB–gH interaction (Fig. 5F). These results support a tight association within CMV gH/gL:gH/gL homodimer.
Next, we tested if MSL-109 had an effect on gB:gH/gL or gH/gL:gH/gL interactions. Addition of MSL-109 did not disrupt the gB:gH/gL interaction (Fig. 6A). Similarly, this interaction was not disrupted by 10F8 or by the neutralizing anti-gB antibody ITC-88 (Fig. S5B). Although ITC-88 did not disrupt the gB–gB interaction, MSL-109 greatly perturbed the TR-FRET signal resulting from the gH/gL:gH/gL association (Figs. S5C and 6B). The antibody 10F8 was also able to perturb this interaction, but to a lesser extent than MSL-109 (Fig. S5D). A similar TR-FRET signal was measured for gH/gL harboring the point mutations: P171H, W168C, and D446N (referred to as gH HCN in Fig. 6C), in the presence of either MSL-109 or the isotype control, indicating that the decrease of TR-FRET is a result of MSL-109 binding (Fig. 6C). With the goal of disrupting gH/gL:gH/gL dimerization, we engineered multiple mutations in gH in the proposed dimerization interface. Unfortunately, none of these mutations allowed for cell-surface expression of gH/gL, strongly suggesting that this domain of gH plays a critical role in gH complex stability and function, possibly by playing a role in dimerization.
Fig. 6.
Effect of MSL-109 binding on glycoprotein interactions as monitored by TR-FRET. (A–C) FRET ratios calculated after addition of control IgG (white bar) or MSL-109 (black bar). FRET measured between (A) ST-gB and ACP-gH/gL, (B) ST-gH/gL and ACP-gH/gL, or (C) between ST-(HCN)gH/gL and ACP-(HCN)gH/gL. Data are means ± SEM of triplicates from three independent experiments.
Discussion
In this study, we have described two findings. First, by generating MSL-109–resistant strains and identifying the resistant mutations, we revealed the epitope of MSL-109 and mapped it on a structural model of the CMV gH/gL heterodimer. These results are supported by the engineered His-tag in the putative MSL-109 epitope that allows for an anti-His antibody to neutralize infection. Second, we have shown by two complementary methods that CMV gH/gL self-associates to form gH/gL:gH/gL homodimers. MSL-109 disrupts the FRET signal between gH/gL homodimers, suggesting that MSL-109 binds at or near the homodimerization interface. gH/gL homodimerization may be conserved between alpha- and betaherpesviruses, given that both CMV and HSV gH/gL demonstrate self-association in the FRET system. We speculate that the potent neutralizing activity of MSL-109 may result from the perturbation of the gH/gL:gH/gL homodimer.
Our data show that resistance against MSL-109 occurs in epithelial cells after a significant time in culture (>2 mo) and that the resulting resistant mutants are attenuated for viral entry. These data suggest that there is a significant barrier to developing resistance against MSL-109, although the barrier in vivo may be lower. The W168C/R mutation yielded the greatest resistance to MSL-109, most likely because of complete loss of MSL-109 binding. This finding is in contrast to that reported by Manley et al. (30), in which resistance to MSL-109 occurred rapidly and did not involve genetic alteration of the virus. This disparity may result from our use of primary epithelial cells, which differs from the cell type used by Manley et al. (30). In addition, fibroblasts yielded a single resistance mutation only after prior mutagenesis of the virus. Importantly, the resistant viruses derived from epithelial cells are resistant to MSL-109 upon infection of fibroblasts and vice-versa, suggesting that the mechanism of MSL-109 neutralization is independent of cell type.
The structure of CMV gH has yet to be elucidated. However, we derived a structural model of CMV gH based on the available structures for gH from HSV, EBV, and PRV. There is very little sequence conservation or structural homology for gL and the first part of domain H1 (H1A); therefore, we were unable to model CMV H1A or gL. Fortunately, the MSL-109 resistance mutations fell within the domain of CMV gH that was amenable to modeling. The MSL-109 Fab antigen-binding domain was able to span the mutations and, in effect, formed a putative binding “footprint” on the structure. This “footprint” on gH/gL corresponds with a known neutralizing epitope on PRV gH/gL, suggesting that this site on gH plays a conserved role in viral entry.
Using the TR-FRET method, we monitored the interactions between glycoprotein extracellular domains in real time at the cell surface. To our knowledge, this is the first report of the TR-FRET method used to demonstrate interactions among viral proteins. For proof of principle, we demonstrated two well-documented interactions: CMV gB association of monomers and the interaction between HSV gH and gD. In addition, we were able to confirm the interaction of CMV gB and gH, which had been previously detected from cell lysates (35). This interaction between CMV gB and gH was disrupted by soluble gB and gH/gL, suggestive of a transient interaction. In contrast, MSL-109 was unable to disrupt this interaction, suggesting that MSL-109 binds to a face of gH/gL not involved with gB binding.
The TR-FRET method also revealed a novel homodimer of gH/gL that is resistant to the additional gH/gL heterodimer but is sensitive to MSL-109. We find that this homodimerization is mediated by gH ectodomains, evidenced by the ability of soluble gH/gL lacking the transmembrane domain to exclusively form homodimers. gH/gL from HSV-1 also yielded a TR-FRET signal, suggestive of dimerization. Although this finding with HSV gH/gL has yet to be confirmed biochemically, it appears that homodimerization of gH/gL dimers may be conserved between alpha- and betaherpesviruses. Given that MSL-109 reduces the FRET signal between gH/gL homodimers, we propose a mechanism of action for MSL-109 neutralization via this perturbation. It is interesting to speculate whether a higher affinity antibody with binding similarities to MSL-109 would be more successful in the clinic. Moreover, by analogy to MSL-109, it would be of interest to evaluate whether specific anti-HSV neutralizing mAbs, such as 52S, perturb HSV gH/gL homodimerization.
Given that MSL-109 forms a soluble co-complex with gH/gL, we envision that MSL-109 is perturbing the gH/gL homodimer rather than abolishing dimerization. MSL-109 may be splaying the dimer apart at sufficient distance to reduce FRET and ultimately prevent proper gH/gL function. We believe that there is structural flexibility at the interface between the homodimers given that MSL-109 is able to access this interface and increase the distance between gH/gL dimers, and that this region can sustain a 6xHis tag. The insertion of the 6xHis epitope tag into the MSL-109 epitope provides evidence that the region can withstand a significant insertion, but sustains the idea that an antibody directed at the region neutralizes virus by perturbing gH/gL homodimerization. We do not know whether MSL-109 neutralizes viral entry by inhibiting receptor engagement or by preventing gH/gL from regulating viral fusion through gB. However, because the identity of such a receptor is unknown, this hypothesis remains challenging to address. In fact, it has been postulated that gH itself may not bind to a receptor and may instead directly interact with membranes and induce a strong, destabilizing local curvature, similar to the role of synaptotagmins in vesicle fusion (18). Perhaps MSL-109 prevents gH/gL from undergoing a critical conformational change necessary for stabilizing such membrane interactions. An available crystal structure of gH/gL complexed with MSL-109 may shed more light on the precise mechanism of MSL-109.
In summary, we have proposed a mechanism for neutralization of viral entry that involves perturbation of gH/gL:gH/gL homodimers and suggests that dimerization is important for viral entry. Multimerization of viral entry glycoproteins is a common feature among enveloped viruses, such as Sindbis virus, measles virus, Newcastle Disease virus, and influenza. Within the herpesvirus family, there is also considerable evidence for multimerization of glycoprotein entry complexes, such as gD, gC, and gB. We speculate that gH/gL dimerization may increase the avidity of these viral entry complexes for their cellular receptors, ultimately promoting infection. Perhaps interfering with glycoprotein multimerization may provide a novel mode of therapeutic intervention for enveloped viruses.
Materials and Methods
Cells, Viral Strains, and Neutralization Assays.
Epithelial cells and fibroblast cells (ARPE-19 and MRC-5, respectively, from American Type Culture Collection) were grown, using the manufacturer’s suggested conditions. HUVEC cells (Lonza) obtained at passage 0 were grown as per the manufacturer’s instruction in EBM supplemented with EGM bullet kit. VR1814 strain was obtained from Maria Grazia Revello (Servizio di Virologia, Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy) and expanded in HUVECs grown without heparin and viral supernatant was pelleted, resuspended in complete media [DMEM supplemented with 10% (vol/vol) FCS, penicillin/streptomycin, l-glutamine; Invitrogen] and 20 mM Hepes (Cellgro), and frozen (36). Viral titer and neutralization were performed as Fouts et al. (37), using MSL-109 or 10F8 (described in SI Materials and Methods), or anti-6xHIS (Clontech). Briefly, following 18 h of infection, cells were stained with either mouse monoclonal anti-CMV IE antibody Mab810 (Millipore) or rabbit anti-CMV IE (Johnson Laboratory, Oregon Health Sciences University, Portland, OR) then incubated with the appropriate Alexa Fluor 488 and Hoechst (Invitrogen) (37). Cells were imaged and counted using the ImageXpress Micro and MetaXpress software (Molecular Devices). Data were graphed and effective inhibitory concentrations (EC) calculated using Prism EC50-curve fitting algorithm. (GraphPad Software).
Selection of Antibody-Resistant Mutant Virus.
Briefly VR1814 virus was passaged weekly in the presence of increasing concentrations of MSL-109 in ARPE-19 or MRC-5 cells. Typically, mutants emerged as single viral plaques by around passage 9. To generate additional resistance mutations, extracellular virus was treated with N-ethyl-N-nitrosourea (ENU; Sigma) or UV light (254λ-Stratalinker; Stratagene) and allowed to infect ARPE-19 or MRC-5 cells before weekly passage with increasing amounts of antibody. For full methods see SI Materials and Methods.
TR-FRET Measurements Between SNAP- and ACP-Tagged Proteins.
COS-7 cells were cotransfected with SNAP-tagged or ACP-tagged constructs (SI Materials and Methods) using Lipofectamine 2000 (Life Technologies) and seeded in a white-bottom 96-well plate (Costar) at 100,000 cells per well. Forty-eight hours later, cells were labeled with 100 nM of donor-conjugated benzyl-guanine SNAP-Lumi-4Tb (Cisbio) for 1 h at 37 °C, 5% (vol/vol) CO2. Cells were then washed twice and subsequently labeled with 3 µM of acceptor-conjugated CoA CoA-A647 (New England Biolabs) in DMEM, 10 mM MgCl2, 50 mM Hepes, 1 µM Sfp (New England Biolabs) for 1 h at 37 °C, 5% (vol/vol) CO2. After three washes, the Lumi4-Tb and the FRET signal was recorded at 620 nm and 665 nm, respectively, for 400 µs after a 60-µs delay following laser excitation at 343 nm using a Safire (38) plate reader (Tecan). The emission signal of the A647 was detected at 682 nm after excitation at 640 nm using the same plate reader. FRET intensity was calculated as follows: (signal at 665 nm from cells labeled with SNAP-donor and ACP-acceptor) – (signal at 665 nm from the same batch of transfected cells labeled with SNAP-donor only). When soluble proteins or antibody were tested, FRET signal was measured before and after 15-min reagent incubation. The TR-FRET signal and the fluorescence emission of each fluorophore (ExpST and ExpACP), which reflects the expression of the associated protein, were recorded and expressed as the TR-FRET ratio plotted against the ExpACP/ExpST.
SI Methods.
Additional methods are contained in the SI Materials and Methods. These describe resistant virus generation, BAC modification (Table S1) and viral stock production, viral entry assay, plasmid construction, surface-expression of glycoproteins, soluble protein expression, antibody purification, CMV gH homology model construction, crystallization, and structural determination and SEC-MALS.
Supplementary Material
Acknowledgments
We thank Gerry Nakamura, Jo-Anne Hongo, and Rajesh Vij for the isolation, cloning, and expression of the anti-CMV antibodies used in this study; Maria Grazia Revello for the VR1814 virus and FIX-bacterial artificial chromosome; Bill Britt for his generous gift of the mouse anti-gB, 27-156, monoclonal antibody; Christine O’Connor (Cleveland Clinic) for advice regarding working with CMV bacterial artificial chromosome; Dick Vandlen for expression and purification of MSL-109 Fab; Alberto Estevez for generating the baculoviruses that express soluble gH/gL and gB and aiding in subsequent protein purification; Michelle Chan for the construction of the vector backbone for use in FRET; Jiabing Ding for constructing the CMV glycoprotein-expressing plasmids; Erin Dueber for help with gH/gL expression and subsequent size-exclusion chromatography-multiangle light scattering; and Eric Brown and Fernando Bazan for helpful discussions. This research was funded by Genentech, Inc./Hoffmann La-Roche.
Footnotes
Conflict of interest statement: All authors of this work are employees of, and hold stock interests in, Genentech, Inc./Hoffmann La-Roche.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4LRI).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1404653111/-/DCSupplemental.
References
- 1.Snydman DR. Cytomegalovirus immunoglobulins in the prevention and treatment of cytomegalovirus disease. Rev Infect Dis. 1990;12(Suppl 7):S839–S848. doi: 10.1093/clinids/12.supplement_7.s839. [DOI] [PubMed] [Google Scholar]
- 2.Stagno S. Cytomegalovirus. In: Remington JS, Klein JO, Wilson CB, Baker CJ, editors. Infectious Diseases of the Fetus and Newborn. Philadelphia: WB Saunders; 2001. [Google Scholar]
- 3.Stagno S, Whitley RJ. Herpesvirus infections of pregnancy. Part I: Cytomegalovirus and Epstein-Barr virus infections. N Engl J Med. 1985;313(20):1270–1274. doi: 10.1056/NEJM198511143132006. [DOI] [PubMed] [Google Scholar]
- 4.Ross DS, Dollard SC, Victor M, Sumartojo E, Cannon MJ. The epidemiology and prevention of congenital cytomegalovirus infection and disease: Activities of the Centers for Disease Control and Prevention Workgroup. J Womens Health (Larchmt) 2006;15(3):224–229. doi: 10.1089/jwh.2006.15.224. [DOI] [PubMed] [Google Scholar]
- 5.Maine GT, Lazzarotto T, Landini MP. New developments in the diagnosis of maternal and congenital CMV infection. Expert Rev Mol Diagn. 2001;1(1):19–29. doi: 10.1586/14737159.1.1.19. [DOI] [PubMed] [Google Scholar]
- 6.Nigro G, Adler SP, La Torre R, Best AM. Congenital Cytomegalovirus Collaborating Group Passive immunization during pregnancy for congenital cytomegalovirus infection. N Engl J Med. 2005;353(13):1350–1362. doi: 10.1056/NEJMoa043337. [DOI] [PubMed] [Google Scholar]
- 7.Revello MG, et al. CHIP Study Group A randomized trial of hyperimmune globulin to prevent congenital cytomegalovirus. N Engl J Med. 2014;370(14):1316–1326. doi: 10.1056/NEJMoa1310214. [DOI] [PubMed] [Google Scholar]
- 8.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. Fusing structure and function: A structural view of the herpesvirus entry machinery. Nat Rev Microbiol. 2011;9(5):369–381. doi: 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg RJ, et al. Herpes virus fusion and entry: A story with many characters. Viruses. 2012;4(5):800–832. doi: 10.3390/v4050800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacson MK, Compton T. Human cytomegalovirus glycoprotein B is required for virus entry and cell-to-cell spread but not for virion attachment, assembly, or egress. J Virol. 2009;83(8):3891–3903. doi: 10.1128/JVI.01251-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J Virol. 2010;84(23):12292–12299. doi: 10.1128/JVI.01700-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chowdary TK, et al. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol. 2010;17(7):882–888. doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gianni T, Amasio M, Campadelli-Fiume G. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J Biol Chem. 2009;284(26):17370–17382. doi: 10.1074/jbc.M109.005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Avitabile E, Forghieri C, Campadelli-Fiume G. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: The interaction between gB and gH/gL does not necessarily require gD. J Virol. 2009;83(20):10752–10760. doi: 10.1128/JVI.01287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci USA. 2005;102(50):18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J Virol. 2010;84(5):2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol. 2006;80(2):710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backovic M, et al. Structure of a core fragment of glycoprotein H from pseudorabies virus in complex with antibody. Proc Natl Acad Sci USA. 2010;107(52):22635–22640. doi: 10.1073/pnas.1011507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc Natl Acad Sci USA. 2010;107(52):22641–22646. doi: 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stampfer SD, Heldwein EE. Stuck in the middle: Structural insights into the role of the gH/gL heterodimer in herpesvirus entry. Curr Opin Virol. 2013;3(1):13–19. doi: 10.1016/j.coviro.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gompels UA, et al. Characterization and sequence analyses of antibody-selected antigenic variants of herpes simplex virus show a conformationally complex epitope on glycoprotein H. J Virol. 1991;65(5):2393–2401. doi: 10.1128/jvi.65.5.2393-2401.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galdiero M, et al. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J Virol. 1997;71(3):2163–2170. doi: 10.1128/jvi.71.3.2163-2170.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plate AE, Smajlović J, Jardetzky TS, Longnecker R. Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J Virol. 2009;83(15):7678–7689. doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omerović J, Lev L, Longnecker R. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J Virol. 2005;79(19):12408–12415. doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu L, Borza CM, Hutt-Fletcher LM. Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J Virol. 2005;79(17):10923–10930. doi: 10.1128/JVI.79.17.10923-10930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nokta M, Tolpin MD, Nadler PI, Pollard RB. Human monoclonal anti-cytomegalovirus (CMV) antibody (MSL 109): Enhancement of in vitro foscarnet- and ganciclovir-induced inhibition of CMV replication. Antiviral Res. 1994;24(1):17–26. doi: 10.1016/0166-3542(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 27.Boeckh M, et al. Randomized, placebo-controlled, double-blind study of a cytomegalovirus-specific monoclonal antibody (MSL-109) for prevention of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2001;7(6):343–351. doi: 10.1016/s1083-8791(01)80005-7. [DOI] [PubMed] [Google Scholar]
- 28.Borucki MJ, et al. AACTG 266 Team A phase II, double-masked, randomized, placebo-controlled evaluation of a human monoclonal anti-Cytomegalovirus antibody (MSL-109) in combination with standard therapy versus standard therapy alone in the treatment of AIDS patients with Cytomegalovirus retinitis. Antiviral Res. 2004;64(2):103–111. doi: 10.1016/j.antiviral.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 29.Jabs DA, et al. Studies of Ocular Complications of AIDS Research Group HIV and cytomegalovirus viral load and clinical outcomes in AIDS and cytomegalovirus retinitis patients: Monoclonal Antibody Cytomegalovirus Retinitis Trial. AIDS. 2002;16(6):877–887. doi: 10.1097/00002030-200204120-00007. [DOI] [PubMed] [Google Scholar]
- 30.Manley K, et al. Human cytomegalovirus escapes a naturally occurring neutralizing antibody by incorporating it into assembling virions. Cell Host Microbe. 2011;10(3):197–209. doi: 10.1016/j.chom.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 31.Murphy E, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci USA. 2003;100(25):14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurel D, et al. Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tag technologies: Application to GPCR oligomerization. Nat Methods. 2008;5(6):561–567. doi: 10.1038/nmeth.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monnier C, et al. Trans-activation between 7TM domains: Implication in heterodimeric GABAB receptor activation. EMBO J. 2011;30(1):32–42. doi: 10.1038/emboj.2010.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stengel KF, et al. Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering. Proc Natl Acad Sci USA. 2012;109(14):5399–5404. doi: 10.1073/pnas.1120606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J Virol. 2008;82(23):11837–11850. doi: 10.1128/JVI.01623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grazia Revello M, et al. In vitro selection of human cytomegalovirus variants unable to transfer virus and virus products from infected cells to polymorphonuclear leukocytes and to grow in endothelial cells. J Gen Virol. 2001;82(Pt 6):1429–1438. doi: 10.1099/0022-1317-82-6-1429. [DOI] [PubMed] [Google Scholar]
- 37.Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J Virol. 2012;86(13):7444–7447. doi: 10.1128/JVI.00467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, et al. Quantitative analysis of neutralizing antibody response to human cytomegalovirus in natural infection. Vaccine. 2011;29(48):9075–9080. doi: 10.1016/j.vaccine.2011.09.056. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.