Significance
LINE-1 (L1) retrotransposons have been shown to mediate various regulatory effects and can affect the transcription of neighboring genes. Thus, novel insertions can potentially result in phenotypic differences that, in turn, could be acted on by selection. We found that a standard Illumina SNP array did not efficiently capture L1s, so that their phenotypic effects might have been missed by previous genome-wide association studies. However, we also found that using whole genome sequencing data, tag SNPs can be identified for a majority of L1s, which opens the way for SNP-based genetic association studies of L1 effects. Moreover, we detected common and unusually long haplotypes around several L1s, which suggests that these insertions might have undergone recent, positive selection in humans.
Keywords: human genetics, population genetics, evolution, L1-seq, extended haplotype homozygosity
Abstract
Insertions of the human-specific subfamily of LINE-1 (L1) retrotransposon are highly polymorphic across individuals and can critically influence the human transcriptome. We hypothesized that L1 insertions could represent genetic variants determining important human phenotypic traits, and performed an integrated analysis of L1 elements and single nucleotide polymorphisms (SNPs) in several human populations. We found that a large fraction of L1s were in high linkage disequilibrium with their surrounding genomic regions and that they were well tagged by SNPs. However, L1 variants were only partially captured by SNPs on standard SNP arrays, so that their potential phenotypic impact would be frequently missed by SNP array-based genome-wide association studies. We next identified potential phenotypic effects of L1s by looking for signatures of natural selection linked to L1 insertions; significant extended haplotype homozygosity was detected around several L1 insertions. This finding suggests that some of these L1 insertions may have been the target of recent positive selection.
LINE-1 retrotransposons are mobile genetic elements that comprise almost 20% of the human genome (1). Most LINE-1 elements are either mutated or truncated and are retrotranposition incompetent. However, a human-specific subfamily of LINE-1 elements (L1Hs, referred to as “L1” below) is currently active in humans.
Over the last years, the application of genome-wide approaches to identify mobile genetic elements has shed new light on retrotransposition in humans. Thousands of new polymorphic insertions have been identified, highlighting the differences in retrotransposon content across individual genomes (2, 3). There are an estimated 12,000 polymorphic L1 insertions with allele frequencies above 0.05 in humans (4) and L1 insertions represent a major source of structural variation between individuals (5, 6). Furthermore, polymorphic L1 elements can be highly active and retrotransposition thus continues to be an ongoing source of genetic variation in today’s populations (7). The genome-wide search for genetic determinants of common human traits and diseases has been largely based on single-nucleotide polymorphisms (SNPs) and has sometimes failed to explain heritability of complex traits (8). In contrast, the association of phenotypic variability and disease susceptibility with structural variation remains relatively less explored (9). The recent realization of the extent and polymorphism of L1 insertions in humans thus make them a particularly interesting source of genetic variation.
Mobile genetic elements were initially proposed to have no impact on phenotype and to be evolutionarily neutral (10, 11). The ongoing retrotransposition activity of L1, however, was shown to cause various genetic diseases by way of insertional mutagenesis (4). Such insertions are expected to be under strong purifying selection. In addition to these deleterious effects, the L1 sequence has been shown to contain a variety of regulatory elements and to be able to critically modify the transcriptional architecture and thereby modulate the function of neighboring genes (12). In particular, the L1 sequence includes promoters in both its 3′ and 5′ UTRs, polyadenylation signals, and splice signals (13–17). Strikingly, Faulkner et al. showed on a genome-wide scale that nearly 20% of all transcriptional start sites originate in LINE-1 elements (18), demonstrating the important regulatory role played by this element in the genome. Novel insertions can thus generate genetic diversity that could potentially result in phenotypic differences that, in turn, could be acted on by selection. However, despite the abundant evidence for the regulatory effects of L1, very little is known about its potential phenotypic effects. Finally, the recent discovery of L1 somatic retrotransposition in tumors and their potential functional involvement in cancer (19) further highlights the importance of understanding potential L1 phenotypic effects.
We hypothesized that L1 insertions could represent genetic variants determining important human phenotypic traits and undergoing evolutionary pressure. We first asked whether L1 insertions were tagged by surrounding SNPs. Indeed, such SNPs could be used as L1 proxies in SNP-phenotype association studies and might help reveal phenotypic effects of specific L1 insertions. Upon integrated analysis of L1 insertions and SNPs in an Asian cohort and in public data of the 1000 Genome Project (1000GP), we found that the majority of L1 insertions were in high linkage with their surrounding genomic region. However, we found that they were not efficiently captured by SNPs on the standard Illumina SNP array. As an additional approach to looking for potential phenotypic effects of L1 insertions, we tested for specific signals of selection around these elements, both within and across several human populations. Our results indicate that a fraction of L1 insertions might have undergone recent natural selection. This finding further suggests that L1 insertions may have important phenotypic effects and provides interesting candidates for functional tests.
Results
L1 Insertion Profiling.
We aimed to analyze linkage disequilibrium (LD) around L1 elements. We thus performed parallel genome-wide profiling and analysis of L1 elements and SNPs in a group of Asian individuals (SI Appendix, Fig. S1). We constructed L1-seq libraries for 20 individuals (Dataset S1) using the method developed by Ewing et al. (2) and identified a total of 1,574 L1 elements (824 L1s per individual on average) with different levels of polymorphism (Dataset S2 and Samples and Construction of L1-Seq Libraries and Computational Pipeline for L1 Calling in SI Appendix, Methods).
The number of L1s detected in these samples was in line with previous L1-seq studies: Ewing et al. (2) detected 1,139 L1s in 25 samples (comprising 15 unrelated individuals) whereas Evrony et al. reported 796 and 773 L1s in two unrelated individuals (see table S2 in ref. 20). Following Evrony et al. (20) we looked up how many of the detected L1s were present in the human reference genome (“known reference” or KR) or had been identified in previous studies [“known nonreference” or KNR, i.e., L1s in dbRIP (21) and refs. 2, 3, 5, 22, and 23, according to table S5 in ref. 20). We found an average of 552 KR and 121 KNR L1s per sample. On the other hand Evrony et al. detected 689 KR and 113 KNR L1s per sample and Ewing et al. (2) detected 628 KR L1s. The lower number of KR insertions in our study could be a consequence of the fact that L1s can show frequency differentiation across populations and that some KR L1s might be more common in samples of Caucasian compared with Asian origin.
Using locus-specific PCR, we validated 91 L1s (mostly novel L1s, i.e., not KR or KNR; see PCR Validation of L1 Elements in SI Appendix, Methods, Datasets S3 and S4, and SI Appendix, Gel Electrophoresis Analyses of Site-Specific PCR Validations for 91 L1s Identified in our Data) in all 20 samples and estimated the specificity and sensitivity of our L1-seq procedure to be around 94% and 78%, respectively (SI Appendix, Fig. S2). The frequency spectrum of all 1,574 L1s showed a remarkable shape, with an overabundance of L1s present in 1 or all 20 individuals (Fig. 1A). Our L1 calling method was agnostic to L1 frequency in the population and we verified that detection sensitivity was approximately constant across the whole frequency range and, in particular, not lower for L1s with intermediate frequencies (SI Appendix, Fig. S3). The abundance of rare L1s could thus potentially reflect recent insertional events or ongoing purifying selection whereas the abundance of fixed L1s could result from demographic effects (e.g., population bottleneck) or selection (e.g., positive selection).
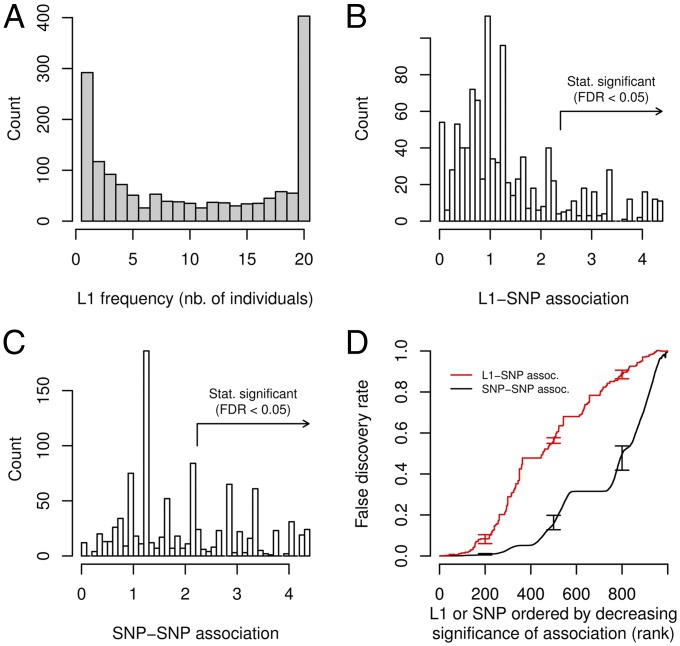
Fig. 1.
L1 frequency spectrum and association between L1 and surrounding SNPs in our Asian cohort. (A) Number of L1 elements detected that were present in a particular number of individuals (x-axis). (B) Distribution of L1-SNP association. Association was measured by the −log10(minimal P value) obtained from a series of Fisher’s exact tests between an L1 and SNPs in the 100 kb surrounding window. A total of 1,005 L1s were polymorphic across the 17 samples and had a least one testable (nonmonoallelic) surrounding SNP (on average 18 testable SNPs per L1). The arrow indicates the association corresponding to FDR ≤ 0.05 (estimated from 100 permutations). (C) Example distribution of SNP–SNP association. We selected an identically sized set of random, frequency-matched SNP and calculated association with neighboring SNPs and FDR in the same way as for L1s. The fraction of SNPs with FDR ≤ 0.05 is much larger than for L1s. (D) Significance of L1–SNP versus SNP–SNP association. L1s were ranked by decreasing association to surrounding SNPs and false discovery rate (FDR) was estimated by a permutation approach (red). The average FDR for the association of identically sized, frequency-matched sets of random SNPs with their surrounding SNPs is shown for comparison (black). Error bars indicate 10th and 90th percentile (100 permutations for L1-SNP, 10 permutation of each of 100 sets of random SNPs for SNP–SNP association).
SNP Array Does Not Efficiently Capture L1 Insertions.
We had previously obtained SNP genotypes for 17 of the individuals in our Asian cohort using Illumina Human610-Quad BeadChips (24). We aimed to test whether L1 presence was efficiently tagged by SNPs on the array and performed LD analysis around L1 elements (Integrated L1-SNP Analysis of our L1-seq Data in SI Appendix, Methods). Such tagging SNPs could be used to uncover potential phenotypic effects of L1s via genetic association studies. Since L1-seq only determines presence or absence of a particular L1 element in an individual, we could not use an allelic measure of L1-SNP association (e.g., R2). We thus measured the association between the presence or absence of a particular L1 and the genotype of surrounding SNPs using Fisher’s exact test and calculated −log10(P value) for all surrounding SNPs. The highest value was then recorded as the tagging score. The distribution of tagging scores for 1,005 L1s that were polymorphic across the 17 samples is shown in Fig. 1B. To assess the significance of the observed scores and address the multiple testing issue arising from testing many L1s, we repeatedly permuted L1 presence/absence labels and repeated the same analysis (100 permutations). For any value of the tagging score, the ratio between the number of L1s with identical or higher score using the permuted data and the number of L1s with identical or higher score using the original data estimated the false discovery rate (FDR). FDR at a particular score value thus measured the fraction of L1s that reached an equal or higher score under the null assumption of no association between L1 and surrounding SNPs.
We found that tagging scores were highly significant for a minority of L1s only. For instance, 154 (15%) of L1s had an FDR ≤ 0.05 (Fig. 1B). For comparison, we calculated tagging scores for SNP-SNP association genome-wide by selecting identically sized sets of random SNPs with the same derived allele frequency spectrum as L1s (100 SNP sets) and calculating FDR for each set in the same way as for L1s (see SI Appendix, Methods for details). The distribution of tagging scores obtained for one random SNP set is shown in Fig. 1C. Overall, we found that tagging scores obtained for SNPs were much more significant than for L1s (on average 34% of random SNPs had an FDR ≤ 0.05; Fig. 1D and see also SI Appendix, Fig. S4A for the corresponding maximal R2 distribution). For instance, the top 500 L1s with highest scores had an FDR of 0.56 (i.e., 280 L1s obtained the same or higher scores under the null assumption of no association) whereas the top 500 random SNPs had an average FDR of 0.16 (i.e., only 80 SNPs obtained the same or higher scores under the null assumption of no association). A small fraction of L1s were nevertheless as efficiently tagged as the most efficiently tagged SNPs as the top 23 L1s reached a significance level that was at least as high as for the corresponding top SNPs.
We investigated whether the low L1 taggability observed in our data could be of technical origin. For instance, errors in L1 calls could lead to seemingly low tagging scores, similar to what was initially observed with imperfectly genotyped structural variants like copy number variations. To assess the potential effects of erroneous calls, we increased the stringency of our L1-calling algorithm and identified a subset of 564 higher confidence L1s (with an estimated specificity of 98%; PCR Validation of L1 Elements in SI Appendix, Methods). We repeated the analysis with this smaller, higher confidence L1 set and observed the same qualitative deficit in L1 taggability compared with random SNPs (SI Appendix, Fig. S4B). We then used 44 polymorphic L1s with presence/absence calls assessed by site-specific PCR (Dataset S4) and directly assessed how errors in L1 calls influenced association scores and their significance. The genotyping error rate was 2% and tagging scores obtained with PCR-based and L1-seq-based genotypes were highly correlated (c = 0.94; SI Appendix, Fig. S5A). Moreover, the differences in scores obtained with PCR-based genotypes did not critically influence significance of tagging scores (SI Appendix, Fig. S5B). We also verified that low L1 taggability was not caused by the abundance of rare and very common L1s by repeating the same analysis without L1s present in 1 or 16 individuals and obtaining the same qualitative result (SI Appendix, Fig. S4C). We conclude that the low L1 taggability observed with these data are unlikely to be caused chiefly by L1 genotyping errors or the specific L1 frequency spectrum. Finally, we also verified that we had sufficient power to detect well tagged L1s by testing a small set of L1s that we independently predicted to be well tagged by SNPs assayed on the array (SI Appendix, Fig. S4D).
L1-Tagging SNPs in the 1000GP Panel.
We next investigated whether the low L1 taggability observed here resulted from the specific set of SNPs assayed on the array and if additional tagging SNPs could be found in a more comprehensive SNP panel. We thus analyzed L1 and SNP genotyping data obtained in the pilot phase (25) of the 1000GP. Based on whole-genome sequencing data obtained from 179 samples representing three continental populations, the 1000GP identified 15 million SNPs (25) as well as several hundred L1s (23). This extensive SNP panel allowed us to test L1 taggability in an unbiased manner. The availability of allelic information obtained through sequencing data allowed us to directly quantify the association between L1 elements and surrounding SNPs using the R2 metric. R2 is akin to a correlation coefficient and ranges from 0 (no association between two loci) to 1 (complete association). We used the maximal R2 observed between a particular L1 and surrounding SNPs to assess taggability (Integrated L1-SNP Analysis of the 1000GP Data in SI Appendix, Methods). We observed that only a small fraction of L1s was tagged compared with random, frequency-matched SNPs (SI Appendix, Figs. S6 and S7). For individuals of European origin (CEU samples) for instance, the fraction of perfectly tagged SNPs (max. R2 = 1) was about 0.8, whereas the fraction of perfectly tagged L1s was about 0.4 (SI Appendix, Fig. S6A).
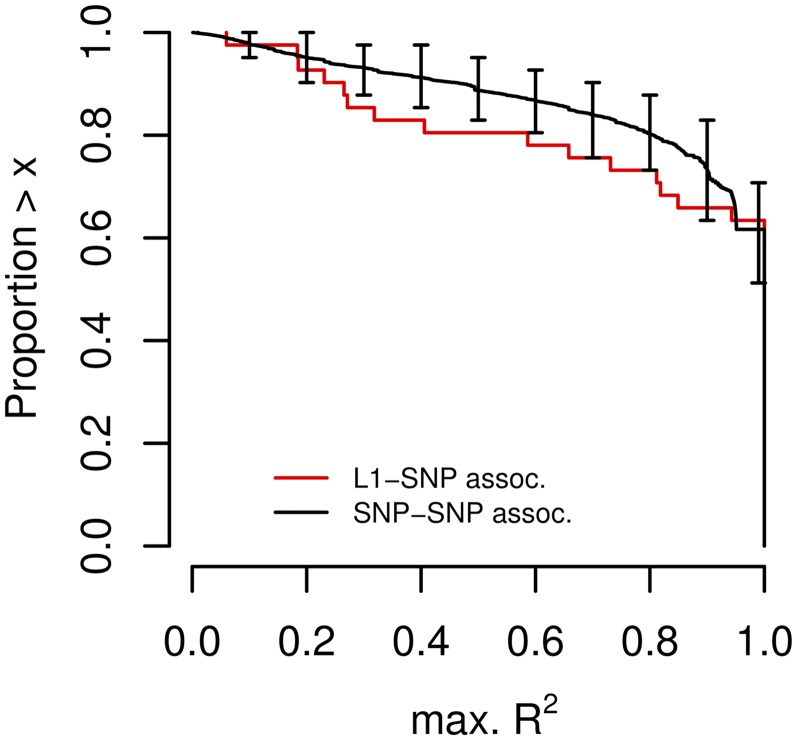
To determine whether low L1 taggability was caused by genotyping errors, we designed locus-specific PCR primers for 42 L1s with varying taggability levels (see PCR Validation in the 1000GP in SI Appendix, Methods, Datasets S5 and S6, and SI Appendix, Gel Electrophoresis Analyses of Site-Specific PCR Validations for 47 L1s from the 1000GP). For this set of L1s, the median and mean error rates (per L1) were 7% and 15%, respectively. We found that genotyping errors dramatically lowered taggability estimates for the majority of these L1s (SI Appendix, Fig. S8). Based on PCR-based genotypes of this validated L1 set, the fraction of perfectly tagged L1s (max. R2 = 1) was not significantly different from the fraction of perfectly tagged SNPs in identically sized sets of frequency-matched SNPs (Fig. 2). The fraction of well tagged L1s (max. R2 > 0.8) was only about 20% less than for SNPs. The major difference compared with the taggability of SNPs was a slight excess of untaggable L1s (max. R2 < 0.4). We tested whether untaggable L1s were associated with particular L1 or genomic features (including L1 length, distance to gene, GC content, and number of repetitive elements in surrounding regions and distance to chromosome ends and centromeres; see Association of L1 LD with Genomic Features in SI Appendix, Methods), but we did not find any significant association.
Fig. 2.
L1 taggability by SNPs of the 1000GP panel. The plot shows the inverse cumulative distribution of the maximal R2 observed between 42 PCR-validated L1s and SNPs in their surrounding 20 kb region (red line), or random SNP sets (1,000 sets of 42 SNPs) and SNPs in their surrounding 20 kb region (black line). L1 genotypes were assessed using site-specific PCR in 40 CEU samples. Error bars show 10th and 90th percentile obtained from 1,000 SNP sets.
Taken together, these results suggest that a majority of L1s might be efficiently tagged by neighboring SNPs so that tagging SNPs could be used to assess potential phenotypic effects of L1s. However, previous genome-wide association (GWA) studies might not have captured these effects since SNP panels assayed on standard arrays might only contain a fraction of L1 tagging SNPs, as shown above by the analysis of our Asian cohort on the Illumina Omni SNP array. We thus used the 1000GP data to identify perfect tagging SNPs (max. R2 = 1). We expect the identification of perfect tagging SNPs to be reliable using the 1000GP genotypes, in contrast to our experience above with imperfectly tagged L1s, for two reasons: (i) random genotyping errors are much more likely to decrease taggability than to increase it, so that perfectly tagged L1s are unlikely to occur by chance; (ii) our PCR validations showed that none of the eight (three in CEU and five in CHB) perfectly tagged L1s we tested contained any genotyping errors (SI Appendix, Fig. S8). We identified 1,903 tagging SNPs corresponding to 106 L1s (Dataset S7). We then tested whether any of these SNPs had been identified in previous GWA studies using the catalog of published GWA studies (26), but none of the SNPs tagging the 106 L1s were linked with a phenotype.
Unusual Haplotypic Structure and Potential Positive Selection of L1s.
We also sought to identify potential phenotypic effects of L1s by looking for signatures of natural selection linked to L1 insertions. In particular, positive selection has been shown to leave a distinct signature in the haplotypic structure around targeted loci: Haplotype diversity is expected to be lower and haplotype length is expected to be greater around a recently selected allele compared with the corresponding ancestral allele (27). This effect was used to design long-range haplotype tests that have been successful at detecting loci under natural selection in the human genome (27–29). We asked whether we could identify such signals of selection for L1s and calculated extended haplotype homozygosity (EHH) statistics (27) around L1 elements. Using the 1000GP data, we focused on L1s for which we had homozygous individuals so that the phase between each L1 and surrounding SNPs was determined (Extended Haplotype Homozygosity Around L1s SI Appendix, Methods). We identified nine candidate L1s with enough homozygous individuals in the CEU population to conduct the analysis. We successfully validated eight of these L1s in 40 CEU samples by PCR and used PCR-based genotypes (Dataset S6) to calculate EHH scores. We calculated significance by generating a large number of identically-sized, frequency-matched random SNP sets and repeating the same analysis. Given the EHH score obtained by a particular L1, the ratio of the number of SNPs (per set) that obtained the same or higher score and the number of L1s that obtained the same or higher score yielded an FDR: it takes a value of 1 if the number of L1s that obtained a given EHH score is identical to random SNPs and lower values if there is an excess of L1s with EHH scores that are higher compared with random SNPs.
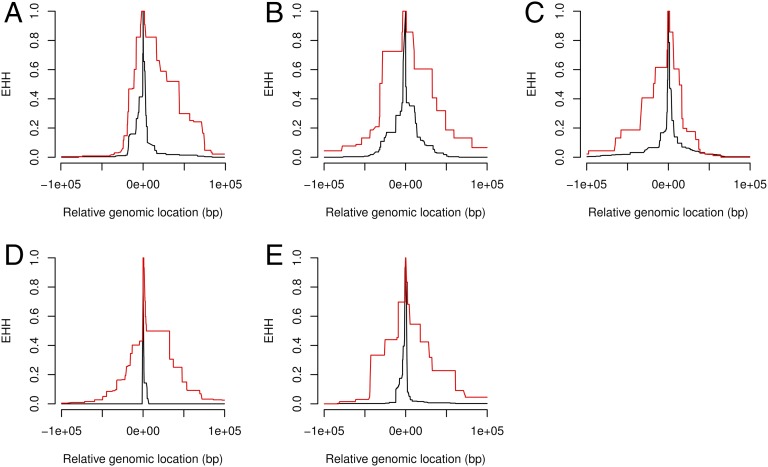
Surprisingly, we found that three of eight L1s had unusually high EHH scores (Fig. 3 A–C). The top three L1s obtained an FDR = 0.28 which means that, on average, we observed less than 1 SNP per set with an identical or higher EHH score (in 1,000 frequency-matched SNP sets). We asked whether similar signatures could be detected in another population and turned to individuals of Chinese descent (CHB samples). We aimed to screen L1s based on EHH scores obtained with 1000GP genotypes and then assess high quality PCR-based genotypes and associated EHH scores for a smaller subset comprising the best candidates. We focused on 18 L1s that had enough homozygous individuals to allow for EHH calculation and significance testing. Based on EHH scores obtained using 1000GP genotypes, we selected the top 6 L1s for PCR validation (FDR = 0.4 suggesting that 3–4 out of 6 had unusual EHH scores). We recalculated their EHH scores and corresponding significance level using PCR-based genotypes and found that the top L1 showed highly significant EHH scores (FDR = 0.01, Fig. 3D). The other 5 also showed unusually high EHH scores compared with random SNPs (FDR = 0.5 for the 6 L1s collectively; see Fig. 3E for the second most significant L1). In conclusion, despite the small number of L1s considered here, we detected unusually high EHH scores around several L1 insertions in both the CEU and CHB populations. This finding is compatible with a fraction of L1s having undergone rapid positive selection.
Fig. 3.
Unusual extended haplotype homozygosity (EHH) in the 100-kb region around L1s in two human populations. (A–C) EHH signals around the top three L1s with highest EHH scores in the CEU population. EHH calculated for the L1-bearing allele (red) is higher and extends further compared with EHH calculated for the allele that does not carry the specific L1 (black). P1_M_061510_1_185 (frequency 0.49; A), P1_M_061510_4_354 (frequency 0.44; B), and P1_MEI_1280&P2_MEI_1388 (frequency 0.37; C) are shown. (D and E) Same for 2 L1s in the CHB population. P1_MEI_2516&P2_MEI_1951 (frequency 0.69; D) and P1_MEI_539&P2_MEI_776 (frequency 0.34; E) are shown.
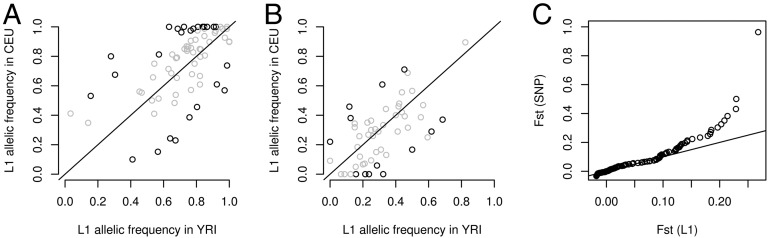
We finally sought to detect L1s that might have been the target of selection by looking at the genetic differentiation between populations. In particular, high levels of population differentiation at specific loci might be interpreted as evidence for adaptive selection and might, in turn, reveal important phenotypic effects. We compared L1 allele frequencies between samples of European (40 CEU samples) and African (37 YRI samples) descent and used the fixation index (Fst) to detect stratification. Based on our set of 42 PCR-validated L1s, we found that the median absolute difference between allelic frequencies estimated using 1000GP and PCR-based genotypes in the CEU population was 0.08 (1000GP genotypes thus systematically overestimated allelic frequencies). We reasoned that this difference would not prevent us from detecting L1 with medium to strong differentiation, and we relied on 1000GP genotypes for this analysis. We found 130 L1s that were not fixed in either population, but none of the 130 L1s exhibited extreme differentiation i.e., fixed in one population and absent from the other (Fig. 4 A and B). We went on to identify L1s with the highest differentiation based on a permutation approach (L1 Frequency Stratification in the 1000GP in SI Appendix, Methods) and found 38 (out of 130) L1s with significant population differentiation. Nine of the 38 L1 insertion sites are within genes (introns), raising the likelihood that they might confer phenotypic effects (SI Appendix, Table S2). We noticed a subset of the significantly differentiated L1s that were almost fixed in the CEU samples but were present at lower frequencies in YRI samples (Fig. 4A). Interestingly, the fraction of intragenic L1s (all of them intronic) was almost twice as high (Fisher’s exact test P = 0.013) in this group (57%) compared with all L1s (30%), consistent with a functional role for these elements and suggesting that positive selection might have contributed to drive (some of) these L1s to fixation. We asked how L1 differentiation compared with the differentiation observed with other variants in the genome. We generated 1000 sets of identically sized, frequency-matched SNPs and compared their Fst with what we obtained for L1s (Fig. 4C). The distribution of Fst values observed with SNPs was in agreement with what was found in previous studies (30), with a small fraction of SNPs reaching very high values. The tail of the SNP distribution was heavier than the sample distribution obtained from the 130 L1s, showing systematically higher Fst values. This result suggests that, if positive selection pressure acts on particular L1s, it might not be as strong as for some SNPs.
Fig. 4.
L1 allele frequency in CEU versus YRI samples. (A) L1s from the 1000GP “deletion” set: 77 L1 elements that are not fixed in both populations are shown. Black dots indicate 26 L1 elements with fixation index (Fst) significantly (FDR < 0.05) different from 0. (B) L1s from the 1000GP “insertion” set: 53 L1s that are not fixed in both populations. Black dots indicate 13 L1 elements with fixation index (Fst) significantly (FDR < 0.05) different from 0. (C) QQ plot comparing the distribution of Fst values for L1s (“deletion” and “insertion” sets combined) and genome-wide SNPs.
Discussion
We performed a comprehensive survey of L1 polymorphism in an Asian cohort and identified an abundance of novel L1 insertions, including many rare and fixed L1s. Integrating L1 and array-based SNP data, we found that SNPs on the Illumina genotyping array used here did not efficiently tag L1s. We relied on a general test of association (Fisher’s exact test) to assess taggability and found that only 15% of detected L1s were significantly (FDR < 0.05) tagged. For comparison, 34% of frequency-matched SNPs were tagged by neighboring SNPs at the same significance level. We verified that low L1 taggability was not caused by genotyping errors. We also used a set of control L1s to check that our approach was able to detect SNPs that were independently predicted to be well tagged. Importantly, these results indicate that potential phenotypic effects of L1s would have been missed by SNP array-based GWA studies and that L1s might contribute to the problem of missing heritability (8).
However, validation of SNPs from a larger, unbiased SNP panel obtained by the 1000GP indicated that LD around L1s might not be quantitatively different from the rest of the genome and that tagging SNPs could in principle be identified for a majority of L1s. This finding opens the way to the use of SNPs as L1 proxies to test potential phenotypic effects of L1 in large populations. Here we identified two sets of L1-tagging SNPs from the CEU and CHB samples of the 1000GP. From the 1,102 SNPs tagging 63 L1s in the CEU population, 55 SNPs (tagging 35 L1s) were present on the standard Illumina Omni array used in this study and 57 (tagging 21 L1s) were present on a comparable Affymetrix array (Human SNP Array 5.0). This result highlights limitations in the ability of SNP arrays of the types that were used for many GWA studies so far to capture L1s genome-wide. Nevertheless, we asked whether any of these SNPs had been identified in previous GWA studies, but we did not find any. Existing evidence for important and varied transcriptional effects of L1s, however, appears to argue for comprehensive, genome-wide assessment of L1 phenotypic effects by association studies. Such studies would require the identification of a custom SNP panel capturing the majority of L1s in a population. Existing population-scale sequencing data like the 1000GP might, in principle, be used to this end; however, this will require L1 genotyping accuracy to be improved, as shown by our analyses (SI Appendix, Fig. S8). Our set of PCR-validated L1 genotypes (Dataset S6) might provide a useful benchmark for the development of improved genotype calling algorithms.
Previous genome-wide studies of other types of structural variants [including copy-number polymorphisms (31), common deletions (32), and short insertion-deletions < 50 bp (33)] generally found a high degree of LD with surrounding SNPs. However, particular classes of structural variants have been shown to be in weak LD with their surrounding, for instance copy number polymorphisms in segmental duplications (34). On the other hand, a minority of SNPs have been found to be untaggable, most likely due their proximity to recombination hotspots (35). L1 insertions have also been shown to provide substrate for genomic rearrangements (36). However, our findings of high LD around most L1s indicate that the majority of L1s do not represent highly active recombination hotspots.
We have also looked for signatures of selection as another way of identifying L1s with potential phenotypic effects. We found evidence for common and unusually long haplotypes around several L1s, and the number of such L1s was surprisingly high compared with random, frequency-matched SNPs. This signature has been previously used to successfully detect recent selection, and our results suggest that a fraction of L1s might have undergone rapid positive selection. Several L1s in the limited set of L1 insertions identified by the 1000GP also showed differentiation between the two human populations. It has been argued, however, that methods based on extended haplotypes or allelic differentiation do not formally test against neutrality (37). As with previous applications of EHH or Fst, alternative explanations including demographic effects cannot be completely ruled out. For instance, a small effective population size (out of Africa), followed by drift and the population explosion might lead to the emergence of higher frequency L1s with large blocks of LD, similarly to the haplotypic structure we detected around several L1s.
Retrotransposon insertions have generally been considered neutral or under purifying selection (e.g., in the case of gene-disrupting insertions) (11, 38, 39). Few previous studies have assessed the role of L1s in recent human evolution. In particular, Stewart et al. (23) have used L1 allele frequency and heterozygosity information derived from the 1000 GP data to test a neutral model of L1 evolution. Their results were consistent with a neutral model, but with some signs of deviations for the Asian population.
L1s with potential phenotypic effects, identified as targets of selection or by association studies, can be further confirmed by functional studies linking their genomic position, transcriptional impact and cellular effect.
Methods
The full description of material and methods is provided in SI Appendix, Methods. In short, we obtained 20 DNA samples (Dataset S1) from individuals of multiethnic Asian origin (40, 41) and constructed L1-seq libraries based on the protocol of Ewing et al. (2). After quality control and trimming, reads were aligned to the human genome and read pile-ups marking L1 3′ flanks were detected using a custom computational pipeline. For each peak, we recorded four parameters: width, mean coverage, maximal coverage, and number of unique reads. L1s were called by setting thresholds on these four parameters. The sensitivity and specificity of L1 was adjusted by comparing L1 calls with the results of PCR validation experiments for 91 L1s (PCR Validation of L1 Elements in our Samples in SI Appendix, Methods). SNP genotype data were previously obtained for 17 samples using Illumina Human610-Quad BeadChips. We also used SNP and L1 genotype data (23) for the three populations studied in the pilot phase of the 1000GP. For validation purposes, we obtained 40 CEU and 29 CHB HapMap (DNA) samples used by the 1000GP. All computational analyses were implemented in R (42) using Bioconductor (43) packages including ShortRead (44), GenomicRanges (45), pROC (46), VariantAnnotation (47), and biomaRt (48).
Supplementary Material
Acknowledgments
We thank Dmitri Petrov for critical discussions and comments on the manuscript. This work was supported by a Visiting Investigator Programme grant from the Joint Council Office of the Agency for Science, Technology and Research in Singapore (to S.R.Q) (project no. 092 110 0080); National Medical Research Council, Singapore, Grants 0796/2003, IRG07nov013, IRG09nov014, STaR/0003/2008, and CG/SERI/2010; and Biomedical Research Council, Singapore Grants 08/1/35/19/550 and 09/1/35/19/616 (to C.-Y.C. and T.Y.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequences have been deposited in the database of Genotypes and Phenotypes (dbGAP) (accession no. phs000732.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401532111/-/DCSupplemental.
References
- 1.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10(10):691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20(9):1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iskow RC, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141(7):1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149(4):740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang CRL, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141(7):1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidd JM, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143(5):837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck CR, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141(7):1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat Rev Genet. 2013;14(2):125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 11.Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. 2001;18(6):926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 12.Tang W, et al. Secreted and membrane attractin result from alternative splicing of the human ATRN gene. Proc Natl Acad Sci USA. 2000;97(11):6025–6030. doi: 10.1073/pnas.110139897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34(5):1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35(4):363–366. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 15.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429(6989):268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 16.Mätlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006(1):71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: For better or worse, in sickness and in health. Genome Res. 2008;18(3):343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 18.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41(5):563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 19.Lee E, et al. Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evrony GD, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151(3):483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, et al. dbRIP: A highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27(4):323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewing AD, Kazazian HH., Jr Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21(6):985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stewart C, et al. 1000 Genomes Project A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7(8):e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cornes BK, et al. Identification of four novel variants that influence central corneal thickness in multi-ethnic Asian populations. Hum Mol Genet. 2012;21(2):437–445. doi: 10.1093/hmg/ddr463. [DOI] [PubMed] [Google Scholar]
- 25.Abecasis GR, et al. 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 28.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabeti PC, et al. International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40(3):340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 31.McCarroll SA, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40(10):1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 32.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet. 2006;38(1):82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 33.Lu JT, Wang Y, Gibbs RA, Yu F. Characterizing linkage disequilibrium and evaluating imputation power of human genomic insertion-deletion polymorphisms. Genome Biol. 2012;13(2):R15. doi: 10.1186/gb-2012-13-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell CD, et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am J Hum Genet. 2011;88(3):317–332. doi: 10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han K, et al. L1 recombination-associated deletions generate human genomic variation. Proc Natl Acad Sci USA. 2008;105(49):19366–19371. doi: 10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- 38.Cordaux R, Lee J, Dinoso L, Batzer MA. Recently integrated Alu retrotransposons are essentially neutral residents of the human genome. Gene. 2006;373:138–144. doi: 10.1016/j.gene.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 39.Boissinot S, Davis J, Entezam A, Petrov D, Furano AV. Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci USA. 2006;103(25):9590–9594. doi: 10.1073/pnas.0603334103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foong AWP, et al. Rationale and methodology for a population-based study of eye diseases in Malay people: The Singapore Malay eye study (SiMES) Ophthalmic Epidemiol. 2007;14(1):25–35. doi: 10.1080/09286580600878844. [DOI] [PubMed] [Google Scholar]
- 41.Lavanya R, et al. Methodology of the Singapore Indian Chinese Cohort (SICC) eye study: Quantifying ethnic variations in the epidemiology of eye diseases in Asians. Ophthalmic Epidemiol. 2009;16(6):325–336. doi: 10.3109/09286580903144738. [DOI] [PubMed] [Google Scholar]
- 42.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; Available at: http://www.R-project.org. [Google Scholar]
- 43.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morgan M, et al. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25(19):2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawrence M, et al. Software for computing and annotating genomic ranges. PLOS Comput Biol. 2013;9(8):e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robin X, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obenchain V, et al. VariantAnnotation: a Bioconductor package for exploration and annotation of genetic variants. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durinck S, et al. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.