Significance
Creating artificial enzymes that catalyze arbitrary chemical reactions is challenging. Although computational approaches to this problem hold great promise, starting designs typically exhibit low efficiency and require extensive optimization through directed evolution. In this study, we chronicle the evolution of a modestly active, computationally designed Diels-Alderase into a proficient biocatalyst for an abiological [4+2] cycloaddition reaction. Biochemical and structural characterization of the evolved enzyme reveals the molecular origins of its enhanced efficiency. The close match between the experimental structure, which changed only subtly over the course of evolution, and the original design model is particularly notable. In addition to enhancing our understanding of the principles of enzymatic catalysis, these findings should aid future efforts to produce active enzymes more reliably.
Keywords: biocatalysis, computational enzyme design, Diels–Alder reaction, laboratory evolution, enzyme mechanism
Abstract
By combining targeted mutagenesis, computational refinement, and directed evolution, a modestly active, computationally designed Diels-Alderase was converted into the most proficient biocatalyst for [4+2] cycloadditions known. The high stereoselectivity and minimal product inhibition of the evolved enzyme enabled preparative scale synthesis of a single product diastereomer. X-ray crystallography of the enzyme–product complex shows that the molecular changes introduced over the course of optimization, including addition of a lid structure, gradually reshaped the pocket for more effective substrate preorganization and transition state stabilization. The good overall agreement between the experimental structure and the original design model with respect to the orientations of both the bound product and the catalytic side chains contrasts with other computationally designed enzymes. Because design accuracy appears to correlate with scaffold rigidity, improved control over backbone conformation will likely be the key to future efforts to design more efficient enzymes for diverse chemical reactions.
The Diels–Alder reaction, one of the most powerful transformations in organic chemistry, generates two carbon–carbon bonds and up to four stereogenic centers in a single concerted step. Although putative enzymes for unimolecular [4+2] cycloadditions have been reported (1–4), no naturally occurring enzyme is known to catalyze a bimolecular Diels–Alder reaction (5, 6). The generation of artificial Diels-Alderases has therefore been a longstanding and alluring goal for protein engineers.
The first noteworthy successes toward this end were achieved with catalytic antibody technology. A range of biocatalysts for both normal and inverse electron-demand Diels–Alder reactions has been elicited in response to immunogenic transition-state analogs (7–12). Analogous nucleic acid-based catalysts have been isolated from large random libraries with powerful in vitro selection techniques (13–16). In a different line of attack, metal ion catalysis has been combined with diverse biomacromolecular scaffolds to promote several cycloaddition reactions (17, 18). Although such catalysts often exhibit significant regio- and stereoselectivity, their activities generally are orders of magnitude lower than those typical of natural enzymes.
More recently, computational design has proven effective at generating novel catalytic activities in proteins (19, 20). This approach was used, for example, to create DA_20_00, an enzyme that catalyzes the enantio- and diastereoselective Diels–Alder reaction between 4-carboxybenzyl-trans-1,3-butadiene-1-carbamate (1) and N,N-dimethylacrylamide (2) (21). The active site of a β-propeller scaffold was altered to bind the diene and dienophile for productive reaction and to stabilize the cycloaddition transition state electronically with appropriately placed hydrogen bond donors and acceptors (Fig. 1). Judging from its catalytic proficiency, [kcat/(Kdiene·Kdienophile)]/kuncat, which reflects the enzyme’s ability to lower the activation barrier for reaction (22), the starting design is a poorer catalyst than analogous catalytic antibodies (9, 11). However, its low initial activity was increased substantially by targeted mutagenesis of active site residues (21) and installation of a lid element through further computational refinement (23).
Fig. 1.
Diels–Alder reaction between 4-carboxybenzyl-trans-1,3-butadiene-1-carbamate (1) and N,N-dimethylacrylamide (2). The theozyme for promoting formation of the 3R,4S endo cyclohexene product (3) is shown in brackets. The phosphorylated product analog (4) was used for inhibition and crystallization experiments.
Systematic laboratory evolution is a powerful tool for boosting the efficiency of artificial enzymes (24). Here, we exploited this approach to further augment the activity of the computationally designed Diels-Alderase, applying extensive random mutagenesis and screening to sculpt a more effective active site for the target reaction. The most advanced catalyst, which exhibits the highest catalytic proficiency reported for any natural or artificial Diels-Alderase, was characterized biochemically and structurally. X-ray crystallographic analysis of its complex with a phosphorylated product analog provides direct insight into the origins of catalysis in this system as well as a basis for understanding the structural changes underlying its enhanced efficiency.
Results
Optimization of the Original Computational Design.
As previously described, the first steps toward optimization of DA_20_00 involved systematic mutation of all residues predicted to interact with the substrates or the designed catalytic residues (21). Six mutations, when combined, improved catalytic efficiency 100-fold [compare kcat/(Kdiene·Kdienophile) for DA_20_00 and DA_20_10, Table 1] (21). Toward further enhancement, we carried out eight consecutive rounds of directed evolution, introducing sequence diversity at random positions in the DA_20_10 gene by error-prone PCR (epPCR). The resulting libraries were screened for variants exhibiting improved production and/or catalytic activity.
Table 1.
Steady-state kinetic parameters
| Catalyst* | kcat, h−1 | Kdiene, mM | Kdienophile, mM | kcat/(Kdiene·Kdienophile), M−1⋅M−1⋅s−1 | EM†, M | 1/KTS‡, M−1 |
| DA_20_00§ | 0.10 ± 0.02 | 3.5 ± 1.5 | 146 ± 2.5 | 0.06 ± 0.03 | 4 | 8.7 × 103 |
| DA_20_10§ | 2.1 ± 0.2 | 1.3 ± 0.1 | 73 ± 5 | 6.1 ± 0.9 | 94 | 1.0 × 106 |
| DA_20_20 | 4.5 ± 0.6 | 1.0 ± 0.2 | 37 ± 4 | 34 ± 9 | 199 | 5.4 × 106 |
| CE6¶ | 2.2 ± 0.1 | 0.2 ± 0.03 | 34 ± 2 | 87 ± 14 | 97 | 1.4 × 107 |
| CE11 | 4.4 ± 0.3 | 0.53 ± 0.06 | 46 ± 4 | 50 ± 8 | 194 | 7.7 × 106 |
| CE20 | 10.8 ± 0.5 | 0.29 ± 0.03 | 19 ± 2 | 540 ± 70 | 478 | 8.4 × 107 |
Measured at 25 °C in PBS, pH 7.4. The estimated errors reflect the SD of three independent measurements. Under the same conditions, kuncat = 2.26 ± 0.02 × 10−2 M−1⋅h−1, in good agreement with the previously reported value (21).
The sequences of all catalysts are provided in Fig. S2.
Effective molarity (EM = kcat/kuncat) (27).
Chemical proficiency (1/KTS = [kcat/(Kdiene·Kdienophile)]/kuncat) (22).
Values reported by Siegel et al. (21).
Values reported by Eiben et al. (23).
In each evolutionary round, one to five mutations were introduced per gene, and the activity of 720 variants was assessed in microtiter plates by a direct MS-MS assay (SI Materials and Methods and Fig. S1). Typically, 1–5% of the total population had activity equal to or higher than either DA_20_10 or the most active clone from the previous round, and these variants were used as input for the following cycle of mutagenesis and screening. Numerous mutations were found that had no effect on activity, either beneficial or deleterious, and these frequently disappeared in subsequent rounds. At the end of the eighth round of evolution, DA_20_20, the most active DA_20_10 descendant, contained five substitutions at surface sites (R50H, V96I, T197R, E288D, and L309S) and two mutations at positions remote from the active site in the hydrophobic core (D232V and H274L) (yellow spheres in Fig. 2A; sequence in Fig. S2). The evolved DA_20_20 showed a 5.5-fold higher catalytic efficiency than DA_20_10 because of an increase in turnover number (kcat) and a simultaneous decrease in KM for the dienophile (Table 1).
Fig. 2.
Optimization of an artificial Diels-Alderase. (A) The computational design DA_20_00 was optimized by cassette mutagenesis of active site residues (green balls) and epPCR (yellow balls) to afford DA_20_20. Mutations are mapped onto the DA_20_00 structure (PDB ID code 3I1C). (B) Combining these mutations with a computationally designed helix-turn-helix lid element (cyan backbone), followed by epPCR of the entire protein (red balls) and then the lid element (blue balls) yielded the proficient Diels-Alderase CE20. Mutations are mapped onto the CE6 structure (PDB ID code 3U0S). The active sites containing manually docked substrates (black carbons) are shown as transparent gray surfaces.
In parallel with these laboratory evolution experiments, Eiben et al. (23) submitted DA_20_10 to additional rounds of computational refinement. They hypothesized that the activity of the catalyst was limited by its very open active site and with the help of players of the online game Foldit, designed CE6, a variant of DA_20_10 containing a 24-residue helix-turn-helix motif (shown in cyan in Fig. 2B) that functions as a lid to constrain the substrates in a productive orientation for reaction (23). This structural element improved the enzyme’s affinity for both substrates, judging from the lower KM values, but left kcat unchanged (Table 1). Compared with DA_20_20, CE6 exhibits a twofold lower turnover number but a fivefold higher chemical proficiency. Directed evolution and computational redesign thus provided complementary optimization pathways for DA_20_10.
Further Evolutionary Refinement.
To generate even better Diels-Alderases, we combined all mutations found during the laboratory evolution of DA_20_10 with the lid structure designed in CE6. Despite targeting different regions of the protein, the results of the two optimization strategies were not additive. Because of compensating effects on kcat and KM, the new variant, CE11, has a kcat/(Kdiene·Kdienophile) value intermediate between that of its precursors (Table 1). It was therefore subjected to further directed evolution. As in the earlier optimization rounds, the gene encoding CE11 was diversified by epPCR. All variants that showed improved activity after four rounds of diversification and screening shared two mutations in the engineered helix-turn-helix motif: T43I in direct contact with the substrates and R56S on the solvent-exposed supporting helix. Subsequent random mutagenesis consequently focused exclusively on this structural element. Over four additional rounds of laboratory evolution, mutations were found at most positions in the targeted segment. In the helix designed to interact with the substrates (residues 36–44), conservative substitutions were found at three sites (S39T, E40D, and K44R/N). The solvent-exposed supporting helix and the interhelical loop proved even more tolerant to mutation. Residues 45–56 were all substituted at least once and often by two or three different amino acids. Only four mutations—T43I, P48L, K53E, and R56S—persisted over the entire evolutionary trajectory. The best variant to emerge after eight rounds of mutagenesis and screening, CE20 (Fig. 2B, red and blue spheres), contained these substitutions as well as an additional mutation in the helix that interacts with the substrates (K44N) and two mutations in the supporting helix (S55R and G57D).
Characterization of CE20.
CE20 was characterized kinetically by measuring the dependence of reaction velocity on dienophile concentration at several fixed diene concentrations. The pattern of intersecting lines in the double-reciprocal plot (Fig. 3A) indicates that the diene and dienophile bind independently to the enzyme active site (25). The steady-state kinetic parameters obtained by globally fitting the data to a random binding mechanism are summarized in Table 1. As a consequence of increases in kcat and decreases in the KM values, this Diels-Alderase is 6 times more efficient than CE6 and 16 times more efficient than DA_20_20 (Table 1). Overall, CE20 has a 100-fold higher effective molarity (kcat/kuncat) and a 9,700-fold higher chemical proficiency than the original DA_20_00 computational design (Table 1).
Fig. 3.

Characterization of Diels-Alderase CE20. (A) Dependence of reaction velocity on dienophile concentration at different fixed diene concentrations. (B) pH dependence of the enzymatic reaction. (C) Stereoselectivity of the uncatalyzed (background) and CE20-catalyzed reactions; authentic standards (control) are shown for reference. All error bars reflect the SD of three independent measurements.
As expected for concerted cycloaddition of diene 1 and dienophile 2, the rate of the CE20-catalyzed reaction is pH independent in the range 5–10 (Fig. 3B). As per design (21), the evolved catalyst only promotes stereoselective formation of the 3R,4S endo product; none of the other possible diastereomers could be detected by chiral HPLC analysis of the reaction mixture (Fig. 3C). Finally, although product inhibition often hampers enzyme-catalyzed bimolecular reactions, this is minimal in the case of CE20. The half-maximal inhibitory concentration (IC50 = 0.42 mM) of the phosphorylated product analog 4, which has enhanced water solubility compared with 3, is similar in magnitude to the value of Kdiene, facilitating efficient substrate processing (Fig. S3A). As a result, CE20 can conduct >770 turnovers at room temperature without signs of inactivation (Fig. S3B).
The optimized features of CE20 enabled preparative-scale synthesis of the reaction product in aqueous buffer and at room temperature. Greater than 90% conversion was observed, yielding 30 mg of the 3R,4S endo cyclohexene product isomer after isolation (SI Materials and Methods). By comparison, refluxing the substrates in toluene for several days in the absence of catalyst affords a 66:34 mixture of racemic endo and exo products (26). The artificial Diels-Alderase thus provides a mild and practical alternative to standard chemical synthesis, and is notable with respect to its efficiency and stereoselectivity.
Crystal Structure of CE20.
The structure of CE20 in complex with the phosphorylated product analog 4 was determined by X-ray crystallography to a resolution of 2.9 Å (Table S1). Overall, it is remarkably similar to the original DA_20_00 computational design (21) and the computationally optimized CE6 intermediate (23). Pairwise comparison of the respective proteins gives Cα atom rms deviations of ∼0.4 Å (Table S2). The β-propeller scaffold used for the design is evidently quite rigid, as neither the appended helix-turn-helix motif nor extensive mutation alters its fold (Fig. S4). It is particularly noteworthy that the side chains of the catalytic residues Tyr134 and Gln208 adopt virtually identical orientations in all three structures (Fig. 4A). The observed rotameric preferences of these side chains are consistent with the predictions of the original design model (21).
Fig. 4.
Crystal structure of CE20 with bound product. (A) The structures of CE20 (orange), CE6 (gray), and the DA_20_00 computational design model (black) are very similar. The catalytic amino acids Tyr134 and Gln208 (shown as sticks) adopt nearly identical conformations. (B) The largest differences between CE20 and CE6 are localized in the appended lid element. The helix designed to interact with the substrates (left in the figure) overlays well with its counterpart in CE6, whereas the interhelical loop and the supporting helix (right) are largely disordered in CE20. (C) The reaction product (black carbons) binds deeply in a shape-complementary pocket (transparent gray surface) containing a buried water molecule (red sphere). (D) The catalytic residues interact with the ligand as designed. The side-chain phenol of Tyr134 donates a hydrogen bond to the carboxamide carbonyl group, whereas the side-chain amide of Gln208 accepts a hydrogen bond from the carbamate NH of the product and donates a hydrogen bond to the carboxamide carbonyl group. The latter interactions are offset by an unfavorable interaction between the Gln208 amide and the carbamate ether oxygen (red dashes). (E) Superposition of the product and catalytic residues shows that the crystal structure (black and yellow carbons) closely matches the original design model (gray and brown carbons). Hydrogen bonds in the design are depicted as black dashes. An unfavorable interaction between Gln208 amide and the carbamate ether oxygen is also evident in the design (3.2 Å in the design vs. 3.0 Å in the crystal structure).
The largest differences between the CE20 and CE6 structures are localized in the appended lid element (Fig. 4B). In CE6, the two helices of the helix-turn-helix motif are resolved and assume a packing angle of 180°, somewhat larger than the 150° predicted by design (23). In CE20, the helix designed to interact with the substrates (residues 36–44) overlays well with its counterpart in CE6 (Cα rmsd = 0.2 Å; Table S2), but the interhelical loop and the second supporting helix are largely disordered. Residues 51–53 in the solvent-exposed helix completely lack electron density, whereas the side chains of the flanking amino acids at positions 45–50 and 54–58 could not be resolved (Table S2). Crystal contacts in the vicinity of the helix-turn-helix of CE6 and their absence in CE20 may account for the different geometries observed for the second helix. The crystal structure of CE11 supports this hypothesis (Table S2): the helix-turn-helix motif in one of the proteins in the asymmetric unit engages in crystal contacts, and the supporting helix is well resolved (packing angle ∼130°), whereas such interactions are absent in the second protein chain, and this helix is disordered. The high mobility of the solvent-exposed helix compared with the active-site helix rationalizes its relative tolerance to mutation.
Visualization of the phosphorylated product analog 4 at the CE20 active site provides direct insight into catalytically relevant interactions as well as a structural basis for rationalizing the evolutionary improvements. The ligand docks snugly in a deep hydrophobic pocket within the protein. Excluding the 2-aminoethylphosphate moiety, which was introduced to increase solubility and points out into solution, 86% of its solvent-accessible surface is buried upon complex formation (Fig. 4C and Fig. S5). Ligand recognition is mediated by 88 van der Waals contacts with the peptide backbone, the side chains of many aliphatic and aromatic residues, and a buried water molecule. Roughly a sixth of these interactions are provided by the appended helical element, which functions as a lid and effectively shields the ligand from bulk solvent (Fig. S6). The cyclohexene ring of the product sits at the bottom of the active site pocket, largely filling the available space. Although small interfacial cavities are discernable, implying slightly imperfect shape complementarity, the ligand is fixed in place by extensive hydrophobic contacts, on the one hand, and specific hydrogen bonding interactions with the two catalytic residues on the other (Fig. 4D). This binding mode agrees quite well with the original design model of the product complex (21) (Fig. 4E).
The high stereoselectivity observed for the catalyst suggests that the diene and dienophile substrates must engage in similar interactions with the protein. As programmed, the side chain of Tyr134 is located at the bottom of the binding pocket, effectively positioned to donate a hydrogen bond to the carbonyl group of the dienophile; the importance of this interaction is underscored by mutagenesis studies with CE11 showing that replacement of Tyr134 by any other amino acid causes complete loss of activity under the screening conditions. The buried water molecule, although somewhat distant in the product complex (Fig. 4 C and D), may further stabilize the expanded transition state by providing a second hydrogen-bonding interaction to the dienophile carbonyl group. Placement of the Gln208 side chain amide also is consistent with the design model. It is poised to accept a hydrogen bond from the carbamate NH of the diene and simultaneously donate a hydrogen bond to the dienophile. Nevertheless, an unfavorable electronic interaction between the amide oxygen of Gln208 and the ether oxygen of the diene carbamate (Fig. 4 D and E) may explain why substitution of this residue with methionine decreased the activity of the CE11 variant only twofold.
Discussion
Because Diels–Alder reactions proceed through highly ordered transition states, complementarily shaped protein binding sites may function as “entropy traps” to increase reaction rate (27). Utilization of binding energy to preorganize the substrates for reaction at an active site represents a simple strategy for overcoming large translational and rotational entropic barriers. Further rate enhancements can be achieved by reducing the energy gap between the highest occupied molecular orbital of the diene and the lowest unoccupied molecular orbital of the dienophile through specific hydrogen bonding interactions (28).
The computational design process that yielded DA_20_00 relied on both strategies to transform a β-propeller scaffold incapable of catalyzing cycloadditions into an artificial Diels-Alderase (21). Its success reflects both the choice of catalytic groups for stabilizing the transition state, in this case Tyr134 and Gln208, and effective experimental installation of the idealized active site, or “theozyme,” in a suitable protein host. The structure of the CE20–product complex attests to the accuracy of the computational modeling of the catalytic residues. The conformations of the Tyr134 and Gln208 side chains, which donate a hydrogen bond to the dienophile and accept a hydrogen bond from the diene, respectively, closely match the design model and changed negligibly over the entire evolutionary trajectory. Because the design algorithms assume an immobile protein backbone, the relative rigidity of the β-propeller fold undoubtedly was conducive to the accurate placement of the theozyme. When more flexible scaffolds have been used for other computationally designed enzymes, significant discrepancies between model and experiment have been observed (29–31).
Despite effective positioning of the catalytic groups in the original DA_20_00 design, considerable refinement of the active site was required to achieve respectable levels of activity. Multiple rounds of directed evolution and further computational refinement helped sculpt a more effective binding pocket without substantially altering the core fold. Tolerance to extensive mutation in this case may also be linked to the robustness of the β-propeller scaffold. The largest leaps in activity were achieved in the early optimization rounds, in which small residues at the bottom of the catalytic pocket were replaced by bulkier amino acids (21) and a lid element was grafted onto the structure to shield the reactants from bulk solvent (23). These changes, together with the subtle modifications caused by T43I and more distant mutations (Fig. 2), resulted in gradual contraction of the active site around the highly ordered transition state (Fig. 5 A–C), enhancing the ability of the Diels-Alderase to constrain the substrates in a reactive orientation with respect to each other and the two catalytic residues. The resulting improvements are manifest in a 102-fold higher “effective molarity” (EM = kcat/kuncat) for CE20 compared with the original computational design and an ∼104-fold increase in catalytic proficiency (∼1/KTS). The difference in proficiency reflects the greater affinity of the evolved enzyme for the cycloaddition transition state (22). Although several mechanistic factors likely contribute to improved molecular recognition of this transient species, the 5.4 kcal/mol of additional stabilization achieved during optimization exceeds the 4.7 kcal/mol predicted by quantum mechanics (21) for electronic stabilization of the transition state by the hydrogen bonding interactions provided by the catalytic residues.
Fig. 5.
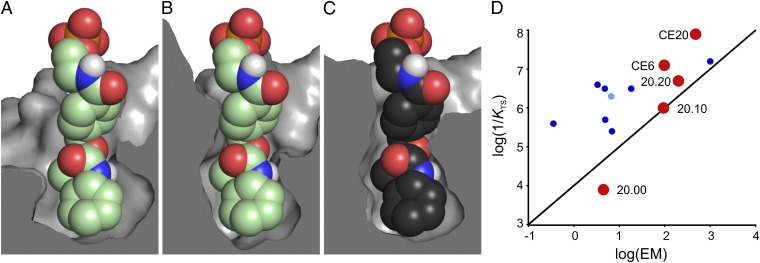
Evolution of a shape-complementary pocket. Cut-away views of the active sites of DA_20_00 variants illustrate the gradual contraction of the binding pocket around the reaction product over the course of evolution. (A) DA_20_00, the original computational design, (B) the evolutionary intermediate CE6, and (C) CE20, the most evolved variant. The ligand depicted in the DA_20_00 and CE6 pockets (green carbons) was docked manually in the same orientation as the product seen in the crystal structure of the CE20 complex (black carbons). (D) Plot of effective molarity (EM = kcat/kuncat) vs. catalytic proficiency (1/KTS = [kcat/(Kdiene·Kdienophile)]/kuncat) for kinetically characterized catalytic antibodies (dark blue), a ribozyme (cyan), and descendants of the computationally designed DA_20_00 Diels-Alderase (red).
As a consequence of these improvements, CE20 is currently the most proficient Diels-Alderase known. It is 30- to 300-fold more active than catalytic antibodies that promote the same [4+2] cycloaddition (9, 11), and it outperforms the best artificial enzymes for other Diels–Alder reactions, including antibody catalysts (7–12), ribozymes (13–16), and artificial metalloenzymes (17, 18) (Fig. 5D and Table S3). For example, antibody 1E9, which accelerates the inverse electron-demand cycloaddition of tetrachlorothiophene dioxide to N-ethylmaleimide (7) and exhibits nearly perfect shape complementarity with its transition-state analog (32), has a sixfold lower catalytic proficiency. Other artificial Diels-Alderases are one to three orders of magnitude less effective. The catalytic proficiency of CE20 is also 63 times higher than that of SpnF, the only natural Diels-Alderase to be characterized kinetically so far, which promotes a transannular [4+2] cycloaddition in the biosynthesis of the natural product spinosyn A (4).
Compared with other natural enzymes, though, the absolute efficiency of CE20 is modest. It turns over only 10 substrate molecules per hour, and the kcat/Kdiene and kcat/Kdienophile parameters are four to five orders of magnitude lower than the kcat/KM values of moderately efficient natural enzymes (33). It is possible that additional rounds of directed evolution might improve this Diels-Alderase further. Directed evolution of other computational designs, such as the Kemp eliminase HG3 (34) and the retroaldolase RA95 (29), has afforded 1,000-fold higher chemical proficiencies. Because active site shape complementarity is not yet fully optimal (Figs. 4C and 5), filling the remaining cavities at the bottom of the pocket might further constrain the substrates and optimize their interactions with the catalytic residues. However, the gains per round of laboratory evolution significantly diminished over the course of evolution, and additional large increases in activity may prove practically elusive. The rigidity of the scaffold may well constrain CE20 evolvability in this instance, precluding the subtle conformational changes of the backbone that might be necessary for attaining perfectly snug transition-state binding. It is also possible that the mechanistic choices implicit in the theozyme intrinsically limit the activities that may be achieved, in which case, different catalytic side chains, or perhaps even metal ion cofactors (35), will be required for more effective substrate activation and transition-state stabilization.
Computational enzyme design has made substantial strides in recent years (19, 20). Despite impressive accomplishments, though, most structurally characterized designs differ significantly from the original computational prediction, especially with respect to ligand binding and catalytic residue placement (29–31, 34, 36). The assumption that the protein backbone stays fixed upon installation of a theozyme into a scaffold, made for computational tractability, is a clear weakness of this approach. The good agreement between the original design model of the catalytic site and the experimentally determined structure in the case of the computationally designed Diels-Alderase studied here shows that when the scaffold does not change significantly upon introduction of mutations, the calculations may be quite accurate. This finding suggests that controlling backbone conformation will be a key determinant of future progress in this area. The drawback with rigid scaffolds is that the backbone cannot readily be tuned to the reaction of interest. Properly modeling the reconfigurations of flexible regions of protein scaffolds upon sequence perturbation—and, going further, the custom design of new backbones with geometry optimal for the desired catalytic site—will be important steps toward a next generation of highly active designed catalysts for any desired chemical reaction.
Materials and Methods
Full details regarding materials and methods are provided in SI Materials and Methods.
In Vitro Evolution.
Gene libraries of the most active variants from each round were generated by epPCR using the GeneMorph PCR mutagenesis kit (Stratagene). The Diels–Alder activity of 720 variants per round was assayed in 96-well plates. A solution containing substrates and hexadeuterated product as an internal standard was added to cleared cell lysates and allowed to react for 3.5 h at room temperature before quenching with 50 mM HCl. The reaction mixture was extracted with water-saturated l-butanol, and the product concentration was determined by a direct MS-MS analysis on a Bruker ESI-(Qq)TOF mass spectrometer (maXis) equipped with an Agilent HPLC system (1200). The clones with the largest increase in activity, typically corresponding to about 1% of the screened population, were picked from replica plates for plasmid isolation, sequencing, and further diversification.
Biochemical Characterization.
All Diels-Alderase variants were produced as C-terminally His-tagged proteins in Escherichia coli BL21(DE3) and purified by affinity chromatography (Ni-NTA). Catalytic activity was assayed by varying the dienophile concentration at several fixed concentrations of diene at 25 °C in PBS buffer containing 4% DMSO and 40 µM hexadeuterated product as an internal standard. Reactions were incubated for 2.5 h at room temperature before acid quenching, extraction, and MS-MS measurement. Steady-state parameters were obtained by fitting the data to the Michaelis–Menten equation. The pH-rate profiles were determined under subsaturating conditions by using acetate buffer for pH 4–6 and Bis-Tris propane for pH 6–10, both containing 100 mM NaCl. Product enantiomers were separated by analytical chiral normal phase HPLC.
Crystallization and Structure Determination.
The proteins were crystallized using the hanging and sitting drop diffusion methods, and their structures were solved by molecular replacement. See SI Materials and Methods for detailed procedures and a summary of crystal parameters, data collection, and refinement statistics for the CE11 structure (PDB ID code 4O5S) and the CE20 structure with product analog (PDB ID code 4O5T).
Supplementary Material
Acknowledgments
We thank Xiangyang Zhang and Louis Bertschi [Mass Spectrometry Service Facility, Laboratory of Organic Chemistry, Eidgenössische Technische Hochschule (ETH) Zurich] for running all MS-MS assays and the beamline staff at the Swiss Light Source for support during data collection. This work was supported by the Swiss National Science Foundation (D.H.), ETH Zurich (D.H.), and the Defense Advanced Research Projects Agency (D.B. and D.H.). T.B. also gratefully acknowledges financial support from Marie Curie Action within the FP7-PEOPLE program (IEF-2011-299400).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4O5S and 4O5T).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401073111/-/DCSupplemental.
References
- 1.Katayama K, Kobayashi T, Oikawa H, Honma M, Ichihara A. Enzymatic activity and partial purification of solanapyrone synthase: First enzyme catalyzing Diels-Alder reaction. Biochim Biophys Acta. 1998;1384(2):387–395. doi: 10.1016/s0167-4838(98)00040-5. [DOI] [PubMed] [Google Scholar]
- 2.Auclair K, et al. Lovastatin nonaketide synthase catalyzes an intramolecular Diels-Alder reaction of a substrate analogue. J Am Chem Soc. 2000;122(46):11519–11520. [Google Scholar]
- 3.Stocking EM, Williams RM. Chemistry and biology of biosynthetic Diels-Alder reactions. Angew Chem Int Ed Engl. 2003;42(27):3078–3115. doi: 10.1002/anie.200200534. [DOI] [PubMed] [Google Scholar]
- 4.Kim HJ, Ruszczycky MW, Choi SH, Liu YN, Liu HW. Enzyme-catalysed [4+2] cycloaddition is a key step in the biosynthesis of spinosyn A. Nature. 2011;473(7345):109–112. doi: 10.1038/nature09981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ose T, et al. Insight into a natural Diels-Alder reaction from the structure of macrophomate synthase. Nature. 2003;422(6928):185–189. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 6.Serafimov JM, Gillingham D, Kuster S, Hilvert D. The putative Diels-Alderase macrophomate synthase is an efficient aldolase. J Am Chem Soc. 2008;130(25):7798–7799. doi: 10.1021/ja8017994. [DOI] [PubMed] [Google Scholar]
- 7.Hilvert D, Hill KW, Nared KD, Auditor MTM. Antibody catalysis of a Diels-Alder reaction. J Am Chem Soc. 1989;111(26):9261–9262. [Google Scholar]
- 8.Braisted AC, Schultz PG. An antibody-catalyzed bimolecular Diels-Alder reaction. J Am Chem Soc. 1990;112(20):7430–7431. [Google Scholar]
- 9.Gouverneur VE, et al. Control of the exo and endo pathways of the Diels-Alder reaction by antibody catalysis. Science. 1993;262(5131):204–208. doi: 10.1126/science.8211138. [DOI] [PubMed] [Google Scholar]
- 10.Meekel AAP, Resmini M, Pandit UK. First example of an antibody-catalyzed hetero-Diels-Alder reaction. Chem Commun. 1995;(5):571–572. doi: 10.1016/0968-0896(96)00087-9. [DOI] [PubMed] [Google Scholar]
- 11.Ylikauhaluoma JT, et al. Anti-metallocene antibodies—a new approach to enantioselective catalysis of the Diels-Alder reaction. J Am Chem Soc. 1995;117(27):7041–7047. [Google Scholar]
- 12.Bahr N, Guller R, Reymond JL, Lerner RA. A nitroxyl synthase catalytic antibody. J Am Chem Soc. 1996;118(15):3550–3555. [Google Scholar]
- 13.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389(6646):54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 14.Seelig B, Jäschke A. A small catalytic RNA motif with Diels-Alderase activity. Chem Biol. 1999;6(3):167–176. doi: 10.1016/S1074-5521(99)89008-5. [DOI] [PubMed] [Google Scholar]
- 15.Seelig B, Keiper S, Stuhlmann F, Jäschke A. Enantioselective ribozyme catalysis of a bimolecular cycloaddition reaction. Angew Chem Int Ed Engl. 2000;39(24):4576–4579. [PubMed] [Google Scholar]
- 16.Chandra M, Silverman SK. DNA and RNA can be equally efficient catalysts for carbon-carbon bond formation. J Am Chem Soc. 2008;130(10):2936–2937. doi: 10.1021/ja7111965. [DOI] [PubMed] [Google Scholar]
- 17.Roelfes G, Feringa BL. DNA-based asymmetric catalysis. Angew Chem Int Ed Engl. 2005;44(21):3230–3232. doi: 10.1002/anie.200500298. [DOI] [PubMed] [Google Scholar]
- 18.Reetz MT. Artificial metalloenzymes as catalysts in stereoselective Diels-Alder reactions. Chem Rec. 2012;12(4):391–406. doi: 10.1002/tcr.201100043. [DOI] [PubMed] [Google Scholar]
- 19.Hilvert D. Design of protein catalysts. Annu Rev Biochem. 2013;82:447–470. doi: 10.1146/annurev-biochem-072611-101825. [DOI] [PubMed] [Google Scholar]
- 20.Kiss G, Çelebi-Ölçüm N, Moretti R, Baker D, Houk KN. Computational enzyme design. Angew Chem Int Ed Engl. 2013;52(22):5700–5725. doi: 10.1002/anie.201204077. [DOI] [PubMed] [Google Scholar]
- 21.Siegel JB, et al. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329(5989):309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267(5194):90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 23.Eiben CB, et al. Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat Biotechnol. 2012;30(2):190–192. doi: 10.1038/nbt.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jäckel C, Kast P, Hilvert D. Protein design by directed evolution. Annu Rev Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 25.Segel IH. 1993. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems (Wiley, New York), p 984.
- 26.Cannizzaro CE, Ashley JA, Janda KD, Houk KN. Experimental determination of the absolute enantioselectivity of an antibody-catalyzed Diels-Alder reaction and theoretical explorations of the origins of stereoselectivity. J Am Chem Soc. 2003;125(9):2489–2506. doi: 10.1021/ja020879d. [DOI] [PubMed] [Google Scholar]
- 27.Page MI, Jencks WP. Entropic contributions to rate accelerations in enzymic and intramolecular reactions and the chelate effect. Proc Natl Acad Sci USA. 1971;68(8):1678–1683. doi: 10.1073/pnas.68.8.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blake JF, Lim D, Jorgensen WL. Enhanced hydrogen-bonding of water to Diels-Alder transition-states—ab-initio evidence. J Org Chem. 1994;59(4):803–805. [Google Scholar]
- 29.Giger L, et al. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat Chem Biol. 2013;9(8):494–498. doi: 10.1038/nchembio.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, et al. Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design. J Mol Biol. 2012;415(3):615–625. doi: 10.1016/j.jmb.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter F, et al. Computational design of catalytic dyads and oxyanion holes for ester hydrolysis. J Am Chem Soc. 2012;134(39):16197–16206. doi: 10.1021/ja3037367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu JA, et al. Evolution of shape complementarity and catalytic efficiency from a primordial antibody template. Science. 1999;286(5448):2345–2348. doi: 10.1126/science.286.5448.2345. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Even A, et al. The moderately efficient enzyme: Evolutionary and physicochemical trends shaping enzyme parameters. Biochemistry. 2011;50(21):4402–4410. doi: 10.1021/bi2002289. [DOI] [PubMed] [Google Scholar]
- 34.Blomberg R, et al. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature. 2013;503(7476):418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- 35.Birney DM, Houk KN. Transition structures of the Lewis acid-catalyzed Diels-Alder reaction of butadiene with acrolein—the origins of selectivity. J Am Chem Soc. 1990;112(11):4127–4133. [Google Scholar]
- 36.Khersonsky O, et al. Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59. Proc Natl Acad Sci USA. 2012;109(26):10358–10363. doi: 10.1073/pnas.1121063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.